Background: RGS-PX proteins are regulators of signaling and trafficking within the endosomal system.
Results: A structural basis for membrane interactions of RGS-PX proteins is established.
Conclusion: The four mammalian paralogues display different membrane interaction properties.
Significance: RGS-PX proteins possess a conserved functional architecture in all eukaryotes.
Keywords: Cell Signaling, Endosome, G Protein-coupled Receptor (GPCR), Membrane Trafficking, Phosphatidylinositol, Regulator of G Protein Signaling (RGS), Scaffold protein, X-ray Crystallography, Phox Homology Domain, Sorting Nexin
Abstract
Sorting nexins (SNXs) or phox homology (PX) domain containing proteins are central regulators of cell trafficking and signaling. A subfamily of PX domain proteins possesses two unique PX-associated domains, as well as a regulator of G protein-coupled receptor signaling (RGS) domain that attenuates Gαs-coupled G protein-coupled receptor signaling. Here we delineate the structural organization of these RGS-PX proteins, revealing a protein family with a modular architecture that is conserved in all eukaryotes. The one exception to this is mammalian SNX19, which lacks the typical RGS structure but preserves all other domains. The PX domain is a sensor of membrane phosphoinositide lipids and we find that specific sequence alterations in the PX domains of the mammalian RGS-PX proteins, SNX13, SNX14, SNX19, and SNX25, confer differential phosphoinositide binding preferences. Although SNX13 and SNX19 PX domains bind the early endosomal lipid phosphatidylinositol 3-phosphate, SNX14 shows no membrane binding at all. Crystal structures of the SNX19 and SNX14 PX domains reveal key differences, with alterations in SNX14 leading to closure of the binding pocket to prevent phosphoinositide association. Our findings suggest a role for alternative membrane interactions in spatial control of RGS-PX proteins in cell signaling and trafficking.
Introduction
The sorting nexin (SNX)6 or phox homology (PX) domain-containing proteins are a large and diverse family of molecules with many different roles in cellular trafficking and signaling (1–3). The defining feature of these molecules is the presence of a PX domain, which is coupled to a variety of different functional modules. This core PX structure is generally able to bind membrane phosphatidylinositol (PtdIns) phospholipids or phosphoinositides, and thus acts to recruit different functional activities including membrane remodeling, protein scaffolding, lipid breakdown and synthesis, and GTPase activating (GAP) domains to membrane-bound compartments of the endocytic system.
The PX domain is one of a number of cytoplasmic lipid-binding modules that have been shown to bind to specific phosphoinositide lipids for membrane recruitment (1, 2, 4–6). Phosphoinositides are generated by phosphorylation and dephosphorylation of PtdIns by phosphoinositide kinases and phosphatases at the 3-, 4-, and 5-hydroxyls of the inositol headgroup, and serve as spatially restricted markers of intracellular organelles by acting as stereospecific platforms for protein recruitment. For example, the plasma membrane is enriched in phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), whereas early or sorting endosomes are enriched with PtdIns3P. The PX domain most commonly binds to the endosomal lipid marker PtdIns3P, although other specificities have been observed (3, 6).
A subset of homologous proteins in mammals, SNX13, SNX14, SNX19, and SNX25, play important roles in cellular signaling, trafficking, and development (3). Despite their importance, however, no structural information is available on this subfamily of SNX molecules. All but SNX19 possess a regulator of G-protein signaling (RGS) domain. RGS domains are small α-helical structures that act as GAPs for Gα subunits of heterotrimeric G-proteins, and thus play critical roles in attenuating G-protein coupled receptor (GPCR) signaling (7, 8). SNX13 (also called RGS-PX1) was identified as a specific GAP and regulator of the trimeric G-protein subunit Gαs involved in cAMP signaling from GPCRs such as the serotonin and β-adrenergic receptors (9). Initial work isolated SNX13 following searches for a potential GAP for Gαs G-protein isoforms, which at the time had not been identified (9). SNX13 was shown to both bind and promote GTPase activity of Gαs, and attenuate Gαs-mediated signaling. Subsequent work found that SNX13 null mice were embryonically lethal, and yolk sac endoderm cells of these embryos display highly disrupted endosome morphology, suggesting that it plays a role in endosomal function that is essential for normal development (10). SNX13 was also shown to form a heteromeric complex on endosomes with both Gαs and hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), a component of the ESCRT pathway (11), and influences the lysosomal targeting of the epidermal growth factor receptor, suggesting a role both in signal attenuation and receptor trafficking (12). These findings regarding SNX13 may be particularly important given the increasing recognition that endosomes are critical sites of signal regulation for many types of GPCRs and other signaling receptors (13–16).
Whether the RGS domains of SNX14 and SNX25 share this GAP activity remains unknown. Recently Hao and colleagues (17) demonstrated a role for SNX25 as a regulator of transforming growth factor β (TGF-β) signaling, binding to and promoting the lysosomal trafficking and degradation of TGF-β receptors (TβRs). Its increased expression in the temporal cortex has also been correlated with temporal lobe epilepsy, although its direct role in the disease has not been confirmed (18). The SNX14 protein is expressed in motor neurons (19) and recent studies suggest that the Snx14 gene is paternally imprinted in neuronal cells, where it promotes synaptic transmission through unknown mechanisms (20). SNX19 is the only member of the family lacking an RGS domain (see below). It plays a critical role in cartilage formation (chondrogenesis), and may be a protective factor against the progression of cartilage degradation in osteoarthritis, but like SNX14 its precise cellular function is unknown (21).
In this study we have examined the evolutionary and detailed structural relationships between this subfamily of sorting nexins, and characterized the specific phosphoinositide membrane binding activities of their PX domains. The proteins comprise a novel modular protein family, with each member possessing a predicted N-terminal transmembrane anchor, and four conserved structural domains called the PXA, RGS, PX, and PXC domains, except for SNX19, which appears to lack the RGS module. These proteins also display a deep evolutionary heritage, being present in all eukaryotes from humans to yeast. However, our biochemical, structural, and cellular studies of the PX domains of the mammalian proteins demonstrate an unexpected diversity in phosphoinositide binding specificity, with potential implications for their role in signal transduction and membrane trafficking.
EXPERIMENTAL PROCEDURES
Molecular Biology
The cDNA encoding mouse SNX19-(528–664) was cloned into the pMCSG10 vector for bacterial expression with an N-terminal GST tag (22). The pMCSG10 vector encodes a tobacco etch virus (TEV) cleavage site. Synthetic genes encoding the PX domains of human SNX13-(558–677), SNX14-(561–686), and SNX25-(506–628) optimized for Eschericia coli expression were purchased from Geneart (USA), and subsequently cloned into the pGEX-4T-2 vector for expression as N-terminal GST fusion proteins with thrombin cleavage sites. For cellular localization studies, genes encoding full-length mouse SNX19 (residues 1–997) and the PX-PXC domains (residues 528–997) were cloned into the pEGFP-N1 vector (Clontech) containing a C-terminal GFP tag. The SNX19 mutants were generated using the QuikChange II site-directed mutagenesis protocol (Stratagene).
Recombinant Protein Expression and Purification
The plasmids encoding GST-PX domain fusions were transformed into BL21(DE3)/pLysS E. coli cells, and expressed in LB broth at 37 °C until A600 reached 0.8. The cultures were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and allowed to grow at 20 °C overnight, and cells were harvested by centrifugation (6000 × g, 10 min, 4 °C). The cell pellet was resuspended in lysis buffer (20 mm Tris (pH 8.0), 200 mm NaCl, 50 μg/ml of benzamidine, 2 tablets of complete EDTA-free protease inhibitor, DNase I, 1 mm β-mercaptoethanol). The purification was performed using affinity chromatography with glutathione-Sepharose, and when required the GST tags were cleaved by TEV protease or thrombin while still bound to the column. The proteins were eluted in 20 mm Tris (pH 8.0), 200 mm NaCl, 10% glycerol, and 1 mm DTT, with the addition of 15 mm glutathione for non-cleaved GST fusions. For NMR experiments, uniformly 15N-labeled SNX19 and SNX14 PX domains were expressed in minimal media containing 15NH4Cl and [13C]glucose as the sole nitrogen and carbon sources.
SNX19 PX Domain Crystal Structure Determination
The affinity purified and TEV-cleaved mouse SNX19 PX domain (residues 528–664) was purified on a Superdex 200 gel filtration column in 10 mm Tris (pH 8.0), 200 mm NaCl, 5% glycerol, and 1 mm DTT, and then 10 mm DTT was added prior to crystallization. The SNX19 PX domain was then concentrated to 12–15 mg/ml for crystallization experiments. Eight commercially available 96-well crystallization screens were set up using a Mosquito robot at 20 °C, and imaging performed using a Rockimager system. Long needle crystals and plates grew rapidly in many different conditions, totaling nearly 20% of the drops (with some microcrystals appearing within seconds). The common factors for crystallization were the presence of polyethylene glycol (PEG) at concentrations between 15 and 30%, and pH > 7.0. Crystals optimized for diffraction were grown in 16–20% PEG 3350, 0.25–0.5 m NaCl, 5% glycerol, and 0.1 m Tris (pH 8.5).
