Background: The plant small GTPase OsRac1 plays an important role in rice innate immunity.
Results: The crystal structure and NADPH oxidase-binding mode of active-form OsRac1 were determined.
Conclusion: The structure explains the mechanism by which OsRac1 regulates reactive oxygen species production and activates the immune response.
Significance: A new insight into the activation of plant immunity by small GTPase is revealed.
Keywords: Crystal Structure, NADPH Oxidase, Plant Defense, Rac (Rac GTPase), Small GTPase, Rice Immunity
Abstract
Rac/Rop proteins are Rho-type small GTPases that act as molecular switches in plants. Recent studies have identified these proteins as key components in many major plant signaling pathways, such as innate immunity, pollen tube growth, and root hair formation. In rice, the Rac/Rop protein OsRac1 plays an important role in regulating the production of reactive oxygen species (ROS) by the NADPH oxidase OsRbohB during innate immunity. However, the molecular mechanism by which OsRac1 regulates OsRbohB remains unknown. Here, we report the crystal structure of OsRac1 complexed with the non-hydrolyzable GTP analog guanosine 5′-(β,γ-imido)triphosphate at 1.9 Å resolution; this represents the first active-form structure of a plant small GTPase. To elucidate the ROS production in rice cells, structural information was used to design OsRac1 mutants that displayed reduced binding to OsRbohB. Only mutations in the OsRac1 Switch I region showed attenuated interactions with OsRbohB in vitro. In particular, Tyr39 and Asp45 substitutions suppressed ROS production in rice cells, indicating that these residues are critical for interaction with and activation of OsRbohB. Structural comparison of active-form OsRac1 with AtRop9 in its GDP-bound inactive form showed a large conformational difference in the vicinity of these residues. Our results provide new insights into the molecular mechanism of the immune response through OsRac1 and the various cellular responses associated with plant Rac/Rop proteins.
Introduction
Small GTP-binding proteins (small G proteins or small GTPase) act as molecular switches and regulate a wide variety of important physiological functions in cells. Plants possess a specific subfamily of small GTPases called Rac/Rop (Rho-related GTPases from plants) (1, 2), which have attracted recent interest due to their function as molecular switches in the regulation of various cellular responses (3–6). For example, OsRac1, a Rac/Rop from Oryza sativa, plays an important role in the regulation of rice immunity (7–15), and NtRac5, from Nicotiana tabacum, regulates reactive oxygen species (ROS)4 production in pollen tubes (16). The AtRop family members AtRop1, AtRop3, and AtRop5 redundantly regulate pollen tube growth in Arabidopsis thaliana (17), whereas AtRop2, AtRop4, and AtRop6 regulate root hair development (17). Recently, the activation of AtRop2 and AtRop6 by auxin has been reported to regulate the subcellular distribution of auxin transporters PIN1 and PIN2, which control PIN-mediated pattern formation and morphogenesis in leaves and roots (18–20). AtRop10 and AtRop11 are specific negative regulators of abscisic acid responses (21, 22). In addition, AtRop9 functions as a signal integrator of auxin and abscisic acid signaling and plays an important role in embryo development and lateral root formation in A. thaliana (23).
Rac/Rop family proteins are composed of ∼200 amino acids and have masses of 20–24 kDa, similar to the animal small GTPases. They are inactive in the GDP-bound form and are activated by the binding of GTP. Several Rac/Rop structures have been reported, including AtRop5 (GDP-bound form), AtRop9 (GDP-bound form) (24), the AtRop4 (GDP-bound form)-guanine nucleotide exchange factor (GEF) complex (25), and the AtRop7(apo)-GEF complex (26), but all of these structures are of inactive forms. Structural analysis of active-form animal small GTPases has revealed the biological processes associated with carcinogenic mutations and the biochemical mechanisms of carcinogenesis (27). Hence, the structural determination of plant Rac/Rop proteins in their active form should be an important step in clarifying the mechanism of activation of target effectors.
