Background: Farnesoid X receptor (FXR) regulates bile acid (BA) metabolism. High BA levels increase susceptibility to intestinal tumorigenesis.
Results: caudal-related homeobox 2 (CDX2) directly binds the FXR promoter and regulates FXR expression.
Conclusion: CDX2 transcription factor is a positive regulator of FXR in the gut.
Significance: Manipulation of the APC-CDX2-FXR axis could reduce BA toxicity and intestinal tumor susceptibility.
Keywords: Bile Acid, Gene Transcription, Intestine, Lipid Metabolism, Nuclear Receptor
Abstract
Farnesoid X receptor (FXR, NR1H4) is a bile acid-activated transcription factor that belongs to the nuclear receptor superfamily. It is highly expressed in the enterohepatic system, where it senses bile acid levels to consequently reduce their synthesis while inducing their detoxification. Bile acids are intestinal tumor promoters and their concentrations have to be tightly regulated. Indeed, reduced expression of FXR in the intestine increases colorectal cancer susceptibility in mice, whereas its activation can promote apoptosis in genetically modified cells. Notably, despite the broad knowledge of the FXR enterohepatic transcriptional activity, the molecular mechanisms regulating FXR expression in the intestine are still unknown. Herein, by combining both gain and loss of function approaches and FXR promoter activity studies, we identified caudal-related homeobox 2 (CDX2) transcription factor as a positive regulator of FXR expression in the enterocytes. Our results provide a putative novel tool for modulating FXR expression against bile acid-related colorectal cancer progression.
Introduction
The epithelium of the small intestine is organized into two morphologically and functionally distinct compartments: finger-shaped luminal protrusions termed villi and flask-shaped submucosal invaginations referred to as the crypt of Lieberkühn. The villus is a differentiated compartment of the intestinal epithelium. At the crypt bottom reside stem cells that self-renew throughout life to generate progenitors that occupy the lower third of the crypt (1). In the midcrypt region, cells stop proliferating to differentiate into enterocytes. After reaching the epithelial surface, differentiated cells undergo apoptosis and are shed into the lumen. The primary driving force of the constant renewal of the gut epithelium is the WNT signaling (1). Thus, mutations of components of the WNT pathway have been reported to promote colorectal cancer (CRC).5 Among these factors, the APC (adenomatous polyposis coli) gene is believed to be the gatekeeper in CRC development.
APC is a component of a multiprotein destruction complex that in the absence of WNT activators sequesters cytoplasmic β-CATENIN to phosphorylate it and targets it for ubiquitin-proteasome degradation (1). Importantly, while reducing the activity of β-CATENIN, APC also induces the expression of genes that promote differentiation and cell cycle arrest, such as P21 (2) and KLF4 (Krüppel-like factor 4) (3). These effects are mediated by the CDX2 (caudal-related homeobox 2) protein, a downstream mediator of APC tumor suppressor activity in intestinal cells (3, 4).
The human CDX2 gene is one of the three mammalian homologues of the Drosophila homeobox-containing gene caudal, whose encoded transcription factor has a primary role during embryonic development, as assessed by the early lethality of Cdx2-deficient mouse embryos and the skeleton abnormalities observed in Cdx2+/− (5). During embryogenesis, CDX2 determines intestinal identity in the presumptive gut endoderm (6, 7), and it is later required to maintain this identity in the intestinal epithelium throughout adulthood (8). At the molecular level, it controls the transcription of a huge number of intestinal genes (9) including KLF4 (3) and P21 (2). CDX2 appears to be differentially phosphorylated along the crypt-villus axis by the mitogen-activated protein kinases (MAPK), and this modulates CDX2 transcriptional activity and half-life (10–12). The expression of CDX2 has been found to be altered in human CRC in relation to the tumor grade (13–15), and its decrease correlates with poor prognosis (16, 17). Moreover, reduction of CDX2 expression facilitates tumor progression of murine models of genetically or chemically induced CRC (18, 19). On the contrary, restoration of CDX2 in colorectal cancer cells has been shown to reduce cell migration in vitro and dissemination in vivo (12, 20).
Farnesoid X receptor is an adopted member of the nuclear receptor (NR) superfamily of transcription factors. Two FXR (farnesoid X receptor) genes have been identified and are referred to as FXRα (NR1H4) and FXRβ (NR1H5). FXRβ is a pseudogene in humans, whereas it is expressed in rodents, rabbits and dogs. It has been proposed to be a lanosterol sensor, although its physiological function remains to be established (21). FXRα (also referred to FXR) is highly expressed in the enterohepatic system, where bile acids (BAs) are its endogenous ligands (22–24). FXRα encodes four isoforms of transcripts in both humans and mice (FXRα1, FXRα2, FXRα3, and FXRα4) as a result of alternative splicing of exon 5 and the use of two distinct promoters that initiate transcription from either exon 1 (5′ promoter) or exon 3 (3′ promoter) (25, 26). The 5′ or 3′ promoters of the FXR gene regulate the expression of FXRα1 plus FXRα2 or FXRα3 plus FXRα4 transcripts, respectively. Notably, the four FXR isoforms are expressed in a tissue-specific manner along the gut-liver axis, and a few FXR target genes are regulated in an isoform-dependent manner (26, 27).
By mediating the transcriptional activity of BAs, FXR regulates BA metabolism. The ability of FXR to reduce BA synthesis while inducing BA detoxification is of critical importance because high levels of circulating BA can increase hepatic and intestinal susceptibility to tumorigenesis (28–31). Notably, the expression of FXR in the gut is restricted to the differentiated compartment of the intestinal epithelium (29, 32), and its activation by BAs will result not only in BA detoxification but also in increased apoptosis and consequent removal of genetically modified cells (29, 33). Indeed, the protective role of FXR against CRC susceptibility has been demonstrated by our group and others in Fxr knock-out mice, where the first hit for CRC development was made by inactivating Apc mutations (29, 33). We also showed that following Apc mutations, the expression of Fxr was already reduced in aberrant crypt foci of Apcmin/+/Fxr+/+ mice (29). These observations suggest that a downstream mediator of the Apc pathway may modulate the expression of Fxr in the intestine.
Although different FXR target genes have been identified in the intestine, such as FGF19/Fgf15 (fibroblast growth factor 19/15) (34), IBABP (ileal bile acid-binding protein) (35), OSTα/β (organic solute transporter α/β) (36), and those genes that codify the enzymes involved in BA detoxification (37), the mechanisms that regulate FXR expression in the intestine are still unknown. Herein, we identify CDX2 as a positive transcriptional regulator of intestinal FXR expression. Thus, the manipulation of the APC-CDX2-FXR cascade might be useful in the future to reduce BA quantity and toxicity fighting against bile acid-related CRC progression.
EXPERIMENTAL PROCEDURES
Familial Adenomatous Polyposis (FAP) Patients
We obtained tissue samples (tumors and normal intestinal mucosa) from Dr. R. Valanzano (University of Florence, Florence, Italy) and Dr. R. Mariani-Costantini (University of Chieti-Pescara, Chieti, Italy). These unrelated Italian patients with adenomatous polyposis coli were recruited for the study after approval by the Ethical Committee of the University “G. D'Annunzio” of Chieti. Written informed consent was obtained from each patient before mutation analysis and tissue harvesting. All of the included cases presented with a classic FAP phenotype and harbored pathogenetic germ line APC mutations (38). Genomic DNA was isolated from at least two independently drawn whole blood samples using QIAamp DNA Blood (Qiagen, Hilden, Germany). The coding sequence and intron-exon borders of APC were analyzed by a combination of PCR-based techniques that included heteroduplex analysis on agarose minigel for recurring mutations, in vitro synthesized protein assay of exon 15, single strand conformational polymorphism analysis of the remaining exons, and denaturing high performance liquid chromatography (Wave 1100, Transgenomic Inc., Omaha, NE) of APC exons 1–15 followed by sequencing. All mutations were confirmed by sequencing independent PCR products.
Mice
Heterozygous Cdx2+/− mice (5) and relative wild-type mice were kindly provided by Dr. Jean-Noel Freund. Apcmin/+ mice on a pure C57BL/6J background were obtained from the Jackson Laboratory (Bar Harbor, ME). Cdx2+/− and Apcmin/+ and their relative wild-type littermate mice were housed under pathogen-free conditions and fed a standard rodent chow diet in a temperature-controlled room (23 °C) on a 12-h light/dark cycle. Apcmin/+ and relative wild-type littermate mice were sacrificed by the age of 6 months, and colon tumors and adjacent normal areas were harvested. Cdx2+/− mice and relative wild-type littermate mice were sacrificed by the age of 3 months, and small and large intestine were harvested. All of the experiments presented in this study have been carried out in male mice. The Ethics Committee of the Consorzio Mario Negri Sud approved this experimental set-up.