Crystals were cooled to 100 K under the cryostream after immersing in mother liquor plus 20% glycerol. Data were collected at the Australian Synchrotron MX1 Beamline, integrated with iMOSFLM (23), and scaled with SCALA (24). The apo-SNX19 PX domain structure was solved by molecular replacement with PHASER (25) using as an input model the cytokine-independent survival kinase PX domain (PDB code 1XTN) after truncation to polyalanine. The resulting model was rebuilt with COOT (26) and refined with PHENIX (27). Residues with missing electron density suggesting structural disorder include N-terminal residues 528–531, residues 569–577 within the loop between strands β2 and β3, and residues 616–626 within the Pro-rich loop. The SO42−-bound SNX19 PX domain structure was determined after soaking crystals in cryosolution containing 100 mm Li2SO4 for 30 s before flash cooling to 100 K.
SNX14 PX Domain Crystal Structure Determination
The affinity purified and thrombin-cleaved human SNX14 PX domain (residues 561–686) was purified on a Superdex 200 gel filtration column in 10 mm Tris (pH 8.0), 200 mm NaCl, 5% glycerol, and 1 mm DTT, and then 10 mm DTT was added prior to crystallization. Crystallization screens were performed as for the SNX19 PX domain. Large single crystals grew in a number of conditions and the two conditions optimized for x-ray diffraction experiments were 1.8 m ammonium citrate tribasic (crystal form 1) and 20% PEG 4000, 0.1 m sodium citrate tribasic (pH 4.5) (crystal form 2).
Crystals were cooled to 100 K under the cryostream after immersing in mother liquor plus 20% glycerol. Data for crystals in space group I41 (crystal form 1) was collected at the UQ ROCX diffraction facility on a Rigaku FR-E Superbright generator with Osmic Vari-Max HF optics and Rigaku Saturn 944 CCD detector. Data for crystals grown in space group P43212 (crystal form 2) was collected at the Australian Synchrotron MX1 Beamline. All data were integrated with iMOSFLM (23) and scaled with SCALA (24). The SNX14 PX domain structures were solved by molecular replacement with PHASER (25) using as an input model the SNX19 PX domain after truncation to polyalanine and removal of flexible loop regions. The resulting model was rebuilt with COOT (26) and refined with PHENIX (27).
Liposome Binding Assays
Small unilamellar vesicles were prepared by the standard extrusion method. Lipids were dissolved in chloroform, and the solvent was removed under a N2 stream to yield a lipid film, which was left to dry under vacuum overnight. The lipid film was hydrated with the desired buffer upon agitation and eight freeze-thaw cycles. The resulting suspensions of multilamellar vesicles were extruded 21 times through polycarbonate filters (1 μm pore size) to obtain small unilamellar vesicles. Different lipid compositions were used: PC/PE (70/30 molar percentage) composed of 70% of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 30% of 1-palmitoyl-2-oleoyl phosphatidylethanolamine (Avanti Polar Lipids), and PC/PE containing PtdIns3P (Echelon Biosciences) (90:10 molar percentage) were prepared to specifically test the effect of the PtdIns3P lipid. The assay was conducted using 20 mm proteins and 1 mg/ml of small unilamellar vesicles in assay buffer (20 mm Hepes (pH 7.5), 150 mm NaCl) in a total volume of 100 μl (mixture of 30 μl of protein and 70 μl of small unilamellar vesicles) and incubated at room temperature for 20 min followed by centrifugation at 100,000 × g (rotor TLA100) for 15 min at 21 °C using a benchtop ultracentrifuge (Optima TL Ultracentrifuge). The supernatant was removed thoroughly and mixed with 4× SDS sample buffer. Pelleted liposomes were resuspended in 100 μl of assay buffer and mixed with 4× SDS sample buffer. The samples were boiled for 5 min and centrifuged at 9,000 × g for 10 min. Supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and Coomassie Blue staining.
NMR Spectroscopy
Uniformly 15N-labeled proteins were prepared in minimal media supplemented with 15NH4Cl. For NMR chemical shift titration studies, soluble phosphoinositide headgroup analogs with diC8 aliphatic chains (Echelon Biosciences) were added to samples containing 50 μm 15N-labeled SNX19 or SNX14 PX domains in NMR buffer (20 mm Hepes (pH 7.0), 100 mm NaCl, 10% D2O). Two-dimensional 1H-15N HSQC spectra of either 15N-labeled SNX19 or SNX14 PX domains were collected with increasing amounts of the soluble phosphoinositide headgroup analogs. All spectra were collected at 298 K on a Bruker 900-MHz spectrometer equipped with a cryoprobe and Z-axis gradients. Spectra were processed using NMRPipe (28) and analyzed with the program CCPNMR (29).
Bioinformatics
To analyze the domain architecture of the RGS-PX proteins, sequences were first subjected to secondary structure predictions using JPRED (30). Proteins were then subjected to multiple sequence alignments, which were manually adjusted to maximize alignment of predicted secondary structure elements. For phylogenetic analysis, a multiple sequence alignment was carried out using MUSCLE from MEGA 5.1 under default settings. The alignments were then used to construct a Maximum Likelihood Tree. The phylogeny test selected was the Bootstrap method with 100 bootstrap replications. The sequences were obtained from NCBI and UniProt database. Accession numbers for sequences are supplied in Table 1.
TABLE 1.
Accession numbers for RGS-PX protein sequences
The sequences were obtained from NCBI and UniProt databases.
| SNX13 | SNX14 | SNX19 | SNX25 | Snazarus | Mdm1p | |
|---|---|---|---|---|---|---|
| Human | Q9Y5W8 | Q9Y5W7 | Q92543 | This study | ||
| Homo sapiens | ||||||
| Chimpanzee | JAA36856 | JAA40159 | JAA43654 | JAA34955 | ||
| Pan troglodytes | ||||||
| Macaque | AFE64496 | AFH28856 | AFH32382 | AFE66285 | ||
| Macaca mulatta | ||||||
| Mouse | NP_001014973 | NP_766514 | NP_083150 | AAT98626 | ||
| Mus musculus | ||||||
| Rat | NP_00156765 | NP_001101644 | NP_001101601 | EDL78878 | ||
| Rattus norvegicus | ||||||
| Chicken | XP_00123468 | NP_001034378 | XP_003642646 | XP_420506 | ||
| Gallus gallus | ||||||
| Western clawed frog | XP_002933347 | NP_001085023 | XP_002938154 | ENSXETT00000064280 | ||
| Xenopus tropicalis | ||||||
| Zebrafish | XP_003200556 | NP_001038258 | XP_697433 | NP_001156765 | ||
| Danio rerio | ||||||
| Worm | XP_002125747 | XP_002122266 | XP_002120443 | |||
| Ciona intestinalis | ||||||
| Roundworm | NP_503026 | NP_496844 | ||||
| Caenorhabditis elegans | ||||||
| Fruitfly | Q9W3N0 | |||||
| D. melanogaster | ||||||
| Yeast | C8ZE98 | |||||
| S. cerevisiae | ||||||
Immunofluorescence Microscopy
SNX19-GFP, SNX19 PX-PXC-GFP (residues 528–997), SNX19 PX-PXC(R587Q)-GFP, and SNX19 PX-PXC(R634K)-GFP were transfected into HeLa cells using Lipofectamine® 2000 (Invitrogen), according to manufacturer's instructions, and their localization was analyzed by immunofluorescence microscopy. Cells transfected with each construct were immunolabeled against several subcellular markers using the following antibodies: EEA1 (mouse monoclonal anti-EEA1; BD Biosciences), LAMP1 (mouse monoclonal anti-LAMP1; BD Biosciences), and p230 (mouse monoclonal anti-p230; BD Biosciences). Primary antibodies were immunolabeled with donkey anti-mouse or goat anti-rabbit Alexa Fluor 546 secondary antibodies (Invitrogen) and images were collected using a Zeiss LSM 510 META Upright Scanning Laser confocal microscope. Colocalization quantification was performed as previously described (31). Briefly, using ImageJ version 64 the captured images were split into gray-scale formats, threshold settings were applied to each individual channel and Colocalization finder, the ImageJ plug-in, used to obtain a Pearson colocalization coefficient.