A constitutively activated mutant of OsRac1 (OsRac1 G19V, denoted as CA-OsRac1) has been reported to increase resistance to rice bacterial blight disease and subsequent cell death (7, 8). Conversely, a dominant-negative mutant (OsRac1 T24N, denoted as DN-OsRac1) was found to decrease the resistance reaction. Transgenic rice lines expressing CA-OsRac1, but not DN-OsRac1, displayed increased production of a phytoalexin and altered expression of defense-related genes (8). Furthermore, overexpression of CA-OsRac1 induced ROS production in cultured rice cells (7). These data clearly show that OsRac1 acts as a molecular switch during plant innate immunity. CA-OsRac1, but not DN-OsRac1, was also shown to interact directly with an NADPH oxidase, OsRbohB (O. sativa respiratory burst oxidase homolog B) (11). Transient coexpression of OsRac1 and OsRbohB in Nicotiana benthamiana leaves enhanced ROS production, supporting the notion that direct OsRac1-OsRbohB interactions activate NADPH oxidase in plants (11). Although the crystal structure of the N-terminal domain of OsRbohB has been reported (28, 29), the molecular mechanism by which OsRac1 activates OsRbohB for ROS production remains largely unknown.
In this report, the crystal structure of OsRac1 in the active form (GMPPNP-bound) was determined in an effort to elucidate the molecular mechanism of ROS production in rice. Based on the structural information obtained, the OsRbohB-binding site on OsRac1 was predicted, and OsRbohB binding-deficient OsRac1 mutants were designed. The OsRbohB-binding activity of these mutants was evaluated by in vitro pulldown assays and NMR measurements, and the mutants were also analyzed by ROS production assays using rice cells. This study, together with our previous reports (11, 29), demonstrates that OsRac1 regulates ROS production through direct interactions with OsRbohB.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant OsRac1
cDNA encoding OsRac1(8–183) C32S/Q68L (denoted as OsRac1; see “Results and Discussion”) was cloned into the multiple cloning site of the pGEX-6P3 vector (GE Healthcare), and several mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene). The resulting plasmids were used to transform Escherichia coli Rosetta (DE3) cells (Novagen), which were then grown in M9 medium until the cell suspension reached the appropriate turbidity. Chimeric proteins comprising GST fused to the N terminus of OsRac1 or its mutants were then overexpressed by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 12 h at 15 °C, after which the cells were harvested by centrifugation. To obtain target proteins for NMR measurements, 0.5 g/liter [15N]ammonium chloride (99 atom % of 15N) was used as the sole nitrogen source in M9 medium. The overexpressed GST-fused OsRac1 proteins were initially purified by affinity chromatography using glutathione-Sepharose 4B resin (GE Healthcare). After enzymatic cleavage of the GST tag from target proteins using GST-human rhinovirus 3C protease, digestion products were passed through glutathione-Sepharose 4B resin, and the OsRac1 and mutant proteins were further purified by size exclusion column chromatography using Superdex 75 (GE Healthcare).
To exchange nucleotide, OsRac1 proteins were incubated with a 25-fold molar excess of GMPPNP (Sigma) in 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10 mm EDTA, and 2 mm DTT for 12 h at 4 °C. After the addition of 10 mm MgCl2, excess unbound nucleotides were removed using a Superdex 75 column.
For crystallization and NMR measurements, purified proteins were concentrated by ultrafiltration using Amicon Ultra-10 filters to 4 mg/ml with 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 5 mm MgCl2, and 2 mm DTT and to 0.1 mg/ml with buffer A (50 mm BisTris (pH 6.8), 50 mm NaCl, 5 mm MgCl2, 2 mm CaCl2, and 2 mm DTT), respectively.