RNA Extraction and qRT-PCR
Animal tissues were snap-frozen in liquid nitrogen immediately after harvesting and then stored at −80 °C until use. Total RNA from tissues and cells was isolated by TRIzol reagent (Invitrogen), following the manufacturer's instructions. To avoid possible DNA contaminations, RNA was treated with DNase (Ambion, Austin, TX). RNA purity was also checked by spectrophotometer, and RNA integrity was checked by examination on agarose gel electrophoresis. cDNA was synthesized, retrotranscribing 4 μg of total RNA in a total volume of 100 μl using the High Capacity DNA Archive Kit (Applied Biosystems) and following the manufacturer's instructions. To detect mRNA expression levels for the genes investigated in this study, primers were designed using Primer Express software spanning intron/exon boundaries whenever possible. qPCR assays were performed in 96-well optical reaction plates, using the ABI 7500HT machine (Applied Biosystem). qPCR assays were conducted in triplicate wells for each sample. The following reaction mixture per well was used: 10 μl of Power SYBR Green (Applied Biosystems), 2.4 μl of primers at a final concentration of 150 nm, 4.6 μl of RNase-free water, 3 μl of cDNA. For all experiments, the following qPCR conditions were used: denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and then at 60 °C for 60 s. Quantitative normalization of cDNA in each sample was performed using cyclophilin mRNA as an internal control. Relative quantification was done using the ΔΔCT method, and expression values have been represented as -fold change between the groups.
Plasmid Constructs
The human CDX2 expression vector was constructed starting from an ATCC clone containing the human CDX2 coding sequence, which was PCR-amplified and subcloned into the pCDNA4B vector using KpnI and BamHI restriction sites. The region spanning −1195 to +21 in reference to the transcriptional start sites (TSS) of the human FXRα3/α4 promoter was PCR-amplified from genomic DNA of HT29 cells. The quality of the PCR products was checked by sequencing. PCR product was then inserted upstream of the firefly luciferase gene in the multiple-cloning site of the promoterless vector pGL3-basic (Promega), using SacI/NheI restriction sites to create pGL3-FXRα3/α4-luc(−1195/+21) reporter construct. The numbers in parenthesis indicate the nucleotide positions with respect to the TSS. Starting from the pGL3-FXRα3/α4-luc(−1195/+21) plasmid, deletion fragments containing putative CDX2 binding sites (ATAAA, sense; TATTT, antisense) were subcloned into the promoterless vector pGL3-basic using SacI/NheI restriction sites to generate the reporter constructs pGL3-FXRα3/α4-luc(−724/+21), pGL3-FXRα3/α4-luc(−376/+21), pGL3-FXRα3/α4-luc(−118/+21), pGL3-FXRα3/α4-luc(−64/+21). Two mutant reporter vectors were generated from the pGL3-FXRα3/α4-luc(−118/+21) and pGL3-FXRα3/α4-luc(−1195/+21) plasmid, respectively, using a site-directed mutagenesis kit (QuikChange, Stratagene, La Jolla, CA). Site-directed mutagenesis of the putative CDX2 binding site was introduced by substitution of the underlined nucleotides in the sequence ATAAA to obtain the AGCAA sequence.
Adenovirus Constructs
CDX2 adenovirus (AdCDX2) was generated using the Viral Power Adenoviral Expression System (Invitrogen). Briefly, the human CDX2 coding sequence was PCR-amplified and subcloned from the pCDNA4B-CDX2 plasmid into the pENTR4 shuttle vector using SalI and XhoI restriction sites. Then pENTR4-CDX2 plasmid was recombined with the destination vector pAd/CMV/V5-DEST to generate an adenovector containing human CDX2 under the control of the human cytomegalovirus promoter. The adenovector coding β-galactosidase under the control of the human cytomegalovirus promoter (pAd/CMV/V5-GW/LacZ) was purchased from Invitrogen. Short hairpin CDX2 adenovirus (AdshCDX2) was generated based on a previously validated sequence against human CDX2 (39) using the BLOCK-iT U6 RNA entry vector kit (Invitrogen). Short hairpin CDX2 oligonucleotides were synthesized with complementary ends to be subcloned into the pENTR/U6 shuttle vector. Then the pENTR/U6-shCDX2 plasmid was recombined with the destination vector pAd/BLOCK-iT-DEST to generate an adenovector containing short hairpin CDX2 under the control of the U6 promoter. The oligonucleotides to generate the adenovector control AdshLacZ were purchased from Invitrogen. Indeed, AdshLacZ was constructed with the same procedure used to make AdshCDX2. AdCDX2, AdLacZ, AdshCDX2, and AdshLacZ vectors were linearized with PacI restriction enzyme, and 1 μg of digested products, after purification with the Qiaex II gel extraction kit (Qiagen), were used to transfect 5 × 105 293A cells (Invitrogen) with 3 μl of Lipofectamine 2000 (Invitrogen) to generate the corresponding adenoviruses. The viruses were propagated into 293A cells. Crude viral lysate stocks were stored at −80 °C until use. The adenoviral titer was determined by qRT-PCR with specific primers (40).
Cell Cultures, Treatments, and Adenovirus Transduction
HT29, Caco2, HeLa, and 293A cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotic. LS174T were maintained in RPMI medium supplemented with 10% fetal bovine serum and 1% antibiotic. HT29 differentiation was achieved by postconfluence and sodium butyrate (NaBt) treatment. Regarding postconfluence-induced differentiation, 5 × 104 cells/well were seeded in a 6-well plate. Cells were harvested at day 3 (preconfluence), day 7 (confluence), and day 14 (postconfluence) after seeding. Regarding NaBt-induced differentiation, 5 × 104 cells/well were seeded in a 6-well plate and treated with NaBt 1 mm for 48 h. Caco2 differentiation was achieved by supplementing cells with 20% fetal bovine serum after confluence. Cells were harvested at days 4, 8, and 15 after confluence. HT29 and LS174T were transduced with a 100 multiplicity of infection (MOI) of AdCDX2 or control AdLaZ for 48 h. For the functional study, 48 h after transduction, HT29 cells were treated for 24 h with 1 μm GW4064. For the NaBt-induced differentiation study, HT29 cells were first transduced with 200 MOI of AdshCDX2 or control AdshLacZ and then 24 h after transduction, cells were treated for 48 h with 1 mm NaBt.
Transient Transfections and Luciferase Assays
For the luciferase assays, HeLa cells, at 70% of confluence, were seeded in 12-well plates for 24 h. The next day, 1 h before transfection, complete DMEM was aspirated, cells were washed twice with warm PBS, and 1 ml of DMEM free of antibiotic was added to each well. Indeed, cells were co-transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Briefly, for the deletion and mutagenesis experiments, cells were co-transfected using 10 μl of Lipofectamine 2000 with 2 μg of pCDNA4B (control) or pCDNA4B-CDX2 expression vector, 1 μg of pGL3-basic or pGL3-FXRα3/α4 promoter constructs, and 0.1 μg of Renilla reporter plasmid (pRT-TK). 24 h after transfection, cells were lysed, and luciferase activity was measured with the Dual Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. Results were expressed as -fold over pGL3 activation. To investigate the effect of mutant CDX2 on the ability of wild-type CDX2 to transactivate the FXRα3/α4 promoter, HeLa cells were co-transfected with 1 mg of pGL3-FXRα3/α4-luc(−1995/+21), 2 μg of wild-type CDX2, and increasing amounts of mutant CDX2. The total amount of DNA was kept constant at 15 μg by the addition of pCDNA4B plasmid. The relative firefly luciferase activities were calculated by normalizing transfection efficiency according to the Renilla luciferase activities. All experiments were performed in triplicate, and similar results were obtained from at least three independent experiments.