RESULTS
Defining the RGS-PX Protein Family
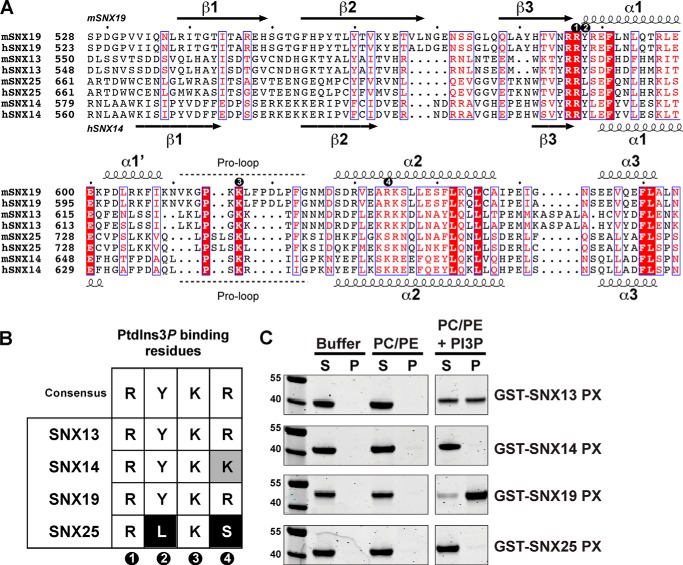
The four mammalian proteins, SNX13, SNX14, SNX19, and SNX25, display a novel and related architecture consisting of a central PX domain, flanked by several conserved structures (Figs. 1 and 2). Although we described these proteins in a recent review (3), here we define their relationships in detail based on combined sequence and structural considerations. The first domain upstream of the PX domain is the RGS module, found in a number of molecules that act as GAPs to attenuate GPCR-mediated G-protein signal transduction (7, 8). This domain is found in SNX13, SNX14, and SNX25, but not in SNX19 where the corresponding region, although similar in length, shows little sequence homology and has no predicted secondary structure. For simplicity, and consistency with previous reports (2, 9, 10, 12), we refer to this family herein as the RGS-PX proteins, and include SNX19 despite lacking the RGS structure.
FIGURE 1.
Secondary structure-based sequence alignment of human RGS-PX proteins SNX13, SNX14, SNX19, and SNX25. Secondary structure predictions calculated with JPRED (30) are indicated for SNX13 (above) and SNX19 (below). For the region encompassing the SNX19 PX domain, secondary structure information is derived from the crystal structure. The alignment was made with ESPript 2.2 (49). In addition to the four conserved domains, SNX13 and SNX25 possess an additional region with the predicted helical structure designated here as PXA′ that lies between the PXA and RGS domains. SNX19 lacks a predicted RGS domain.
FIGURE 2.
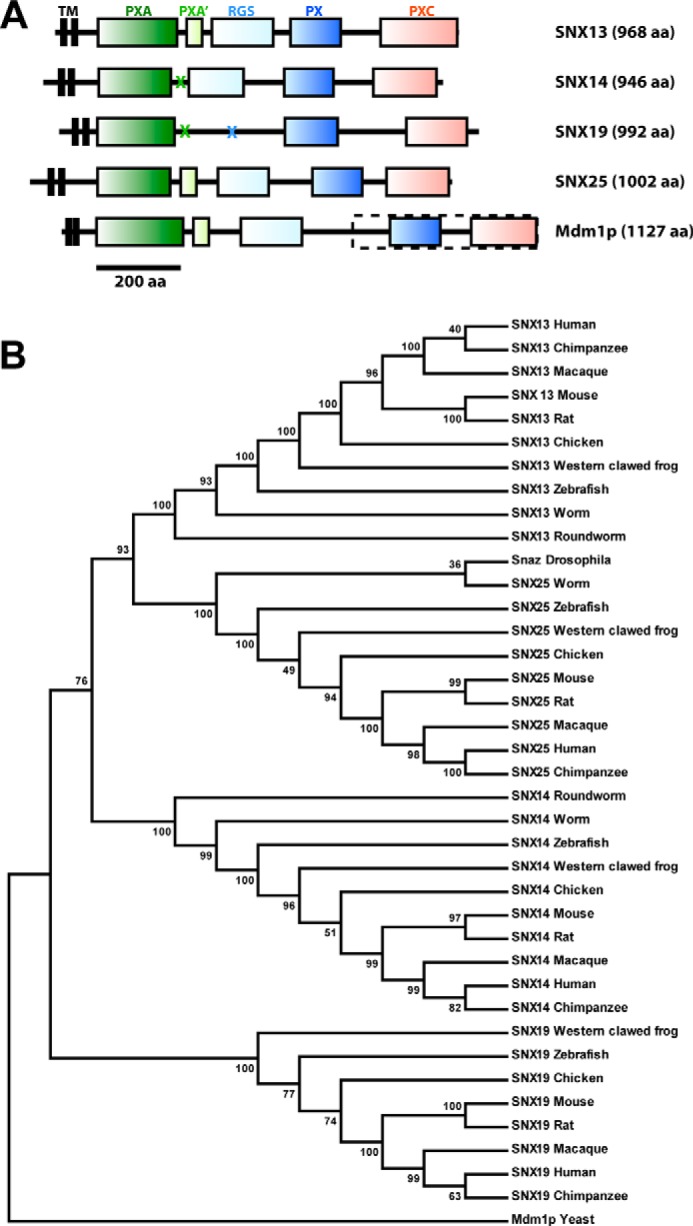
The RGS-PX proteins are modular multidomain proteins conserved in all eukaryotes. A, architectures of the human SNX13, SNX14, SNX19, SNX25, and yeast Mdm1p proteins are shown to scale. Crosses indicate where particular domains are not present. The dashed box indicates the region of Mdm1p that was previously characterized (33–36). A full sequence alignment of the four human proteins is presented in the legend to Fig. 1. B, the maximum likelihood phylogenetic tree for RGS-PX proteins including 39 sequences from vertebrates, invertebrates, and unicellular species (Table 1. The tree was constructed in MEGA 5.1 with the Bootstrap method as Test of Phylogeny (100 bootstrap replications) and was rooted using yeast Mdm1p as the outgroup.
A second conserved but uncharacterized domain (∼200 amino acids in length) has previously been annotated based on sequence homology and named the PX-associated (PXA) domain (1–3). This domain is N-terminal to the RGS and PX domain. Our secondary structure predictions and guided sequence alignments indicate that the PXA domain is entirely α-helical; and in the case of SNX13 and SNX25 is followed by a short (∼50 amino acids long) α-helical region prior to the RGS domain we refer to as the PXA′ segment. Secondary structure comparisons further indicate the presence of a conserved C-terminal domain found in all of these proteins (∼150 amino acids long), which is also predicted to be entirely α-helical in structure. For consistent nomenclature we refer to the N- and C-terminal domains as the PXA and PXC domains, respectively, indicating they are PX-associated domains of unknown structure and function. Note, however, that the PXA and PXC domains are not related to each other, either by sequence homology or secondary structure similarity. Finally, all possess two N-terminal predicted trans-membrane helices. Analysis using the SignalP3.0 server (32) suggests the first N-terminal hydrophobic segment is a transmembrane anchor that is unlikely to be removed by signal cleavage. These molecules are therefore predicted to be two-pass transmembrane proteins with a short cytoplasmic N terminus, a very short extracellular or lumenal loop, and a long and highly modular cytoplasmic C-terminal structure consisting of three to four globular domains interspersed by variable linker regions.
Although human SNX25 is annotated in several protein databases as lacking the N-terminal transmembrane helices, and studies of SNX25 have used constructs lacking such regions (17), our comparisons of the rat, mouse, and human genomes clearly identify conserved N-terminal coding sequences that are transcribed and contain the two predicted transmembrane segments. Thus putative transmembrane anchors are common to all members of the family (Figs. 1 and 2A).
In addition to the mammalian RGS-PX proteins a related protein, Mdm1p, is reported to play a role in mitochondrial inheritance in Saccharomyces cerevisiae (33–36). The original articles outlining Mdm1p cloning and function describe a 443-residue protein that begins with the sequence “MEKV.” Based on this reported sequence Mdm1p has the C-terminal PX and PXC domains, but not the N-terminal PXA or RGS domains (12). However, the current annotated S. cerevisiae genome sequence encodes a protein of 1127 amino acids, where the original published sequence represents a C-terminal subfragment (residues 685–1127) (Fig. 2A). Examination of secondary structure predictions of this longer sequence indicates Mdm1p has essentially identical domain arrangements to mammalian SNX13 and SNX25; i.e. PXA, PXA′, RGS, PX, and PXC domains, and two predicted N-terminal transmembrane helices. The Drosophila melanogaster snazarus protein also possesses all putative domains. Therefore Mdm1p and snazarus are bona fide homologs of the mammalian RGS-PX proteins, and this protein family clearly has a deep eukaryotic evolutionary heritage. A phylogenetic tree of the RGS-PX proteins from vertebrates, invertebrates, and unicellular species confirms their conservation in all eukaryotic organisms (Fig. 2B).