For the prey protein in a GST pulldown assay, cDNA encoding OsRbohB(138–313) was cloned into pET32c (Novagen). Following overexpression of chimeric thioredoxin-His6-OsRbohB(138–313) in E. coli Rosetta (DE3) cells, protein was purified by affinity chromatography using nickel-nitrilotriacetic acid-agarose resin (Qiagen). After enzymatic cleavage of the thioredoxin-His6 tag from the target protein using recombinant enterokinase (Novagen), OsRbohB(138–313) was further purified by anion exchange and size exclusion chromatography using Superdex 75 in buffer A. For GST pulldown assays, purified OsRbohB(138–313) was concentrated to 0.6 mm by ultrafiltration using Ultra-10 filters with buffer A.
Crystallization and X-ray Data Collection
OsRac1 complexed with the GTP analog GMPPNP was crystallized as described (30). In brief, OsRac1 crystals were obtained at 20 °C using the sitting-drop vapor-diffusion method by mixing 0.7 μl of 4 mg/ml purified protein with 0.7 μl of reservoir solution consisting of 100 mm MES (pH 6.0) and 10–30% PEG 6000. For the x-ray diffraction experiments, crystals in a reservoir solution supplemented with 25% (v/v) glycerol as a cryoprotectant were mounted in nylon loops (Hampton Research), flash-cooled using a 100 K dry nitrogen stream, and then kept under the nitrogen stream during data collection. X-ray diffraction data were collected from GMPPNP-bound OsRac1 crystals using a Rayonix MX225HE CCD detector installed on beamline BL44XU at SPring-8 (Harima, Japan). The camera was fixed at a distance of 260 mm from the crystal sample using an x-ray wavelength of 0.9000 Å. Data collection for native crystals was performed using an angular range of 240°, an oscillation step of 1.5°, and an exposure time of 1.0 s for each image. All data were integrated and scaled using HKL-2000 (31). Diffraction and intensity data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics of the OsRac1-GMPPNP-Mg2+ complex (OsRac1(GMPPNP))
Values in parentheses refer to the highest resolution shell.
| OsRac1(GMPPNP) | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell constants (a, b, c) (Å) | 36.8, 59.1, 64.4 |
| Resolution (Å) | 50–1.9 (1.97–1.9) |
| Redundancy | 8.2 (8.3) |
| Completeness (%) | 99.8 (100) |
| I/σ(I) | 19.7 (7.7) |
| Rmerge (%)a | 9.5 (28.4) |
| Refinement | |
| Resolution range (Å) | 50–1.9 |
| No. of reflections | 10,901 |
| Rwork (%)b | 15.7 |
| Rfree (%)b | 20.3 |
| No. of atoms | |
| Protein | 1384 |
| Ligand and ion | 51 |
| Solvent | 90 |
| Average B-factor (Å2) | 16.1 |
| Root mean square deviations | |
| Bond length (Å) | 0.019 |
| Bond angles° | 2.2° |
| Ramachandran analysis | |
| Favored (%) | 98.9 |
| Allowed (%) | 1.1 |
| Disallowed (%) | 0 |
a Rmerge = ΣhΣi|I(h)i − 〈I(h)〉|/ΣhΣiI(h)i, where I(h) is the intensity of reflection h, Σh is the sum of all measured reflections, and Σi is the sum of i measurements of reflection.
b Rwork and Rfree = Σhkl‖Fo|−|Fc‖)/Σhkl|Fo|, where the free reflections (5% of the total used) were held aside for Rfree throughout refinement.
Structure Determination and Refinement
The crystal structure was solved by the molecular replacement method using Phaser (32). The tertiary structure coordinates of A. thaliana Rac7/Rop9 (Protein Data Bank (PDB) code 2J0V) (24) were used as a search model for OsRac1. Crystal structures of OsRac1 were rebuilt using Coot (33) and refined using REFMAC5 (34) and CNS (35). Ramachandran plot analysis was performed using Rampage (36). Final refinement statistics are summarized in Table 1. The solvent accessibility of each amino acid was analyzed using Naccess. Coordinates of the final model and structure factors of OsRac1 have been deposited in the PDB (code 4U5X). All structures in the figures were generated using PyMOL.