Histology and Immunohistochemistry
Tissue specimens were fixed in 10% formalin for 12–24 h, dehydrated, and paraffin-embedded. Standard histology (H&E) and immunohistochemical procedures were performed. Briefly, with respect to immunohistochemistry, 5-μm-thick sections were treated with 3% hydrogen peroxide for 5 min, to quench endogenous peroxidase and subjected to antigen retrieval by boiling the slides in antigen unmasking solution (Vector Laboratories Inc., Burlingame, CA) for 15 min according to the manufacturer's instructions. Sections were sequentially incubated for 1 h at room temperature in 50% non-immune serum (from the host animal in which the secondary antibody was raised) in PBS (to avoid nonspecific signal) and overnight at 4 °C with the primary antibodies (FXR antibody was a generous gift from Phenex Pharmaceutical AG; CDX2 antibody was purchased from Zymed Laboratories, Inc. (San Francisco, CA)). Sections were washed for 10 min in PBS and incubated for 30 min at room temperature with the secondary biotinylated antibody (Vector Laboratories). After several washing steps with PBS (three washes, 5 min each), sections were incubated with the avidin-biotin complex (Vector Laboratories) for 30 min at room temperature. After washing in PBS, the peroxidase reaction was developed by incubation with 3,3-diaminobenzidine (Sigma-Aldrich). Counterstaining was carried out with methylene blue (Sigma-Aldrich). For negative controls, 1% non-immune serum in PBS replaced the primary antibodies.
Western Blot
Cells were lysed with radioimmune precipitation buffer (150 mm NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0; Sigma) containing protease inhibitors (Roche Applied Science). Cells lysed were cleared by centrifugation at 12,000 rpm for 10 min. Protein concentration was determined by DC protein assay reagent (Bio-Rad). Protein lysates (50 μg/sample) were heated at 100 °C for 5 min in Laemmli buffer (60 mm Tris, pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.005% bromphenol blue). Proteins were isolated by 7.5% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Biosciences). The membranes were blocked with a blocking buffer and then incubated at room temperature for 2 h with antibodies against FXR (a gift from Phenex Pharmaceutical AG) and CDX2 (Zymed Laboratories, Inc.). The membranes were extensively washed with PBS, 20% Tween and then incubated at room temperature for 1 h with a horseradish peroxidase-labeled anti-rabbit IgG for FXR or anti-mouse IgG for CDX2. The immune complexes were visualized with the use of Super Signal ECL Plus (Amersham Biosciences) according to the manufacturer's protocol.
Alkaline Phosphatase Measurement
NaBt-treated HT29 cells and preconfluent, confluent, and postconfluent HT29 and Caco2 cells were scraped from 6-well plates in ice-cold PBS and then centrifuged for 5 min at 3000 rpm at 4 °C. Supernatant was discarded, and pellet was resuspended in 200 μl of lysis buffer made by 80% radioimmune precipitation buffer (50 mm Tris-HCl, pH 8.0, with 150 mm sodium chloride, 1.0% Igepal CA-630 (Nonidet P-40), 0.5% sodium deoxycholate, 0.1% SDS; Sigma) and 20% protease inhibitors (Roche Applied Science). After 15 min in ice, samples were centrifuged for 10 min at 12,000 rpm at 4 °C. Then 20 μl of supernatant was added to 100 μl of alkaline phosphatase reagent (Sigma), and the absorbance was measured at 504 nm after a 30-min incubation at 37 °C. Alkaline phosphatase levels were corrected with total proteins, whose levels were evaluated by DC protein assay reagent.
Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared from HeLa cells transduced with 100 MOI of AdCDX2. Cells were harvested 48 h after transduction by scraping and then resuspended in cold Tris-buffered saline (pH 7.4), followed by centrifugation at 4000 rpm for 2 min. Cells were resuspended in 100 μl of buffer A (10 mmol/liter HEPES, pH 7.9, 1.5 mmol/liter MgCl2, 10 mmol/liter KCl, and 0.5 mmol/liter phenylmethylsulphonyl fluoride) and incubated on ice for 15 min. After the addition of 4 μl of 10% Triton X-100, samples were vortexed for 10 s and centrifuged at 13,000 rpm for 2 min. Pellets were resuspended in 50 μl of cold buffer C (20 mmol/liter HEPES, pH 7.9, 1.5 mmol/liter MgCl2, 0.4 mol/liter NaCl, 0.2 mmol/liter EDTA, 25% glycerol, 0.2 mmol/liter DTT, and 0.5 mmol/liter phenylmethylsulphonyl fluoride) and incubated on ice for 30 min. Protein concentrations in nuclear extracts were 0.5–1 mg/ml, as determined by the Bradford method. The sense and antisense oligonucleotides containing the CDX2-binding sites were annealed with a complementary primer and radiolabeled with [32P]dCTP (Amersham Biosciences). The labeled probes were purified by size exclusion chromatography through a 0.5-ml column of Sephadex G-25 (Amersham Biosciences). Nuclear extracts (5 μg) were incubated with radiolabeled DNA probes (1 × 105 cpm) for 20 min at room temperature in 18 μl of binding buffer containing 60 mmol/liter HEPES, 180 mmol/liter KCl, 3 mmol/liter EDTA, 36% glycerol, 15 mmol/liter DTT, and 4 μg of poly(dI-dC). Protein-DNA complexes were separated from free DNA probes by electrophoresis through 5% non-denaturing acrylamide gels in 5× TBE buffer. Gels were dried under vacuum on filter paper and exposed for 16–40 h to Eastman Kodak Co. XAR film at −80 °C for autoradiograph detection. Specificity of binding was ascertained by competition with excess cold consensus oligonucleotides.
Chromatin Immunoprecipitation (ChIP)
HCT116 cells were transfected with pFLAG-CMV2 (Sigma) or pFLAG-CMV2-hCdx2 (41) using JetPrime (Polyplus transfection) as recommended (1 × 106 cells in 3.5-cm plates: 2.4 μg of total DNA and 4.8 μl of JetPrime in 250 μl of JetPrime buffer). After 48 h, transfected cells were fixed with 1% (v/v) formaldehyde for 10 min at room temperature and quenched with 0.125 m glycine for 5 min. ChIP experiments were carried out using the Magna ChIPTM G chromatin immunoprecipitation kit as recommended by the supplier (Millipore). Sheared cross-linked chromatin from ∼106 cells was incubated overnight at 4 °C with 2 μg of normal mouse IgG (Santa Cruz Biotechnology, Inc.) or mouse anti-FLAG (clone M2, Sigma) antibody. Input corresponded to non-immunoprecipitated sheared cross-linked chromatin from ∼105 cells (1%). qPCR analyses were performed with one-twenty-fifth of the immunoprecipitated DNA as template using SYBR Green Master Mix in a 7500Fast PCR detection system (Applied Biosystem). Primers are located in the human FXR promoter (forward, GAGCCAGTGAACAGAAACCC; reverse, AGTGAGAGAGGACAGAGGTTG).
Statistical Analysis
All results are expressed as mean ± S.E. Data distribution and gene expression statistical analysis were performed using NCSS statistical and power analysis software 2007 (Kaysville, UT). Multiple groups were tested by one-way analysis of variance or two-way analysis of variance repeated measures where appropriate, followed by Fisher's least significant difference test for unpaired data. Comparisons of two groups were performed using Student's t test, followed by a Mann-Whitney U test where appropriate. p < 0.05 was considered significant.
RESULTS
Inactivating APC Mutations Result in Both CDX2 and FXR Reduced Intestinal Expression
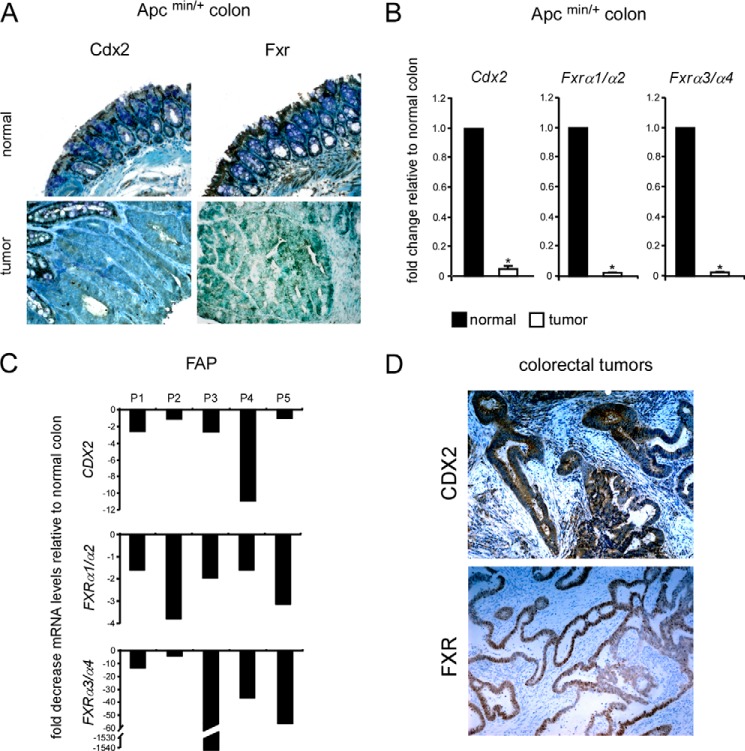
CDX2 is a downstream mediator of APC, and because most colorectal tumors present inactivating mutations of APC protein (42) that result in decreased expression and activity of CDX2 (3, 4), the link between CDX2 and FXR expression was first investigated in the context of APC-dependent tumors. To this end, we used samples of Apcmin/+ mice and FAP patients. Immunohistochemical analysis of colon sections from Apcmin/+ mice confirmed the parallel reduced expression of Cdx2 and Fxr also at the protein level when comparing tumors with normal adjacent regions (Fig. 1A). Also, in the colon of both models of intestinal tumorigenesis, the expression of CDX2 and FXRα1/α2 as well as FXRα3/α4 was significantly reduced at the mRNA level when compared with normal adjacent areas (Fig. 1, B and C). Interestingly, at support of the idea that CDX2 might regulate FXR expression, human differentiated colorectal tumor that retained a certain expression of CDX2 also presented a remarkable FXR expression (Fig. 1D).