Analysis of RGS-PX transcripts in mouse using the BioGPS server (37) suggests a broad pattern of expression for SNX13, SNX14, and SNX19, whereas SNX25 is highly elevated in lung samples. This is consistent with published Northern blots of SNX25, which show the highest level of expression in lung, with lower but significant expression in brain and spleen (17). Previous Northern blot and in situ hybridization analyses and Western blots of mouse tissues suggest higher levels of SNX14 expression in the nervous system (19, 20).
The PX Domains of the RGS-PX Proteins Show Different Phosphoinositide Binding Preferences
The PX domains of yeast PX proteins have been found to interact almost exclusively with the endosomal lipid PtdIns3P, and this includes the RGS-PX family member Mdm1p (38). However, our previous survey of the literature on mammalian PX domains highlighted a broader phosphoinositide binding capability that may drive recruitment to a greater diversity of cellular organelles and membrane domains (3). For the canonical interaction with PtdIns3P, structures of p40phox (39), SNX9 (40, 41), and the yeast Snx3p/Grd19p molecule (42) in complex with the lipid headgroup reveal four side chains required for binding. These are an Arg and Tyr pair found in the loop between β3 and α1, a Lys within an extended Pro-rich loop, and an Arg within helix α2, labeled as residues 1, 2, 3, and 4, respectively in Fig. 3, A and B. These side chains provide critical contacts with the 3-phosphate, inositol ring, 1-phosphate, and 4- and 5-hydroxyls of the PtdIns3P molecule, respectively. Within the RGS-PX subfamily SNX13 and SNX19 possess these conserved residues required for PtdIns3P binding, and SNX14 has a conservative substitution of Arg to Lys at the hydroxyl-coordinating site 4. SNX25 however, has combined substitutions of the key hydroxyl-binding Arg side chain 4 to Ser, and the Tyr at site 2 necessary for inositol ring stacking to Leu, which would alter the pocket significantly and would be predicted to perturb the typical PtdIns3P association (Fig. 3, A and B).
FIGURE 3.
Phosphoinositide binding by the PX domains of the mammalian RGS-PX proteins. A, sequence alignment of the PX domains from human and mouse RGS-PX proteins. The secondary structures of human SNX14 and mouse SNX19 as determined from their crystal structures (see Fig. 5) are indicated at the top and bottom, respectively. The four key consensus PtdIns3P-binding residues are indicated by numbered circles from 1–4 as in B. B, summary table comparing the putative key PtdIns3P-binding residues of the RGS-PX proteins to the canonical PtdIns3P binding side chains identified in previous PX domain complex structures (3). Each of the four consensus residues is numbered from 1 to 4. C, liposome pelleting assay for PX domains of SNX13, SNX14, SNX19, and SNX25. GST fusion proteins were incubated with the indicated artificial liposomes, or buffer as a control. Samples were subjected to ultracentrifugation followed by SDS-PAGE and Coomassie staining of the unbound supernatant S and bound pellet P fractions. Both SNX13 and SNX14 bind liposomes supplemented with PtdIns3P, whereas SNX14 and SNX25 do not.
To test binding of the PX domains from the RGS-PX family to phosphoinositides, we employed a liposome pelleting assay incorporating artificial liposomes of different compositions (Fig. 3C). Using PC/PE membranes as controls, we observe specific binding to liposomes containing the endosomal PtdIns3P lipid. Using this assay both SNX13 and SNX19 showed robust interactions with membranes supplemented with PtdIns3P. This is in line with the conservation of PtdIns3P-coordinating side chains for these proteins (Fig. 3, A and B). Interestingly, despite the relatively minor and conservative change in the SNX14 binding this protein did not bind to membranes. The SNX25 PX domain also does not display significant PtdIns3P, consistent with the idea that its altered binding pocket is unable to coordinate this phosphoinositide in the usual manner.
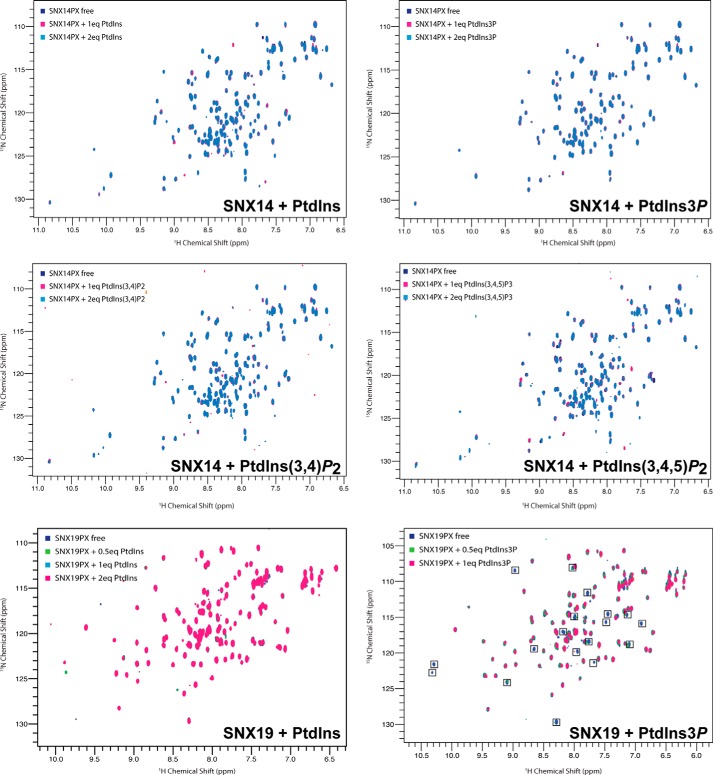
Next, using NMR spectroscopy with soluble phosphoinositide headgroup analogs, we confirmed the results of the liposome pelleting assays for the SNX14 and SNX19 PX domains, which display differential recognition of endosomal PtdIns3P (Fig. 4). Although we have not assigned the backbone spectra for these domains, 1H-15N HSQC spectra for 15N-labeled samples of both proteins are well dispersed and suitable for titration studies. Supporting the liposome pelleting assays of Fig. 3C, we find that SNX14 does not appear to bind specifically to PtdIns3P, or any other phosphoinositide species tested, whereas SNX19 shows a specific interaction with PtdIns3P, with a number of peaks in the 1H-15N HSQC spectrum showing clear line broadening on the addition of the headgroup consistent with direct association.
FIGURE 4.
Phosphoinositide binding of SNX14 and SNX19 PX domains assessed by NMR. 1H-15N HSQC spectra of 15N-labeled SNX14 and SNX19 PX domains are shown in the presence and absence of soluble phosphoinositides. Spectra were recorded at 25 °C using 50 μm PX domains. In line with the liposome pelleting assays (Fig. 3C) SNX14 shows no significant binding to any of the phosphoinositides tested, whereas SNX19 shows clear interaction with PtdIns3P as shown by the number of specific chemical shift perturbations observed.
Crystal Structures of the SNX14 and SNX19 PX Domains
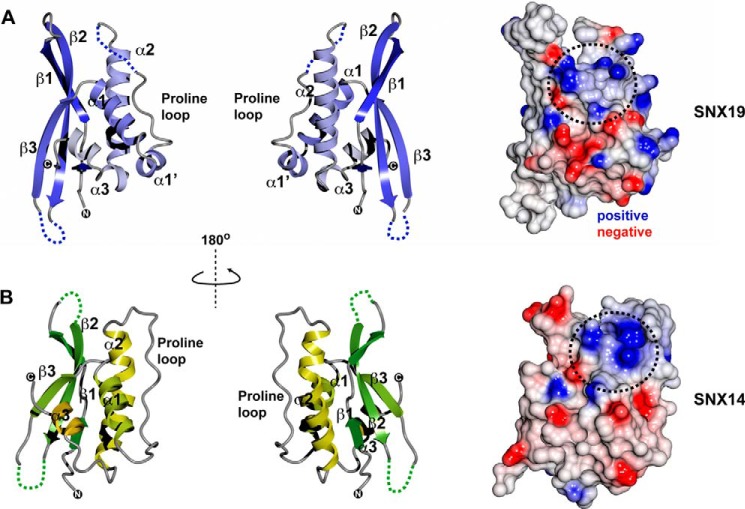
To better understand the molecular basis for why SNX19 binds to PtdIns3P but SNX14 does not we determined their x-ray crystallographic structures (Fig. 5, Table 2). The mouse SNX19 PX domain was crystallized in space group P21 with two copies in the asymmetric unit. The SNX19 PX domain displays the canonical PX domain fold composed of three β-strands (β1-β3) followed by three α-helices (α1-α3), with a long and flexible loop between α1 and α2 that contains a conserved Pro residue that likely contributes to this flexibility. The mouse SNX19 PX domain is 97% identical to human SNX19 and no significant structural differences are expected between the two species. However, the sequence similarity of the PX domains across the four different members of the mammalian RGS-PX family is relatively low, with pairwise identities of only between 17 and 29% (Fig. 3A).