GST Pulldown Assays
Following immobilization of each GST-tagged protein onto a 50-μl gel volume of glutathione-Sepharose 4B resin, 20 μl of solution containing 0.6 mm purified OsRbohB(138–313) was added, and the mixture was incubated at 4 °C for 12 h. After washing the resin several times with fresh buffer solution (50 mm BisTris (pH 6.8), 50 mm KCl, 5 mm MgCl2, 2 mm CaCl2, 1 mm phenylmethylsulfonyl fluoride, and 3% (v/v) dimethyl sulfoxide), bound proteins were eluted and analyzed by SDS-PAGE. The SDS-polyacrylamide gels were stained with Coomassie Brilliant Blue.
NMR Measurements
All NMR spectra were measured using a Bruker AVANCE 800 spectrometer equipped with a TXI cryogenic probe at 303 K, and the collected data were processed and analyzed using NMRPipe (37) and Sparky 3 NMR Assignment and Integration Software (University of California, San Francisco), respectively. All of the two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) experiments were performed using 0.1 mm uniformly 15N-labeled OsRac1 in 50 mm BisTris (pH 6.8), 50 mm KCl, 5 mm MgCl2, and 1 mm DTT in 90% H2O and 10% D2O.
Rice Cell Cultures
To generate rice suspension cells (Kinmaze) expressing CA-OsRac1, CA-OsRac1 Y39A, or CA-OsRac1 D45A, the coding regions of mutated OsRac1 were introduced into the p2K-GW binary vector (for transgenic plants expressing genes under the control of the maize ubiquitin promoter) using the Gateway system (Invitrogen). Agrobacterium tumefaciens-mediated transformation of rice calli was performed according to Hiei et al. (38). Transformants selected by hygromycin resistance were subcultured in 22 ml of R2S medium every week and incubated on a rotary shaker (90 rpm) at 30 °C.
RT-PCR
Total RNA was extracted from rice suspension cultures using the RNeasy plant mini kit (Qiagen), and 1 μg was used as a template for reverse transcription using an oligo(dT) primer and SuperScript II (Invitrogen). PCR analyses were performed using specific primers for OsRac1 (5′-AGATAGGGCCTATCTTGCTGATCATC-3′ and 5′-CTAGAAGTTTCCTCCTAGCTGCAAGC-3′), hygromycin phosphotransferase (5′-GAGCCTGACCTATTGCATCTCC-3′ and 5′-GGCCTCCAGAAGAAGATGTTGG-3′), and ACT1 (5′-CAATCGTGAGAAGATGACCC-3′ and 5′-GTCCATCAGGAAGCTCGTAGC-3′).
Measurement of ROS Production
Rice suspension cells were subcultured for 4 days in fresh medium, and cells (∼20 mg) were placed into each well of 96-well white plates (Greiner Bio-One). Two-hundred microliters of 500 μm L-012 (Wako Chemicals) dissolved in medium was added to each well, and chemiluminescence was detected using an LAS-4000 mini luminescent image analyzer (Fujifilm) at 180 min. Emission intensity from each well was measured using ImageJ(http://rsbweb.nih.gov/ij/), and the following formula: (intensity of each well − intensity of background)/weight of suspension cells in each well.
RESULTS AND DISCUSSION
Structure Determination of OsRac1 in the Active Form
For the crystallization of active-form OsRac1, an OsRac1 mutant and the GTP analog GMPPNP were used to prevent GTP hydrolysis. The GTPase activity of the human small GTPase HsRhoA is significantly attenuated by substitution of glutamine with leucine at position 63 (39). In this study, the corresponding OsRac1 mutant comprising the substitution Q68L was used. To increase the stability of the OsRac1 protein, Cys32 was substituted with serine, and the N and C termini were truncated. Hereafter, OsRac1 refers to this truncated mutant, OsRac1(8–183) C32S/Q68L.