FIGURE 1.
Intestinal down-regulation of CDX2 and FXR expression after inactivating APC mutations. A, paraffin-embedded specimens from normal colonic mucosa and colon tumors of Apcmin/+ mice were immunostained with Cdx2 and Fxr antibodies to determine expression and localization of CDX2 and FXR proteins in normal tissue and tumors (×200 magnification). B, qRT-PCR was used to investigate the relative expression of Cdx2, Fxrα1/α2, and Fxrα3/α4 in tumor samples and adjacent normal areas from the colon of Apcmin/+ mice. Values shown represent -fold change ± S.E. (error bars) (p < 0.05) for five independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and normal samples were used as a calibrator. C, qRT-PCR was used to investigate the relative expression of CDX2, FXRα1/α2, FXRα3/α4 in colon tumors from five FAP patients. Values shown are -fold decrease of mRNA levels in colon tumors relative to normal adjacent tissue. Cyclophilin was used as a housekeeping gene to normalize data. D, immunohistochemical analysis of sections of human colorectal tumors with certain CDX2 expression. Shown are representative fields of sections of human colon tumors stained with CDX2 and FXR antibodies (×200 magnification).
The Intestinal Expression of FXR Is Reduced in Cdx2+/− Mice
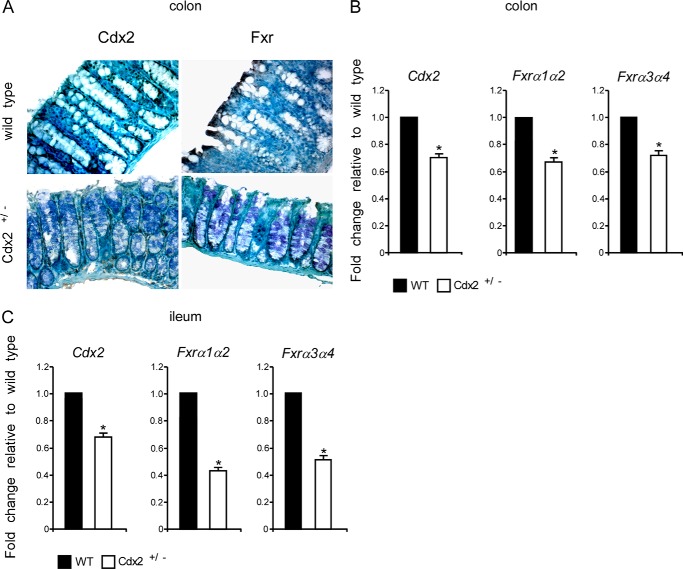
To further corroborate our hypothesis that CDX2 regulates FXR expression in vivo, we assessed mRNA and protein levels of Fxr in Cdx2+/− mice. Indeed, immunohistochemical analysis of morphologically normal colon sections from Cdx2+/− mice revealed a consequent reduced expression of Cdx2 and Fxr (Fig. 2A). This result was confirmed at the mRNA level, where both Fxrα1/α2 and Fxrα3/α4 isoforms displayed a reduced expression in Cdx2+/− mice compared with littermate wild-type mice (Fig. 2B). Notably, the same results were obtained for the small intestine, with an even more significant reduction of Fxr expression (Fig. 2C). Overall, our findings suggest that FXR expression in the intestinal mucosa is directly proportional to Cdx2 allelic quantity.
FIGURE 2.
Intestinal FXR expression is reduced in Cdx2+/− mice. A, paraffin-embedded specimens from normal colonic mucosa of Cdx2+/− mice and relative littermate wild-type mice were immunostained with CDX2 and FXR antibodies to determine expression and localization of CDX2 and FXR proteins (×200 magnification). B, qRT-PCR was used to investigate relative mRNA levels of Cdx2, Fxrα1/α2, and Fxrα3/α4 in the colon of Cdx2+/− mice and relative littermate wild-type mice. Values shown represent -fold change ± S.E. (error bars) (p < 0.05) for five independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and wild-type mice were used as a calibrator. C, qRT-PCR was used to investigate relative mRNA levels of Cdx2, Fxrα1/α2, and Fxrα3/α4 in the ileum of Cdx2+/− mice and relative littermate wild-type mice. Values shown represent -fold change ± S.E. (p < 0.05) for five independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and wild-type mice were used as a calibrator.
FXR Expression Correlates with Increased CDX2 Expression during Enterocyte Differentiation
APC gains a gradient of expression along the crypt-villus axis with lower expression in the proliferative compartment and higher expression in the differentiated compartment. Indeed, CDX2, which is expressed all along the crypt-villus axis, is mainly active in differentiating enterocytes (9, 11, 43), where it induces the transcription of several genes associated with differentiation of the intestinal epithelium. In line with these observations, several in vitro studies have reported that overexpression of CDX2 in gut epithelial cells leads to growth arrest accompanied by up-regulation of several markers associated with intestinal differentiation (44, 45). Because FXR is also expressed in the differentiated compartment of the intestinal epithelium (Fig. 1A), we investigated whether the expression of FXR follows that of CDX2 during differentiation.
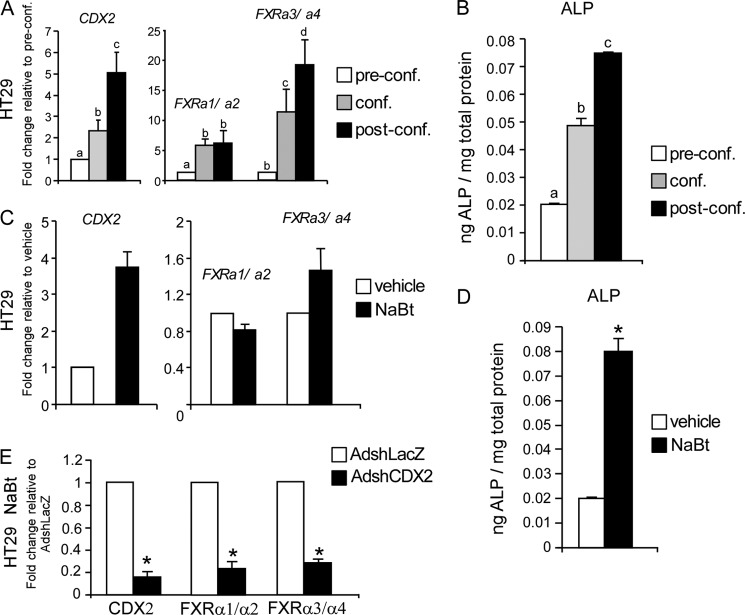
In vitro cell culture experiments have shown that cell-to-cell contact can trigger both cell cycle exit and induction of the differentiation process (46). To investigate the correlation between CDX2 and FXR expression during enterocyte differentiation, we used colorectal cancer HT29 cells, which show morphological features of enterocyte differentiation when maintained in the confluence state. In contrast, in colon tumor specimens with APC mutations (Fig. 1, A and B), we noticed an increased FXR expression (mainly FXRα3/α4) over time along with that of CDX2 at the mRNA level (Fig. 3A). These results were also confirmed in Caco2 cells, which are widely used to study the intestinal differentiation process in vitro (data not shown). In support of our model of intestinal differentiation in vitro, we observed an increase of alkaline phosphatase (ALP) activity from pre- to postconfluence state (Fig. 3B). Indeed, ALP is a well established marker of intestinal differentiation, which is expressed by differentiated but not by undifferentiated enterocytes.