FIGURE 5.
Crystal structures of the mouse SNX19 and human SNX14 PX domains. Left panels show back and front views of the overall structures of the PX domains and right panels show a surface representation of the (A) mouse SNX19 and (B) human SNX14 proteins. The ribbon structure of SNX19 is colored dark blue to gray from the N to C terminus. The ribbon structure of SNX14 is colored dark green to light green from the N to C terminus. Surfaces are colored by electrostatic potential from −0.5 to +0.5 V. Both SNX19 and SNX14 possess a pocket of high positive charge in the approximate location of the putative phosphoinositide binding site.
TABLE 2.
Summary of crystallographic structure determination statistics
Highest resolution shell is shown in parentheses.
| SNX19 PX domain apo | SNX19 PX domain SO42− bound | SNX14 PX domain (crystal form 1) | SNX14 PX domain (crystal form 2) | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.9537 | 0.9537 | 1.54187 | 0.95370 |
| Space group | P21 | P21 | I41 | P43212 |
| Cell dimensions | ||||
| a, b, c (Å) | 63.8, 32.9, 81.9 | 64.2, 33.0, 82.5 | 86.9, 86.9, 74.4 | 121.7, 121.7, 82.4 |
| α, β, γ (°) | 90, 107.5, 90 | 90, 107.2, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 31.7-1.90 (2.00-1.90) | 78.8-2.40 (2.53-2.40) | 22.10-2.55 (2.69-2.55) | 68.2-3.00 (3.16 (3.00) |
| Rmerge | 0.104 (0.561) | 0.097 (0.576) | 0.082 (0.682) | 0.140 (1.049) |
| I/σ(I) | 15.1 (3.3) | 13.4 (3.4) | 15.4 (1.9) | 20.9 (4.7) |
| Completeness (%) | 99.9 (100.0) | 99.4 (99.4) | 99.8 (99.3) | 100.0 (100.0) |
| Number unique reflections | 26,154 (3,782) | 13,235 (1,909) | 9,073 (1,318) | 12,933 (1,839) |
| Redundancy | 7.3 (7.3) | 7.0 (7.1) | 6.5 (3.8) | 27.8 (28.8) |
| Wilson B-factor | 18.3 | 47.7 | 72.3 | 89.5 |
| Refinement | ||||
| Resolution (Å) | 30.44–1.90 (1.98–1.90) | 61.28–2.40 (2.64–2.40) | 22.1–2.55 (2.92 -2.55) | 48.9–3.00 (3.30–3.00) |
| No. reflections/No. Rfree | 25,639/1,300 | 12,772/629 | 9,067/432 | 12,883/629 |
| Rwork/Rfree | 0.213/0.247 | 0.221/0.263 | 0.183/0.209 | 0.234/0.285 |
| No. atoms | ||||
| Protein | 1,896 | 1,892 | 915 | 3475 |
| Solvent | 309 | 84 | 23 | 19 |
| No protein chains per asymmetric unit | 2 | 2 | 1 | 4 |
| Average B-factor (Å2) | 25.2 | 48.3 | 53.6 | 92.7 |
| Root mean square deviations | ||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.010 | 0.011 |
| Bond angles (°) | 0.983 | 1.016 | 1.260 | 1.452 |
| PDB code | 4P2I | 4P2J | 4PQO | 4PQP |
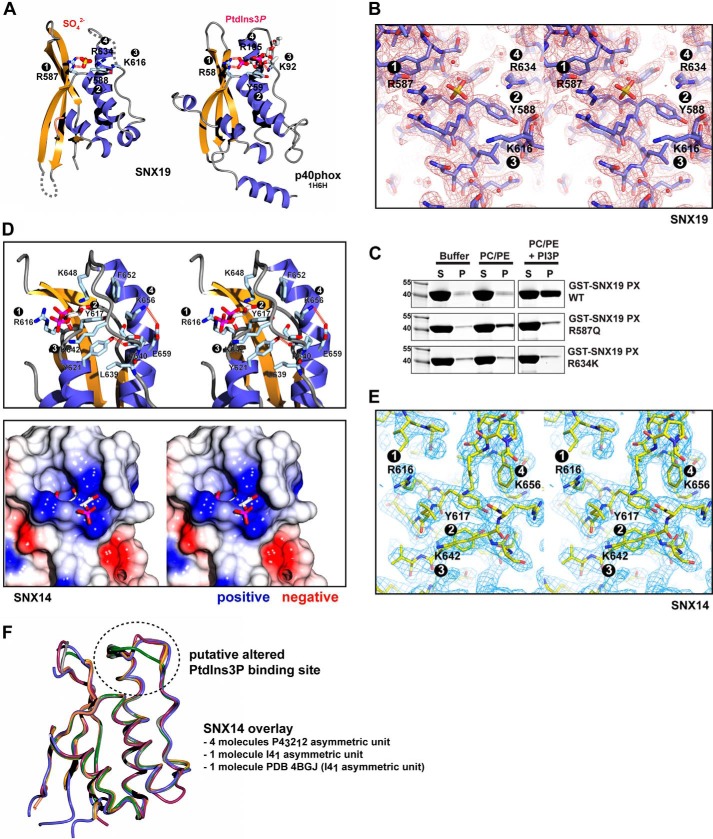
The human SNX14 PX domain crystallized in two distinct crystal forms, and structures were determined in space groups I41 (crystal form 1) and P43212 (crystal form 2) at 2.5- and 3.0-Å resolution, respectively. As this work was underway, a third structure of the human SNX14 PX domain was deposited in the Protein Data Bank (PDB) by the Yue lab (51) of Oxford University and colleagues of the Structural Genomics Consortium (PDB code 4BGJ). The deposited structure is analogous to crystal form 1 reported here, with the same space group (I41) and unit cell parameters and an overall root mean square deviation of 0.98 Å over 102 Cα atoms. Crystal form 2 (P43212) has four copies of SNX14 in the asymmetric unit; and six SNX14 molecules compared across the three crystal structures are very similar with an root mean square deviations varying from 0.8 to 1.1 Å (see Fig. 7F).
FIGURE 7.
Structures of the SNX19 and SNX14 PX domains explain their different PtdIns3P binding affinities. A, structure of SNX19 PX domain bound to SO42−, and comparison with the structure of the PtdIns3P bound PX domain from p40phox (39). The four key PtdIns3P-coordinating side chains are shown for both proteins. The four side chains of SNX19 are poised in the correct orientations to bind a potential PtdIns3P lipid, and the SO42− ion is bound in the same location as the 3-phosphate group of the PtdIns3P headgroup in p40phox. B, electron density of the SNX19 PX domain with bound SO42− ion. The image shows a close up of the putative PtdIns3P-binding pocket. Refined 2Fo − Fc density contoured at 1.0 σ is shown in wall-eye stereo view. C, liposome pelleting assay of SNX19 PX domain and structure-based mutations of the phosphoinositide binding pocket. Mutation of the 3-phosphate binding Arg587 at site 1 to Gln abrogates interaction with PtdIns3P-containing membranes. Mutation of the Arg634 side chain at site 4 to Lys (as in the SNX14 PX domain) also inhibits membrane association. D, in the upper panel the potential phosphoinositide binding pocket of the SNX14 PX domain is shown in wall-eye stereo view, with key side chains indicated. Also included is the PtdIns3P headgroup as it would be expected to bind based on an overlay of SNX14 with the p40phox-PtdIns3P complex. The lower panel shows a similar stereo view of the SNX14 structure in surface representation colored for electrostatic potential (from −0.5 to +0.5 V). As discussed in the text, despite the presence of a negatively charged surface, the required pocket is completely occluded due to a number of key alterations in the surrounding residues. E, electron density of the SNX14 PX domain (chain A in space group I41). The image shows a close up of the occluded region around the putative PtdIns3P binding pocket. Refined 2Fo − Fc density contoured at 1.0 σ is shown in wall-eye stereo view. F, an overlay of the four molecules of the SNX14 PX domain in the asymmetric unit of crystal form 2 is shown, along with the structure of SNX14 in space group I41 (crystal form 1) (this study) and PDB coordinates 4BGJ available from the PDB. Chain A in crystal form 2 adopts an even more closed conformation of the putative PtdIns3P binding pocket than other chains as represented in Fig. 6C (green worms).