The OsRac1-GMPPNP-Mg2+ complex, designated as OsRac1(GMPPNP), was crystallized as described previously (30). The crystals gave strong and high-resolution x-ray diffraction. The diffraction data were collected to 1.9 Å resolution from one crystal (Table 1) and displayed orthorhombic P212121 symmetry with an estimated mosaicity of 0.41–0.62° and a Wilson B-factor of 12.7 Å2. The unit cell (a = 36.8, b = 59.1, and c = 64.4 Å) contains one molecule of OsRac1(GMPPNP) per asymmetric unit. The crystal structure of OsRac1(GMPPNP) was determined at 1.9 Å resolution with clear electron density for all atoms with Rwork = 15.7% and Rfree = 20.3% (Fig. 1A). OsRac1(GMPPNP) comprised a half-β-barrel-shaped structure formed by six β-strands (β1–β6). These β-strands were sandwiched between five α-helices (α1–α5), where two α-helices (α1 and α5) were wrapped inside and three α-helices (α2–α4) were located outside.
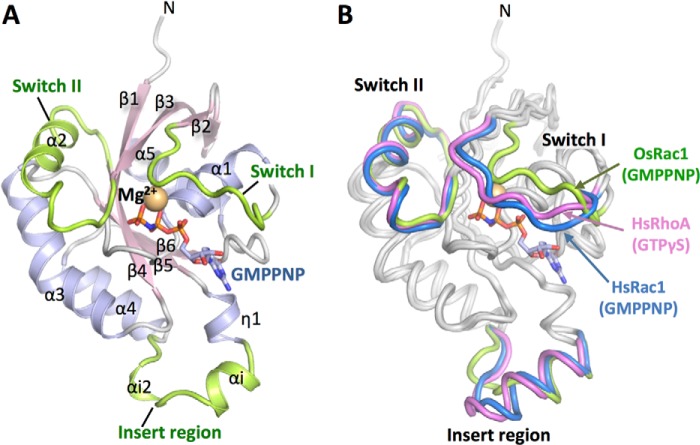
FIGURE 1.
Overall structure of OsRac1. A, ribbon representation of the overall structure of OsRac1 in its GMPPNP-Mg2+-bound form. The Switch I, Switch II, and insert regions are colored green. GMPPNP is shown as a stick model (red, oxygen; blue, nitrogen; orange, phosphorus). The Mg2+ ion is shown as an orange sphere. B, superimposition of the crystal structures of OsRac1(GMPPNP), HsRac1(GMPPNP) (PDB code 1MH1), and HsRhoA(GTPγS) (PDB code 1A2B). The Switch I, Switch II, and insert regions are colored as indicated by the protein labels. Models were generated using PyMOL.
Structural Comparison of OsRac1 with HsRac1 and HsRhoA-Rac/Rop Proteins in the GTP-bound Form
OsRac1 belongs to the Rho family of proteins, which includes HsRac1 and HsRhoA. OsRac1 shares 62% amino acid identity with HsRac1 and 49% identity with HsRhoA. Because the active-form structures of HsRac1 and HsRhoA have been determined (40, 41), these three Rho family protein structures were compared. The overall structures of OsRac1(GMPPNP) and the HsRac1-GMPPNP-Mg2+ complex, designated as HsRac1(GMPPNP) (PDB code 1MH1), were similar (Fig. 1B). The root mean square deviation of 141 Cα atoms was ∼1.4 Å. The overall structures of OsRac1(GMPPNP) and the HsRhoA-GTPγS-Mg2+ complex, designated as HsRhoA(GTPγS) (PDB code 1A2B), were also similar (Fig. 1B), and the root mean square deviation of 140 Cα atoms was ∼1.6 Å. The geometry of the nucleotide- and Mg2+ ion-binding pocket and the hydrogen bond network pattern are essentially the same among these three Rho family proteins. Major structural differences between OsRac1(GMPPNP), HsRac1(GMPPNP), and HsRhoA(GTPγS) were found in Switch I (35FPTDYIPTVFD45 in OsRac1) and the insert region (129DRAYLADHPASSII142 in OsRac1).