FIGURE 3.
Correlation between CDX2 and FXR expression during in vitro confluence- and NaBt-induced differentiation. A, relative mRNA levels of FXRα1/α2, FXRα3/α4, and CDX2 in HT29 cells (preconfluence, confluence, and postconfluence) were investigated by real-time qPCR. Values shown represent -fold change ± S.E. (error bars) of four independent determinations performed in duplicate. Cyclophilin was used as housekeeping gene to normalize data, and preconfluent cells were used as calibrators. Different lowercase letters indicate significant difference (p < 0.05). B, ALP activity was analyzed as marker of differentiation in HT29 cells from pre- to postconfluence. Values shown represent mean ± S.E. (p < 0.05) for four independent determinations performed in duplicate and were normalized to total mg of proteins. C, relative mRNA levels of CDX2, FXRα1/α2, and FXRα3/α4 in HT29 cells treated with 1 mm NaBt for 24 h. Values shown represent -fold change ± S.E. of four independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and vehicle-treated cells were used as calibrators. D, ALP activity was analyzed as marker of differentiation in HT29 cells treated with NaBt 1 mm for 48 h. Values shown represent mean ± S.E. (p < 0.05) for four independent determinations performed in duplicate and were normalized to total mg of proteins. E, down-regulation of CDX2 during enterocyte NaBt-induced differentiation impairs FXR up-regulation. HT29 cells were first transduced with 200 MOI of AdshCDX2 or AdshLacZ and then, 24 h after adenovirus transduction, treated with 1 mm NaBt. CDX2, FXRα1/α2, and FXRα3/α4 mRNA levels were investigated 48 h after NaBt treatment. Values shown represent -fold change ± S.E. (p < 0.05) of four independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and AdshLacZ transduced cells were used as calibrators.
In addition to differentiation induced by cell-to-cell contact, we also investigated a chemical model of differentiation. NaBt is a short-chain fatty acid formed in the gastrointestinal tract of mammals as a result of anaerobic bacterial fermentation of soluble dietary fibers and is avidly absorbed by the colonic epithelium. In vitro studies have indicated that NaBt induces differentiation and apoptosis, effects that might mediate the protective role of dietary fibers against CRC. Treatment of HT29 cells with NaBt at 1 mm concentration was able to induce the expression of CDX2 and FXR (mainly FXRα3/α4) at the mRNA level (Fig. 3C). Again, differentiation was confirmed by the increased levels of ALP following NaBt treatment (Fig. 3D).
To further confirm that the induction of FXR expression during enterocyte differentiation is dependent on CDX2 activity, HT29 cells were first transduced with AdshCDX2 or AdshLacZ and then treated with NaBt to induce differentiation. As shown in Fig. 3E, treatment with NaBt was able to induce FXRα3/α4 expression only in the presence of AdshLacZ and not in the presence of AdshCDX2. These data demonstrate that CDX2 is required to induce FXR expression during enterocyte maturation.
Functional Induction of FXR in the Enterocytes via CDX2
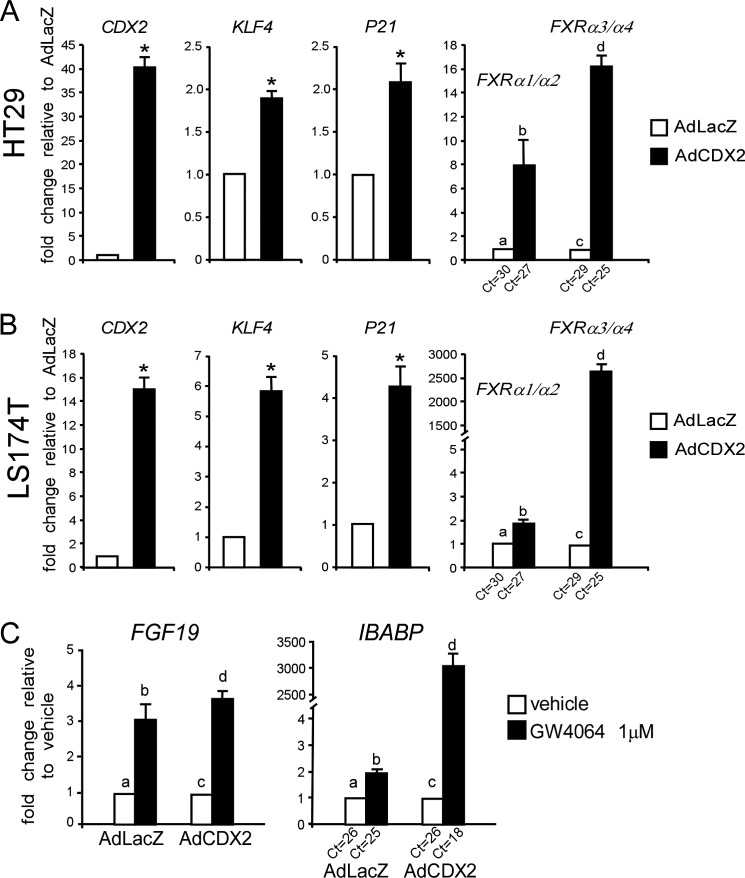
To further demonstrate that CDX2 induces FXR expression, HT29 cells were transduced with AdCDX2 or control AdLacZ. Ectopic expression of CDX2 resulted in higher mRNA levels of FXRα3/α4 and also FXRα1/α2 (Fig. 4A). KLF4 and P21 and CDX2 target genes, were also induced by CDX2 transduction. Interestingly, the same results were obtained in LS174T cells, which do not differentiate into enterocytes (Fig. 4B). Because HT29 cells have a mutated form of APC but a wild type β-CATENIN, whereas LS174T cells have a wild type APC but a mutated β-CATENIN, these results underline the independence of CDX2 transcriptional activity from the cytoplasmic APC and nuclear β-CATENIN-TCF-4 status of the cell.
FIGURE 4.
Functional induction of FXR by CDX2. A, HT29 cells were transduced with 100 MOI of AdCDX2 or AdLacZ, and relative mRNA levels of CDX2, KLF4, P21, FXRα1/α2, and FXRα3/α4 were investigated by qRT-PCR after 48 h. Values shown represent -fold change ± S.E. (error bars) (p < 0.05) for four independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and AdLacZ-transduced cells were used as a calibrator. Different lowercase letters indicate significant difference (p < 0.05). B, LS174T cells were transduced with 100 MOI of AdCDX2 or AdLacZ, and relative mRNA levels of CDX2, KLF4, P21, FXRα1/α2, and FXRα3/α4 were investigated by qRT-PCR after 48 h. Values shown represent -fold change ± S.E. (p < 0.05) for four independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and AdLacZ-transduced cells were used as a calibrator. Different lowercase letters indicate significant difference (p < 0.05). C, 48 h after AdCDX2 or AdLacZ transduction, HT29 cells were treated with 1 μm GW4064 for 24 h, and the expression of two FXR target genes (FGF19 and IBABP) was investigated. Values shown represent -fold change ± S.E. for four independent determinations performed in duplicate. Cyclophilin was used as a housekeeping gene to normalize data, and AdLacZ-transduced cells treated with vehicle were used as calibrator. Different lowercase letters indicate significant difference (p < 0.05).
To investigate whether the induction of FXR by CDX2 was also functional, HT29 cells were first transduced with AdCDX2 or AdLAcZ and then treated with GW4064, a selective FXR ligand. As shown in Fig. 4C, treatment with GW4064 resulted in further induction of FGF19 and IBABP (FXR target genes) after AdCDX2 transduction. These results indicate that CDX2 induction of FXR expression is coupled with augmented FXR transcriptional activation in response to ligand.
In Silico Analysis of FXR Promoters
To identify new candidates for the transcriptional regulation of FXR in the intestine, in silico analysis of both FXR promoters until −3000 bp from their relative TSS was performed using the Mat Inspector software (Genomatix). In particular, we focused on transcription factors that are both specifically expressed in the intestine and under the control of APC. Thus, we found several TTTAT (sense) or ATAAA (antisense) motifs that have been shown to bind to CDX2, a downstream mediator of APC, which is exclusively expressed in the gut.