Although the SNX14 and SNX19 PX domains can be structurally aligned with an root mean square deviation of 1.9 Å over 93 Cα atoms, there are major differences in some structural elements (Figs. 5 and 6). Notably, SNX19 has an extended β-sheet structure compared with SNX14 (and other PX domains) as well as an additional short α-helix we call α1′, whereas the SNX14 α2 helix possesses a significant kink compared with SNX19 and other PX proteins. Both PX domains possess electrostatically polar surfaces, notably retaining a significant basic patch at the site expected to bind to negatively charged phosphoinositide headgroups (Fig. 5). The top three structural matches to both SNX19 and SNX14 using the DALI web server (43) were the PX domains of serum/glucocorticoid-regulated kinase 3, SNX7, and phosphatidylinositol 4-phosphate 3-kinase C2α (PI3K-C2α), although a number of PX proteins gave similar high alignment scores, highlighting the conserved structural profile of the family. Fig. 6 shows a comparison of SNX19 and SNX14 to several of these. Of the known PX domain structures the protein with the most dramatically different architecture is SNX5. SNX5 is a member of the BAR (Bin/amphiphysin/Rvs) domain-containing subfamily (44), and its PX domain (and that of close homologs SNX6 and SNX32) has a large and unique helical hairpin insertion between helices α1 and α2. The functional significance of this insertion still remains unclear, but it has been suggested to confer altered membrane binding and remodeling properties to this subset of SNX-BAR proteins (3). Recently, the structure of the SNX11 PX domain was reported to possess a novel extended α-helical structure (also present in SNX10), which is required for its regulation of endosomal vacuolization (45).
FIGURE 6.
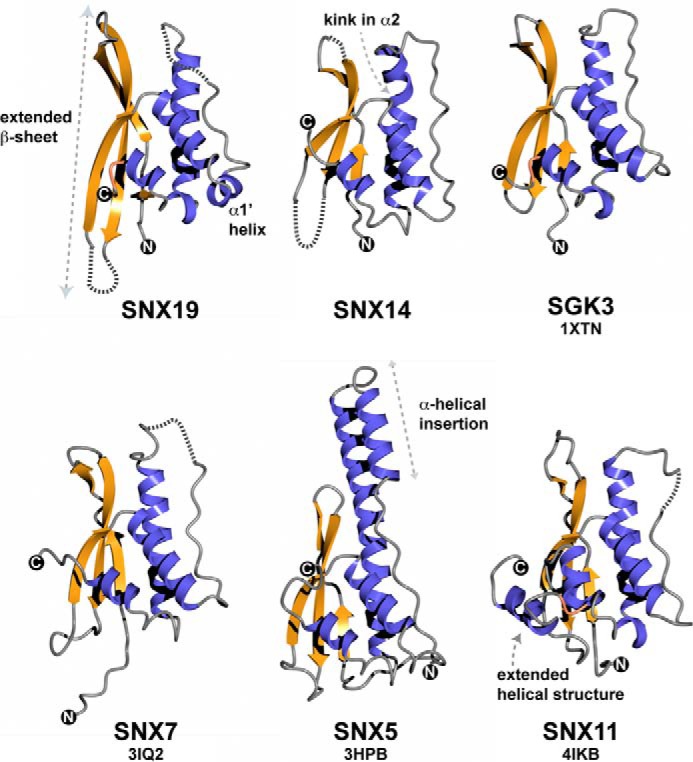
Comparison of SNX14 and SNX19 PX domain structures with other PX domain proteins. The structures of SNX19 and SNX14 are shown in comparison with their two closest structural matches SNX7 and serum/glucocorticoid-regulated kinase 3 (50), as well as the structurally divergent PX domain from SNX5, which has a highly extended α-helical hairpin insertion following the Pro-rich loop (44), and the SNX11 PX domain shown to possess extended α-helical structure at the C terminus important for its function (45). β-Strands are indicated in gold, and α-helices in blue. The SNX19 PX domain has a significantly elongated β-sheet substructure compared with other PX domain proteins, including SNX14. Unique to SNX19 is the presence of a short helical insert we designate as α1′ between helices α1 and α2, and preceding the Pro-containing loop.
Structural Basis for the Different Phosphoinositide Preferences of the SNX19 and SNX14 PX Domains
As shown in Figs. 3C and 4, SNX19 specifically associates with PtdIns3P, whereas SNX14 lacks this binding activity. We attempted to soak SNX19 crystals with soluble PtdIns3P (and other phosphoinositide analogs) but did not observe any lipids bound within the putative binding pocket, most likely due to occlusion by crystal packing. Notably the Pro-rich loop is restricted at its distal C terminus by a crystal contact and therefore may be unable to make the minor conformational alterations necessary for PtdIns3P binding. Nonetheless, the four key side chains normally required to mediate PtdIns3P binding are all present, and approximately poised to support PtdIns3P interaction. Previous reports have shown that SO42− ions can be coordinated by the site 1 Arg side chain where the 3-phosphate group is normally bound (46, 47), so we next soaked crystals with Li2SO4 and observed a clear difference density for a SO42− ion bound to the analogous SNX19 side chain Arg587 (Arg582 in human SNX19) (Fig. 7, A and B). Comparisons with PtdIns3P-bound SNX9 (40, 41) and p40phox (39) confirm specific coordination of the SO42− ion in exactly the same location as the PtdIns3P 3-phosphate group would be expected to bind (Fig. 7A). Overall this is strong evidence of a viable PtdIns3P binding site within the SNX19 PX domain, and given the similar ability of SNX13 to bind PtdIns3P membranes (Fig. 3C), and the conservation of the required binding side chains, it is likely that SNX13 shares a similar PtdIns3P binding pocket. To confirm the importance of the binding site, we generated a mutant SNX19 PX domain (R587Q) and tested its association with membranes in liposome pelleting assays (Fig. 7C). As expected this point mutant no longer binds PtdIns3P-containing membranes above background levels.
In contrast to SNX19, we find that SNX14 does not bind PtdIns3P-containing membranes (Fig. 3C). Closer examination of the putative phosphoinositide-binding site reveals key alterations that explain this lack of phosphoinositide association (Fig. 7D). First, whereas the side chains at sites 1, 2, and 3 are conserved and approximately oriented for binding the headgroup, the fourth side chain at site 4 is altered from Arg to Lys (Lys656), and is pointed away from the binding site, stabilized by interaction with the side chain of Glu659 (density for the Lys656 side chain is shown in Fig. 7E, and the side chain is similarly oriented in all molecules of the asymmetric units in the two crystal forms). Overall the Pro-rich loop is well ordered, unusually for most PX domains, and is in fact stabilized by a hydrogen bond between the backbone carbonyl of Pro640 within the loop and the side chain of Tyr621 within helix α1. A network of core interactions between Leu639, Tyr621, Tyr617, and Phe652 overall contribute to the formation of a relatively closed pocket, and in addition Lys648 is prominently positioned in such a way as to sterically interfere with potential PtdIns3P headgroups. Surface representations of the domain demonstrate that the normal mode of PtdIns3P coordination would be completely excluded (Fig. 7D). In the three combined crystal structures of the SNX14 PX domain there are a total of six identical protein chains within the asymmetric units. All chains adopt a similar closed conformation of the putative PtdIns3P-binding pocket with the exception of chain A in our crystal form 2. In this chain, the pocket adopts a slightly different conformation where the binding pocket is even more severely occluded (green chain in Fig. 7F). To summarize, replacement of the site 4 Arg side chain to Lys, and key alterations in the binding pocket geometry appear to exclude the canonical phosphoinositide association with SNX14.
In support of the key role of Lys656 in the abrogation of PtdIns3P binding we engineered a SNX14 mimicking substitution in the SNX19 PX domain, altering the equivalent site 4 Arg634 to Lys (R634K). This mutant, similarly to R587Q, reduces PtdIns3P liposome interactions to background levels (Fig. 6C). Although it is unlikely that the SNX19 R634K mutant completely mimics the closed binding pocket architecture of SNX14, as this is also promoted by other key side chains, this result shows that simply altering the site 4 Arg to Lys is sufficient to perturb the normal PtdIns3P interactions.