Switch I in OsRac1(GMPPNP) forms a loop structure, as it does in HsRac1(GMPPNP) and HsRhoA(GTPγS); however, the Switch I structures were not superimposed (Fig. 1B). The root mean square deviation of OsRac1(GMPPNP) and HsRac1(GMPPNP) was ∼2.5 Å for the 11 Cα atoms located in Switch I, and that of OsRac1(GMPPNP) and HsRhoA(GTPγS) was 2.2 Å. Switch I in OsRac1(GMPPNP) was located farther away from the guanine nucleotide than it was in the other two proteins (Fig. 1B).
The insert region is conserved in the Rho family, but not in other small GTPase families. HsRac1(GMPPNP) and HsRhoA(GTPγS) contain one α-helix (αi), followed by an extended short loop, whereas OsRac1(GMPPNP) contains two helices (αi and αi2) in this region (Fig. 1B and supplemental Fig. S1). The insert region of OsRac1 is relatively short due to the absence of two amino acids, and helix αi is significantly shorter (supplemental Fig. S1). The 310-helix αi2 (Ala138–Ser140) is absent in other Rho family proteins and seems to represent unique structural features of OsRac1(GMPPNP) (Fig. 1 and supplemental Fig. S1).
Structural Comparison of OsRac1 with Plant Rac/Rop Proteins
In an effort to determine whether unique structural features of OsRac1(GMPPNP) are conserved in the plant Rac/Rop protein family, the structure of OsRac1(GMPPNP) was compared with the reported structures of Rac/Rop proteins, such as AtRop5(GDP), AtRop9(GDP) (24), and the AtRop4(GDP)-GEF (25) and AtRop7(apo)-GEF (26) complexes. Of these plant Rac/Rop proteins, only OsRac1 is the GTP-bound form. The overall structure of OsRac1 was similar to those of the AtRop proteins. However, structural differences were observed in the Switch I and II regions of GTP (GMPPNP)-bound OsRac1 and the GDP-bound AtRop proteins (Fig. 2 and supplemental Fig. S2). In animals, extensive structural analysis revealed that large conformational changes occur between the GDP- and GTP-bound states in two regions, Switches I and II. These regions play important roles in interactions with downstream targets to transduce signals (41).
FIGURE 2.
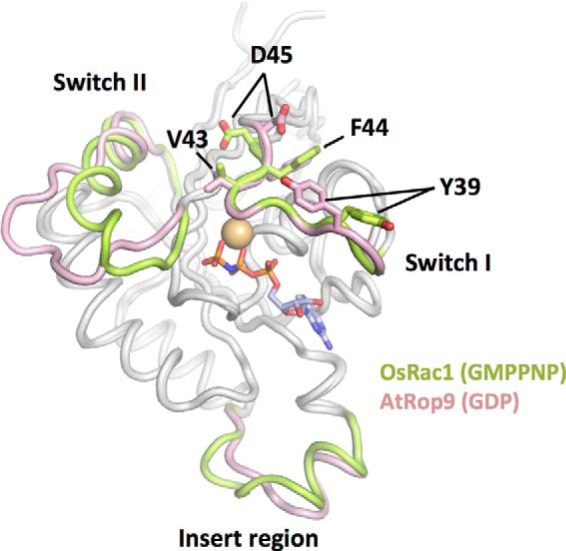
Comparison of the overall structures of OsRac1(GMPPNP) and AtRop9(GDP). The main chains of OsRac1(GMPPNP) and AtRop9(GDP) (chain B, PDB code 2J0V) were superimposed using PyMOL. The Switch I, Switch II, and insert regions of the OsRac1 and AtRop9 proteins are colored green and pink, respectively. The side chains of four key Switch I residues in OsRac1 (Val43, Phe44, Asp45, and Tyr39) and equivalent residues in AtRop9 (Val39, Phe40, Asp41, and Tyr35) are shown in stick representation.