CDX2 Is a Direct Transcriptional Activator of FXRα3/α4 Promoter
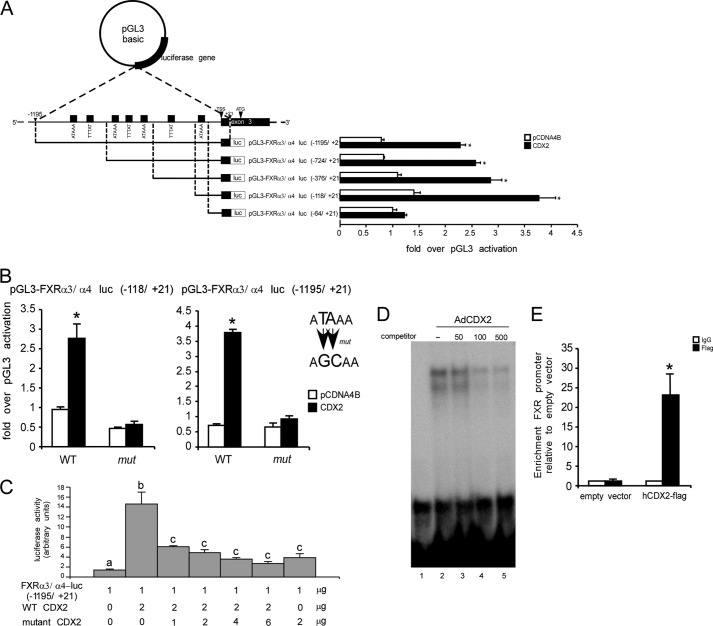
Following our in silico analysis, both FXRα1/α2 and FXRα3/α4 promoter regions were cloned into the pGL3-luc basic vector to evaluate whether FXR is a transcriptional direct target of CDX2 by a reporter assay. Indeed, FXR-luc reporter constructs and pCDNA4B-CDX2 expression vector were co-transfected into HeLa cells, which lack endogenous CDX2. With respect to FXRα1/α2, although we cloned until −3000 bp from its TSS, CDX2 was not able to trans-activate this promoter. On the other hand, when the FXRα3/α4-luc(−1195/+21) reporter along with pCDN4B-CDX2 were co-transfected, CDX2 was able to trans-activate the FXRα3/α4 promoter (Fig. 5A). These data suggest that CDX2 might regulate directly FXRα3/α4 and indirectly FXRα1/α2. To better characterize the putative CDX2 binding sites in this region of the FXRα3/α4 promoter, we generated a series of reporters, including a progressively decreasing number of the putative binding sites of CDX2 (Fig. 5A). As a result, deletions of putative CDX2 binding sites on the 5′ side from −1195 to −118 did not affect the ability of CDX2 to transactivate this promoter, suggesting that the putative CDX2 binding site closer to the TSS is crucial for CDX2 to transactivate the FXRα3/α4 promoter. Indeed, in situ mutagenesis of this putative CDX2 binding site abrogated the ability of CDX2 to transactivate the FXRα3/α4 promoter (Fig. 5B). These results clearly demonstrate that the sequence between −118 and −64 has a key role in inducing FXRα3/α4 expression via CDX2. The role of CDX2 in transactivating the FXRα3/α4 promoter has been further confirmed in a competition study where HeLa cells were co-transfected with the FXRα3/α4-luc(−1195/+21) reporter and increasing ratios of mutant devoid of transcriptional activity over wild-type CDX2. In the presence of mutant CDX2, the ability of wild-type CDX2 to transactivate the FXRα3/α4 promoter was dramatically reduced (Fig. 5C), underlining that a wild-type form of CDX2 is required to induce FXRα3/α4 expression.
FIGURE 5.
CDX2 is a direct transcriptional activator of FXRα3/α4 promoter. A, structure of the pGL3-FXRα3/α4-luc plasmid is shown. The pGL3-FXRα3/α4-luc(−1195/+21), pGL3-FXRα3/α4-luc(−724/+21), pGL3-FXRα3/α4-luc(−376/+21), pGL3-FXRα3/α4-luc(−118/+21), and pGL3-FXRα3/α4-luc(−64/+21) plasmids contain the 1195-, 724-, 376-, 118-, and 64-bp fragments, respectively, of the human FXRα3/α4 gene promoter plus the first 21 bp of exon 3 linked to a luciferase reporter. Seven putative CDX2 binding sites are shown. Luciferase activity was measured in response to expression of CDX2 in HeLa cells co-transfected with the pRL-TK Renilla luciferase reporter and the different pGL3-FXRα3/α4-luc plasmids. Results were normalized to Renilla luciferase activity and expressed as -fold over pGL3-basic activation. Values are expressed as the means ± S.E. (error bars) of triplicate cultures (p < 0.05). B, HeLa cells after transient co-transfection of CDX2 or relative empty vector (pCDNA4B), pGL3-FXRα3/α4-luc(−118/+21), and pGL3-FXRα3/α4-luc(−1195/+21) reporters (WT) or relative mutated plasmids for the candidate CDX2 binding site (mut). Results were normalized to Renilla luciferase activity and expressed as -fold over pGL3-basic activation. The results are expressed as the means ± S.E. of triplicate cultures (p < 0.05). C, the effect of mutant CDX2 on the ability of wild-type CDX2 to transactivate FXRα3/α4 promoter. HeLa cells were co-transfected with pGL3-FXRα3/α4-luc(−1195/+21), pRL-TK Renilla luciferase reporter, pCDNA4B, and either wild-type CDX2 alone, wild-type CDX2 with increasing amounts of mutant CDX2, or mutant CDX2 alone. The total amount of DNA transfected was kept constant at 15 μg. The ratio of firefly luciferase activity to control Renilla luciferase activity is indicated (in arbitrary units). All experiments are expressed as the means ± S.E. of triplicate cultures. Different lowercase letters, significant statistical difference (p < 0.05). D, EMSA shows physical interaction between CDX2 protein and FXRα3/α4 promoter. Synthetic oligonucleotides containing the putative CDX2 binding site between −118 and +21 on the FXRα3/α4 promoter are reported under “Results.” Nuclear extract (5 μg) from HeLa cells transduced with AdCDX2 was incubated with the 32P-labeled FXR oligonucleotide probe alone (lane 2) and in the presence of 50, 100, or 500 unlabeled FXR competitor oligonucleotides (lanes 3–5). In lane 1, only 32P-labeled FXR oligonucleotide was added. Samples were loaded on a 5% acrylamide gel. E, binding of CDX2 to the regulatory region of FXR was confirmed by ChIP-qPCR in the HT116 cell line transfected with pFLAG-CMV-2-hCDX2 plasmid. Results are shown as enrichment of FXR promoter relative to empty vector. We used normal IgG as negative control. Bar plots, mean ± S.E. of two independent experiments. The significance of difference was determined by Student's paired t test.
Finally, to examine the physical interaction between FXRα3/α4 promoter and CDX2 homeodomain protein, we performed an EMSA. The oligonucleotide sequences used in this study were CTTTCCTCATAAACATTTACA (forward) and TGTAAATGTTTATGAGGAAAG (reverse). They were chosen on the base of the deletion mutant analysis, indicating that the portion from −118 to −64 contains the genuine CDX2 binding site. These sequences were labeled, incubated in the presence of nuclear extract from HeLa cells transduced with AdCDX2, and then migrated through a 5% non-denaturating polyacrylamide gel. The autoradiograph in Fig. 5D reveals the position of rapidly migrating unbound probes at the base of the gel and several DNA-protein complexes of slower mobility that were formed after incubation with the HeLa nuclear extract (lane 2), whereas no band shift occurred in lane 1, in which no nuclear extract was added. The specificity of DNA/CDX2 protein complexes was ascertained by competition with a 50–500-fold excess of cold oligonucleotides (lanes 3–5). These results indicate that CDX2 interacts physically with the FXRα3/α4 promoter. In order to further demonstrate that FXR is a CDX2 target gene, we performed chromatin immunoprecipitation (ChIP) experiments in HT116, a colon cancer cell line. We observed a significant enrichment of FXR promoter in HCT116 cell line transfected with pFLAG-CMV-2-hCDX2 over empty vector as well as negative controls as IgG (Fig. 5E). These data show the direct binding of CDX2 on the FXR promoter in vivo, thus further supporting the hypothesis that CDX2 regulates FXR expression in the gut.