Localization of SNX19 to a Subpopulation of Endosomes Requires PtdIns3P Interaction
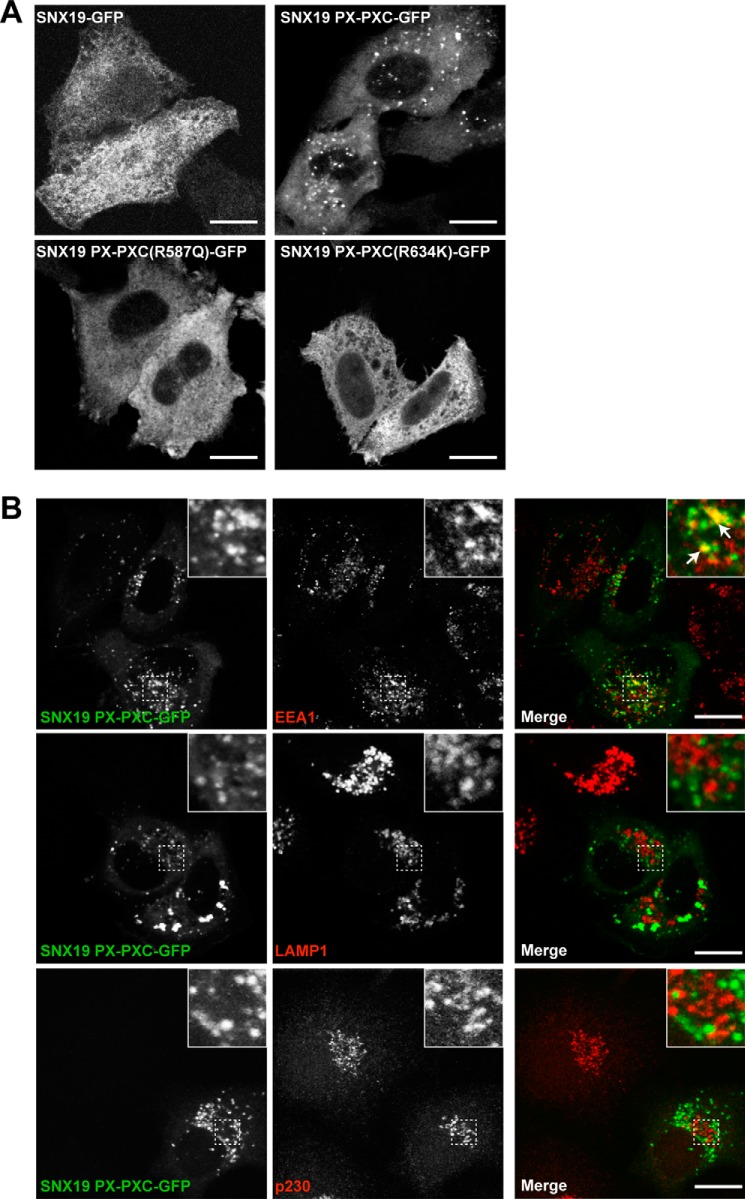
Previous studies of the cellular localization of exogenously expressed and truncated forms of SNX13 (9, 12) and SNX25 (17) have shown members of this family to be on endosomal organelles. In the case of SNX13 a construct containing the C-terminal RGS, PX, and PXC domains is localized to endosomes (9, 12). In the case of mouse SNX25, the PX domain is sufficient for endosomal localization, although the N-terminal PXA domain can also associate with endosomal membranes in isolation (17). To date no studies have reported the localization of a full-length RGS-PX protein, due to poor expression, lack of antibodies to the endogenous proteins, or incompletely cloned sequences. We thus attempted to assess the cellular localization of the full-length SNX19 protein with a C-terminal GFP tag (to avoid interference with putative N-terminal transmembrane domains), however, this construct was not expressed well and showed diffuse labeling (Fig. 8A). Following previous examples of truncated SNX13 (9) and SNX25 (17) we engineered a construct of SNX19 with only the C-terminal PX and PXC domains (SNX19 PX-PXC; residues 528–997). This construct strongly labeled cytoplasmic puncta (Fig. 8A), and showed partial overlap with the early endosomal marker EEA1 (Pearson colocalization coefficient 0.1559 ± 0.0207 S.D.), but no correlation with the late endosomal marker LAMP1 (Pearson colocalization coefficient 0.0167 ± 0.0018 S.D.) or trans-Golgi network marker p230 (Pearson colocalization coefficient 0.0261 ± 0.0176 S.D.) (Fig. 8B), indicating its recruitment to a subset of endosomal organelles similarly to other RGS-PX family proteins. Confirming the importance of PtdIns3P binding for membrane association of this SNX19-C-terminal fragment, we find that the SNX19 PX-PXC (R587A) mutant is no longer localized to punctate membrane structures and shows a diffuse cytoplasmic labeling (Fig. 8A). Similarly, the SNX19 PX-PXC (R634K) mutant, which mimics the substitution of the site 4 Arg to Lys as in SNX14 and blocks PtdIns3P, association is also no longer membrane associated (Fig. 8A). Altogether, the data shows PtdIns3P interaction by the SNX19 PX domain mediates association with endosomal membranes, and suggests the altered binding pocket of SNX14 will not be able to support endosomal recruitment.
FIGURE 8.
Endosomal localization of the SNX19 protein requires PtdIns3P interaction. A, subcellular localization of full-length SNX19-GFP, SNX19 PX-PXC-GFP, SNX19 PX-PXC-GFP R643K, and SNX19 PX-PXC-GFP R643K proteins in HeLa cells. Full-length SNX19-GFP shows a diffuse cellular localization, whereas the C-terminal SNX19 PX-PXC-GFP fragment is found on punctate membranes. SNX19 R587Q PX-PXC-GFP and SNX19 R643K PX-PXC-GFP are both redistributed to the cytoplasm indicating an essential role for these side chains in membrane targeting. B, the punctate membranes positive for SNX19 PX-PXC-GFP overlap with EEA1 (early endosome) but not LAMP1 (late endosomes/lysosomes) or p230 (Golgi) subcellular localization markers. All images represent a 1 absorbance unit single slice captured using a Zeiss LSM 510 META Upright Scanning Laser confocal microscope at ×63 magnification. Scale bar, 10 μm.
DISCUSSION
Members of the RGS-PX protein family are important for GPCR-mediated signaling via Gαs trimeric G-protein subunits (9), signaling by TGFβ (17), endosomal trafficking (12), and normal cellular development in different eukaryotic organisms (10, 35, 48). Our analyses indicate that the proteins are conserved across the entire eukaryotic lineage, with unicellular and invertebrate genomes containing a single isoform, and apparent gene expansion occurring in vertebrates with mice and humans possessing four paralogs. Across all species the RGS-PX proteins possess a similar modular architecture with domains of both known and unknown structure and function.
Although the RGS-PX family members possess highly conserved putative transmembrane domains, the importance of these domains for cellular localization or protein function is still unknown. Expression of a tagged full-length SNX19 in our hands leads to diffuse cellular localization, which is distinct from the punctate endosomal membrane recruitment of various ectopically expressed RGS-PX truncation variants incorporating the PX and PXC domains of SNX13 (9), SNX25 (17), and SNX19 itself (this study). The development of antibodies to the endogenous proteins will likely be necessary to properly characterize membrane localization of the full-length proteins and confirm whether they are truly integral membrane proteins. The RGS domain of SNX13 has been shown to possess GAP activity and mediate subsequent signal attenuation of Gαs-coupled GPCRs (9), but it remains to be determined whether other family members possessing this domain (SNX14 and SNX25) have similar or divergent activities. The PXA and PXC domains of these proteins remain the most poorly characterized domains of all. Whereas they are predicted to possess α-helical structures, they are not identified as belonging to any known protein-fold class, and their functions have not been directly determined to date. The SNX25 PXA domain possesses an intrinsic ability to localize to EEA1-positive early endosomes, and can cooperate with the PX domain to interact with TβRs and promote their lysosomal degradation, suggesting a membrane targeting and/or protein-interaction activity (17). But further studies of these domains will be required to define their functional roles.
In this work we have focused on the membrane targeting activities of the PX domain of the RGS-PX family. The four mammalian RGS-PX paralogs show significant divergence in the sequences of their PX domains, with distinct differences in the side chains that are normally required for binding the early endosomal phosphoinositide PtdIns3P. Our data shows a clear pattern of phosphoinositide binding preference related to the differences in these residues. Notably, SNX25 lacks two of the four side chains required for PtdIns3P association, and does not show interaction with PtdIns3P-containing liposomes in vitro. The mechanism by which the SNX25 PX domain is localized to endosomal membranes (17) therefore remains unclear. Both SNX13 and SNX19 retain the canonical residues at the 1, 2, 3, and 4 sites for coordination of PtdIns3P, both share the ability to recognize membranes enriched in this lipid, and this binding pocket is required for membrane association in vivo. However, our studies show that a relatively minor alteration in SNX14 at site 4, with the typical hydroxyl-coordinating Arg side chain modified to Lys, combined with changes in several key residues surrounding the pocket, leads to a profound alteration of the binding pocket architecture. The pocket is thus occluded and unable to accommodate PtdIns3P in the usual way, explaining the lack of PtdIns3P recognition. The localization of SNX14 in cells remains to be determined, but our prediction is that it will not be stably associated with PtdIns3P-enriched endosomal membranes in the same way as SNX13 and SNX19. This prediction is partly supported by the observation that SNX19 with a substitution in the Arg at position 4 to Lys (thus partly mimicking SNX14) is no longer recruited normally to membranes either in vitro or in cells.
In conclusion, our data shows that small alterations in the core residues of the phosphoinositide binding pocket can significantly alter the membrane specificity of the PX domain. We propose that the highly conserved RGS-PX proteins will coordinate regulatory protein-protein interactions via PXA and PXC domains with GAP activity via the RGS domain to assemble complexes necessary for modulating cell signaling, whereas the coupling of differential phosphoinositide preferences within the PX domain will act to promote the formation of these complexes in different domains of the endocytic and secretory system to guide distinct signaling outcomes.