DISCUSSION
FXR is highly expressed in the enterohepatic system, where BAs, which are its endogenous ligands, circulate. Upon activation by BAs, FXR is engaged in many molecular pathways for the management of BA homeostasis, including reduced synthesis and increased detoxification. Indeed, physiological FXR expression and function is required not only to preserve BA homeostasis but also to temper their deleterious effects in the enterohepatic system, as shown in Fxr-null mice (28–31, 47). Activation of FXR can induce the orphan nuclear receptor SHP (small heterodimer partner) (48) and FGF19/Fgf15 (49) expression in the liver and intestine, respectively, to synergistically reduce BA synthesis. At the same time, activation of FXR in the enterohepatic system induces CYP3A4/Cyp3a11 (cytochrome P450 3A4/3a11), SULT2A1 (cytosolic sulfotransferase 2A1), and UGT2B4 (UDP-glycosyltransferase 2B4) to induce BA detoxification (37).
Although different FXR target genes important to deal with BA levels have been identified in the enterohepatic system, so far the molecular mechanisms regulating FXR expression in the intestine have remained unknown. In the present study, we shed light on this issue, showing that CDX2 positively regulates FXR expression in the intestine. Because we previously showed that upon inactivating the APC mutation, the intestinal expression of FXR is dramatically reduced (29), we focused on a transcriptional downstream mediator of APC activity that was co-expressed with FXR in the differentiated compartment of the intestinal epithelium. Our analysis revealed several putative binding sites for CDX2 in both FXRα1/α2 and FXRα3/α4 promoters. Because of APC induces the expression of CDX2 (4), we found that the expression of both FXRα1/α2 and FXRα3/α4 was reduced not only in Apcmin/+ mice but also in FAP patients. Additional data indicating that FXR is a target gene of CDX2 were obtained in Cdx2+/− mice, where the reduced expression of Cdx2 proceeded with the decreased mRNA levels of both Fxrα1/α2 and Fxrα3/α4. The correlation between CDX2 and FXR expression was also confirmed in an in vitro model of enterocyte differentiation. During the confluence- or NaBt-induced differentiation of HT29 and Caco2 cells, the expression of FXR (mainly FXRα3/α4) resulted increased along with CDX2. As support of the role of CDX2 in inducing the expression of FXR during differentiation, knocking down CDX2 during NaBt treatment resulted in a lack of FXR induction. Moreover, infection with AdCDX2 of HT29 cells and LS174 cells resulted in overexpression of FXR (mainly FXRα3/α4). Interestingly, the induction of FXR by CDX2 was also functional because treatment with GW4064 resulted in up-regulation of FXR target genes, such as FGF19 and IBABP. Finally, a reporter and EMSA assay along with ChIP experiments on the FXRα3/α4 promoter demonstrated a direct regulation of FXRα3/α4 by CDX2. With regard to FXRα1/α2, we could not show a direct regulation of its promoter by CDX2 at the beginning of the region that we investigated (−3000 bp from the TSS). This does not exclude the possibility of a genuine CDX2 binding site even upstream of the region investigated by us. However, the fact that in our gain of function studies CDX2-increased expression correlates mainly with FXRα3/α4 up-regulation when compared with FXRα1/α2 and the fact that CDX2 binding sites are generally close to the TSS of its target genes (as confirmed for FXRα3/α4) suggest that CDX2 might regulate the expression of FXRα1/α2 in an indirect way. Alternatively, the proximal regulatory site identified for FXRα3/α4 may also act as a distant regulatory site for FXRα1/α2. Further studies will be required to shed light on a possible downstream target of CDX2 that would regulate the expression of FXRα1/α2.
Combining our new results with what we previously published (29), we can now speculate that, after inactivating mutations of the APC gene, which is believed to be the primum movens in the sequence from normal colonic mucosa to CRC, FXR expression would be decreased, as a consequence of the reduced expression and activity of the APC downstream mediator CDX2. In this scenario, a long term “Western style” diet would produce high levels of toxic BAs that would not be buffered by the detoxification activity of FXR and would be free to further promote CRC via FXR-independent mechanisms. As a result, a high fat diet would reduce the life span for the manifestation of CRC, contributing as one of the environmental factors responsible for the increased susceptibility to CRC observed in the industrialized world. Finally, because it has been also shown that BAs can induce CDX2 expression (50), one could speculate that BA might stimulate their own detoxification in the intestine by inducing CDX2 expression, which upon activation by APC would stimulate FXR expression that, although promoting apoptosis, would induce the expression of its target genes to induce BA detoxification. Thus, when the APC-CDX2 axis is altered, as in the majority of CRC, the reduced bile acid detoxification would account for the increased risk of CRC development. This finding is concomitant with a report demonstrating that CDX2 induces xenobiotic metabolism enzyme genes in the intestine, an action that could be mediated by FXR after induction by CDX2.
In conclusion, after becoming an adopted NR in 1999 with the discovery of BAs as its endogenous ligands (22–24), herein for the first time we provide evidence that CDX2 is the transcriptional regulator of intestinal FXR. We believe that our findings will provide the impetus to develop novel strategies aiming at increasing intestinal FXR expression and activity to prevent or delay high fat diet-related CRC development.
Acknowledgments
We thank Dr. R. Valanzano (University of Florence) and Dr. R. Mariani Costantini (G. D'Annunzio University of Chieti-Pescara) for providing FAP patient samples. We also thank Dr. F. Beck for providing the CDX2+/− mice.
This work was supported by Italian Association for Cancer Research (AIRC; Milan, Italy) Grant IG 14732 (to A. M.); the Italian Ministry of University and Education (Finanziamenti per la Ricerca di Base IDEAS RBID08C9N7 (to A. M., Programma Operativo Nazionale PON01_01958 (to A. M.), and PRIN 2010FHH32M_002 to (to A. M.)); Italian Ministry of Health Grants GR-2008-1143546 and GR-2010-2314703 (to A. M.); NR-Net Marie Curie ITN (to A. M.)); University of Bari, Italy, Grants ORBA 08WEZJ, 07X7Q1, 06BXVC, and IDEA GRBA0802SJ (to A. M.); and CariSPAQ (L'Aquila, Italy).
- CRC
- colorectal cancer
- NR
- nuclear receptor
- BA
- bile acid
- FAP
- familial adenomatous polyposis
- TSS
- transcriptional start site(s)
- NaBt
- sodium butyrate
- MOI
- multiplicity of infection
- ALP
- alkaline phosphatase
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Clevers H. (2006) Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 2. Bai Y. Q., Miyake S., Iwai T., Yuasa Y. (2003) CDX2, a homeobox transcription factor, upregulates transcription of the p21/WAF1/CIP1 gene. Oncogene 22, 7942–7949 [DOI] [PubMed] [Google Scholar]
- 3. Dang D. T., Mahatan C. S., Dang L. H., Agboola I. A., Yang V. W. (2001) Expression of the gut-enriched Krüppel-like factor (Krüppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene 20, 4884–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. da Costa L. T., He T. C., Yu J., Sparks A. B., Morin P. J., Polyak K., Laken S., Vogelstein B., Kinzler K. W. (1999) CDX2 is mutated in a colorectal cancer with normal APC/β-catenin signaling. Oncogene 18, 5010–5014 [DOI] [PubMed] [Google Scholar]
- 5. Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997) Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84–87 [DOI] [PubMed] [Google Scholar]
- 6. Beck F., Chawengsaksophak K., Waring P., Playford R. J., Furness J. B. (1999) Reprogramming of intestinal differentiation and intercalary regeneration in Cdx2 mutant mice. Proc. Natl. Acad. Sci. U.S.A. 96, 7318–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao N., White P., Kaestner K. H. (2009) Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stringer E. J., Duluc I., Saandi T., Davidson I., Bialecka M., Sato T., Barker N., Clevers H., Pritchard C. A., Winton D. J., Wright N. A., Freund J. N., Deschamps J., Beck F. (2012) Cdx2 determines the fate of postnatal intestinal endoderm. Development 139, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S., Shivdasani R. A. (2010) Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Houde M., Laprise P., Jean D., Blais M., Asselin C., Rivard N. (2001) Intestinal epithelial cell differentiation involves activation of p38 mitogen-activated protein kinase that regulates the homeobox transcription factor CDX2. J. Biol. Chem. 276, 21885–21894 [DOI] [PubMed] [Google Scholar]
- 11. Rings E. H., Boudreau F., Taylor J. K., Moffett J., Suh E. R., Traber P. G. (2001) Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology 121, 1437–1450 [DOI] [PubMed] [Google Scholar]
- 12. Gross I., Duluc I., Benameur T., Calon A., Martin E., Brabletz T., Kedinger M., Domon-Dell C., Freund J. N. (2008) The intestine-specific homeobox gene Cdx2 decreases mobility and antagonizes dissemination of colon cancer cells. Oncogene 27, 107–115 [DOI] [PubMed] [Google Scholar]
- 13. Brabletz T., Spaderna S., Kolb J., Hlubek F., Faller G., Bruns C. J., Jung A., Nentwich J., Duluc I., Domon-Dell C., Kirchner T., Freund J. N. (2004) Down-regulation of the homeodomain factor Cdx2 in colorectal cancer by collagen type I: an active role for the tumor environment in malignant tumor progression. Cancer Res. 64, 6973–6977 [DOI] [PubMed] [Google Scholar]
- 14. Ee H. C., Erler T., Bhathal P. S., Young G. P., James R. J. (1995) Cdx-2 homeodomain protein expression in human and rat colorectal adenoma and carcinoma. Am. J. Pathol. 147, 586–592 [PMC free article] [PubMed] [Google Scholar]
- 15. Subtil C., Guérin E., Schneider A., Chenard M. P., Martin E., Domon-Dell C., Duluc I., Brabletz T., Kedinger M., Duclos B., Gaub M. P., Freund J. N. (2007) Frequent rearrangements and amplification of the CDX2 homeobox gene in human sporadic colorectal cancers with chromosomal instability. Cancer Lett. 247, 197–203 [DOI] [PubMed] [Google Scholar]
- 16. Baba Y., Nosho K., Shima K., Freed E., Irahara N., Philips J., Meyerhardt J. A., Hornick J. L., Shivdasani R. A., Fuchs C. S., Ogino S. (2009) Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin. Cancer Res. 15, 4665–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Sousa E Melo F., Wang X., Jansen M., Fessler E., Trinh A., de Rooij L. P., de Jong J. H., de Boer O. J., van Leersum R., Bijlsma M. F., Rodermond H., van der Heijden M., van Noesel C. J., Tuynman J. B., Dekker E., Markowetz F., Medema J. P., Vermeulen L. (2013) Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 19, 614–618 [DOI] [PubMed] [Google Scholar]
- 18. Aoki K., Tamai Y., Horiike S., Oshima M., Taketo M. M. (2003) Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/− compound mutant mice. Nat. Genet. 35, 323–330 [DOI] [PubMed] [Google Scholar]
- 19. Bonhomme C., Duluc I., Martin E., Chawengsaksophak K., Chenard M. P., Kedinger M., Beck F., Freund J. N., Domon-Dell C. (2003) The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut 52, 1465–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mallo G. V., Soubeyran P., Lissitzky J. C., André F., Farnarier C., Marvaldi J., Dagorn J. C., Iovanna J. L. (1998) Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J. Biol. Chem. 273, 14030–14036 [DOI] [PubMed] [Google Scholar]
- 21. Otte K., Kranz H., Kober I., Thompson P., Hoefer M., Haubold B., Remmel B., Voss H., Kaiser C., Albers M., Cheruvallath Z., Jackson D., Casari G., Koegl M., Pääbo S., Mous J., Kremoser C., Deuschle U. (2003) Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell Biol. 23, 864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. (1999) Identification of a nuclear receptor for bile acids. Science 284, 1362–1365 [DOI] [PubMed] [Google Scholar]
- 23. Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. (1999) Bile acids: natural ligands for an orphan nuclear receptor. Science 284, 1365–1368 [DOI] [PubMed] [Google Scholar]
- 24. Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. (1999) Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553 [DOI] [PubMed] [Google Scholar]
- 25. Huber R. M., Murphy K., Miao B., Link J. R., Cunningham M. R., Rupar M. J., Gunyuzlu P. L., Haws T. F., Kassam A., Powell F., Hollis G. F., Young P. R., Mukherjee R., Burn T. C. (2002) Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene 290, 35–43 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Kast-Woelbern H. R., Edwards P. A. (2003) Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. J. Biol. Chem. 278, 104–110 [DOI] [PubMed] [Google Scholar]
- 27. Lee H., Zhang Y., Lee F. Y., Nelson S. F., Gonzalez F. J., Edwards P. A. (2006) FXR regulates organic solute transporters α and β in the adrenal gland, kidney, and intestine. J. Lipid Res. 47, 201–214 [DOI] [PubMed] [Google Scholar]
- 28. Kim I., Morimura K., Shah Y., Yang Q., Ward J. M., Gonzalez F. J. (2007) Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 28, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Modica S., Murzilli S., Salvatore L., Schmidt D. R., Moschetta A. (2008) Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 68, 9589–9594 [DOI] [PubMed] [Google Scholar]
- 30. Yang F., Huang X., Yi T., Yen Y., Moore D. D., Huang W. (2007) Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67, 863–867 [DOI] [PubMed] [Google Scholar]
- 31. Degirolamo C., Modica S., Palasciano G., Moschetta A. (2011) Bile acids and colon cancer: solving the puzzle with nuclear receptors. Trends Mol. Med. 17, 564–572 [DOI] [PubMed] [Google Scholar]
- 32. Modica S., Gofflot F., Murzilli S., D'Orazio A., Salvatore L., Pellegrini F., Nicolucci A., Tognoni G., Copetti M., Valanzano R., Veschi S., Mariani-Costantini R., Palasciano G., Schoonjans K., Auwerx J., Moschetta A. (2010) The intestinal nuclear receptor signature with epithelial localization patterns and expression modulation in tumors. Gastroenterology 138, 636–648 [DOI] [PubMed] [Google Scholar]
- 33. Maran R. R., Thomas A., Roth M., Sheng Z., Esterly N., Pinson D., Gao X., Zhang Y., Ganapathy V., Gonzalez F. J., Guo G. L. (2009) Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 328, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 35. Grober J., Zaghini I., Fujii H., Jones S. A., Kliewer S. A., Willson T. M., Ono T., Besnard P. (1999) Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene: involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274, 29749–29754 [DOI] [PubMed] [Google Scholar]
- 36. Landrier J. F., Eloranta J. J., Vavricka S. R., Kullak-Ublick G. A. (2006) The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-α and -β genes. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G476–G485 [DOI] [PubMed] [Google Scholar]
- 37. Modica S., Bellafante E., Moschetta A. (2009) Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front. Biosci. 14, 4719–4745 [DOI] [PubMed] [Google Scholar]
- 38. Cama A., Palmirotta R., Curia M. C., Esposito D. L., Ranieri A., Ficari F., Valanzano R., Battista P., Modesti A., Tonelli F. (1995) Multiplex PCR analysis and genotype-phenotype correlations of frequent APC mutations. Hum. Mutat. 5, 144–152 [DOI] [PubMed] [Google Scholar]
- 39. Scholl C., Bansal D., Döhner K., Eiwen K., Huntly B. J., Lee B. H., Rücker F. G., Schlenk R. F., Bullinger L., Döhner H., Gilliland D. G., Fröhling S. (2007) The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Invest. 117, 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas M. A., Lichtenstein D. L., Krajcsi P., Wold W. S. (2007) A real-time PCR method to rapidly titer adenovirus stocks. Methods Mol. Med. 130, 185–192 [DOI] [PubMed] [Google Scholar]
- 41. Hinkel I., Duluc I., Martin E., Guenot D., Freund J. N., Gross I. (2012) Cdx2 controls expression of the protocadherin Mucdhl, an inhibitor of growth and β-catenin activity in colon cancer cells. Gastroenterology 142, 875–885.e3 [DOI] [PubMed] [Google Scholar]
- 42. Fearon E. R., Vogelstein B. (1990) A genetic model for colorectal tumorigenesis. Cell 61, 759–767 [DOI] [PubMed] [Google Scholar]
- 43. Silberg D. G., Swain G. P., Suh E. R., Traber P. G. (2000) Cdx1 and cdx2 expression during intestinal development. Gastroenterology 119, 961–971 [DOI] [PubMed] [Google Scholar]
- 44. Lorentz O., Duluc I., Arcangelis A. D., Simon-Assmann P., Kedinger M., Freund J. N. (1997) Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J. Cell Biol. 139, 1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suh E., Traber P. G. (1996) An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell Biol. 16, 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taupin D., Podolsky D. K. (1999) Mitogen-activated protein kinase activation regulates intestinal epithelial differentiation. Gastroenterology 116, 1072–1080 [DOI] [PubMed] [Google Scholar]
- 47. Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 [DOI] [PubMed] [Google Scholar]
- 48. Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 49. Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., Goodwin B., Jones S. A. (2003) Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kazumori H., Ishihara S., Rumi M. A., Kadowaki Y., Kinoshita Y. (2006) Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut 55, 16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]