Acknowledgments
We acknowledge support from the staff and facilities of the University of Queensland Remote Operation Crystallization and x-ray (UQ ROCX) facility and the Australian Synchrotron. We thank Patricia Walden and Jenny Martin for assistance with x-ray diffraction data collection. Microscopy was performed at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience Dynamic Imaging Facility for Cancer Biology established with the support of ACRF.
This work was supported in part by Australian Research Council Grant DP0985029 and National Health and Medical Research Council Grants APP1042082 and APP1058734.
The atomic coordinates and structure factors (codes 4P2I, 4P2J, 4PQO, and 4PQP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SNX
- sorting nexin
- PX
- phox homology
- GPCR
- G protein-coupled receptor
- PtdIns
- phosphatidylinositol
- GAP
- GTPase activating protein
- RGS
- regulator of G-protein signaling
- TEV
- tobacco etch virus
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- HSQC
- heteronuclear single quantum coherence
- PXA
- PX-associated.
REFERENCES
- 1. Cullen P. J. (2008) Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 [DOI] [PubMed] [Google Scholar]
- 2. Seet L. F., Hong W. (2006) The phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 1761, 878–896 [DOI] [PubMed] [Google Scholar]
- 3. Teasdale R. D., Collins B. M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J. 441, 39–59 [DOI] [PubMed] [Google Scholar]
- 4. Krauss M., Haucke V. (2007) Phosphoinositides: regulators of membrane traffic and protein function. FEBS Lett. 581, 2105–2111 [DOI] [PubMed] [Google Scholar]
- 5. Balla T. (2005) Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 118, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 6. Moravcevic K., Oxley C. L., Lemmon M. A. (2012) Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jean-Baptiste G., Yang Z., Greenwood M. T. (2006) Regulatory mechanisms involved in modulating RGS function. Cell Mol. Life Sci. 63, 1969–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ross E. M., Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 9. Zheng B., Ma Y. C., Ostrom R. S., Lavoie C., Gill G. N., Insel P. A., Huang X. Y., Farquhar M. G. (2001) RGS-PX1, a GAP for Gαs and sorting nexin in vesicular trafficking. Science 294, 1939–1942 [DOI] [PubMed] [Google Scholar]
- 10. Zheng B., Tang T., Tang N., Kudlicka K., Ohtsubo K., Ma P., Marth J. D., Farquhar M. G., Lehtonen E. (2006) Essential role of RGS-PX1/sorting nexin 13 in mouse development and regulation of endocytosis dynamics. Proc. Natl. Acad. Sci. U.S.A. 103, 16776–16781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurley J. H. (2010) The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45, 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng B., Lavoie C., Tang T. D., Ma P., Meerloo T., Beas A., Farquhar M. G. (2004) Regulation of epidermal growth factor receptor degradation by heterotrimeric Galphas protein. Mol. Biol. Cell 15, 5538–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanyaloglu A. C., von Zastrow M. (2008) Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 14. Sadowski L., Pilecka I., Miaczynska M. (2009) Signaling from endosomes: location makes a difference. Exp. Cell Res. 315, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 15. Scita G., Di Fiore P. P. (2010) The endocytic matrix. Nature 463, 464–473 [DOI] [PubMed] [Google Scholar]
- 16. Sorkin A., von Zastrow M. (2009) Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hao X., Wang Y., Ren F., Zhu S., Ren Y., Jia B., Li Y. P., Shi Y., Chang Z. (2011) SNX25 regulates TGF-beta signaling by enhancing the receptor degradation. Cell. Signal. 23, 935–946 [DOI] [PubMed] [Google Scholar]
- 18. Du Y., Zou Y., Yu W., Shi R., Zhang M., Yang W., Duan J., Deng Y., Wang X., Lü Y. (2013) Expression pattern of sorting nexin 25 in temporal lobe epilepsy: a study on patients and pilocarpine-induced rats. Brain Res. 1509, 79–85 [DOI] [PubMed] [Google Scholar]
- 19. Carroll P., Renoncourt Y., Gayet O., De Bovis B., Alonso S. (2001) Sorting nexin-14, a gene expressed in motoneurons trapped by an in vitro preselection method. Dev. Dyn. 221, 431–442 [DOI] [PubMed] [Google Scholar]
- 20. Huang H. S., Yoon B. J., Brooks S., Bakal R., Berrios J., Larsen R. S., Wallace M. L., Han J. E., Chung E. H., Zylka M. J., Philpot B. D. (2014) Snx14 regulates neuronal excitability, promotes synaptic transmission, and is imprinted in the brain of mice. PloS One 9, e98383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kan A., Ikeda T., Saito T., Yano F., Fukai A., Hojo H., Ogasawara T., Ogata N., Nakamura K., Chung U. I., Kawaguchi H. (2009) Screening of chondrogenic factors with a real-time fluorescence-monitoring cell line ATDC5-C2ER: identification of sorting nexin 19 as a novel factor. Arthritis Rheum. 60, 3314–3323 [DOI] [PubMed] [Google Scholar]
- 22. Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 30. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Follett J., Norwood S. J., Hamilton N. A., Mohan M., Kovtun O., Tay S., Zhe Y., Wood S. A., Mellick G. D., Silburn P. A., Collins B. M., Bugarcic A., Teasdale R. D. (2014) The Vps35 D620N mutation linked to Parkinson's disease disrupts the cargo sorting function of retromer. Traffic 15, 230–244 [DOI] [PubMed] [Google Scholar]
- 32. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 33. Fisk H. A., Yaffe M. P. (1997) Mutational analysis of Mdm1p function in nuclear and mitochondrial inheritance. J. Cell Biol. 138, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fisk H. A., Yaffe M. P. (1999) A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McConnell S. J., Yaffe M. P. (1992) Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J. Cell Biol. 118, 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McConnell S. J., Yaffe M. P. (1993) Intermediate filament formation by a yeast protein essential for organelle inheritance. Science 260, 687–689 [DOI] [PubMed] [Google Scholar]
- 37. Wu C., Macleod I., Su A. I. (2013) BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu J. W., Lemmon M. A. (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179–44184 [DOI] [PubMed] [Google Scholar]
- 39. Bravo J., Karathanassis D., Pacold C. M., Pacold M. E., Ellson C. D., Anderson K. E., Butler P. J., Lavenir I., Perisic O., Hawkins P. T., Stephens L., Williams R. L. (2001) The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol. Cell 8, 829–839 [DOI] [PubMed] [Google Scholar]
- 40. Pylypenko O., Lundmark R., Rasmuson E., Carlsson S. R., Rak A. (2007) The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 26, 4788–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Q., Kaan H. Y., Hooda R. N., Goh S. L., Sondermann H. (2008) Structure and plasticity of endophilin and sorting nexin 9. Structure 16, 1574–1587 [DOI] [PubMed] [Google Scholar]
- 42. Zhou C. Z., Li de La Sierra-Gallay I., Quevillon-Cheruel S., Collinet B., Minard P., Blondeau K., Henckes G., Aufrère R., Leulliot N., Graille M., Sorel I., Savarin P., de la Torre F., Poupon A., Janin J., van Tilbeurgh H. (2003) Crystal structure of the yeast phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278, 50371–50376 [DOI] [PubMed] [Google Scholar]
- 43. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koharudin L. M., Furey W., Liu H., Liu Y. J., Gronenborn A. M. (2009) The phox domain of sorting nexin 5 lacks PTDINS(3)P specificity and preferentially binds to PtdIns(4,5)P2. J. Biol. Chem. 284, 23697–23707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu J., Xu T., Wu B., Ye Y., You X., Shu X., Pei D., Liu J. (2013) Structure of sorting nexin 11 (SNX11) reveals a novel extended phox homology (PX) domain critical for inhibition of SNX10-induced vacuolation. J. Biol. Chem. 288, 16598–16605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghai R., Mobli M., Norwood S. J., Bugarcic A., Teasdale R. D., King G. F., Collins B. M. (2011) Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc. Natl. Acad. Sci. U.S.A. 108, 7763–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karathanassis D., Stahelin R. V., Bravo J., Perisic O., Pacold C. M., Cho W., Williams R. L. (2002) Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21, 5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suh J. M., Stenesen D., Peters J. M., Inoue A., Cade A., Graff J. M. (2008) An RGS-containing sorting nexin controls Drosophila lifespan. PloS one 3, e2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 50. Xing Y., Liu D., Zhang R., Joachimiak A., Songyang Z., Xu W. (2004) Structural basis of membrane targeting by the phox homology domain of cytokine-independent survival kinase (CISK-PX). J. Biol. Chem. 279, 30662–30669 [DOI] [PubMed] [Google Scholar]
- 51.Yue, Structural Genomics Consortium