The Switch I regions in OsRac1(GMPPNP) and AtRop9(GDP) were well ordered and formed similar loop structures. In contrast, in other AtRop proteins, this region was partially disordered (Fig. 2 and supplemental Figs. S2 and S3). The Switch II region of OsRac1(GMPPNP) adopted a long helical conformation (Fig. 2 and supplemental Figs. S2 and S3). A similar long helical conformation was observed in AtRop5(GDP) and the animal Rho family. However, most AtRop proteins adopted a loop-like structure without a long helix. These structural differences in the Switch I and II regions were not unexpected because all of the AtRop protein structures reported were of the inactive form. A notable structural difference also occurred in the insert region. As mentioned above, the insert region of OsRac1 comprised an insert α-helix (αi) and the short 310-helix (αi2) (Fig. 2 and supplemental Figs. S2 and S3). Helix αi was present in all AtRop proteins, but helix αi2 was unique to OsRac1. These structural differences in the insert region may affect the interaction with GEF proteins because helix αi within the insert region of AtRop4 is known to be involved in the interaction with the PRONE (plant-specific Rop nucleotide exchanger) domain of PRONE8 (25, 26).
Interaction of OsRac1 with OsRbohB(138–313)
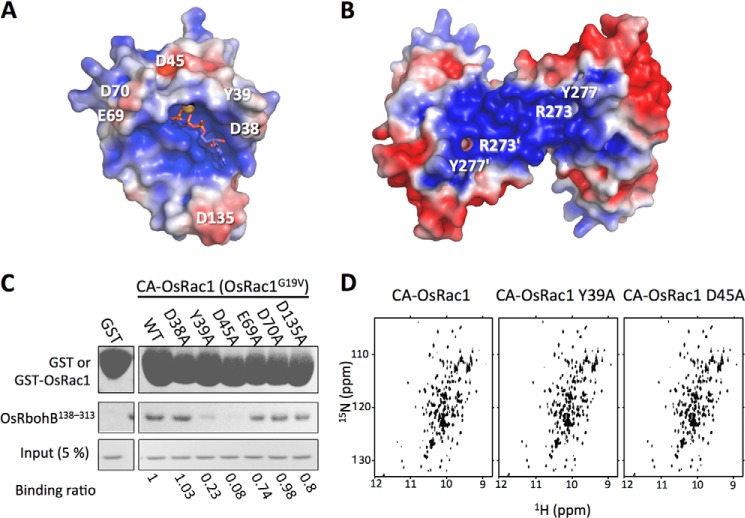
CA-OsRac1, but not DN-OsRac1, interacts directly with OsRbohB(138–313) (11). In structural comparisons of OsRac1(GMPPNP) with inactive-form AtRop proteins, Switch I, Switch II, and the insert region were found to differ. Therefore, the OsRbohB(138–313)-binding site of OsRac1(GMPPNP) was expected to involve Switch I, Switch II, or the insert region. Acidic residues in Switch I (Asp38, Tyr39, and Asp45), Switch II (Glu69 and Asp70), and the insert region (Asp135) were found to be located on the molecular surface of OsRac1(GMPPNP) (Fig. 3A and supplemental Fig. S4). The crystal structure of OsRbohB(138–313) has been solved, and it was suggested that Arg273 and Tyr277 of OsRbohB(138–313) play an essential role in binding to OsRac1 (29). Interestingly, Arg273, Tyr277, and surrounding residues form a positively charged cluster on the molecular surface of OsRbohB(138–313) (Fig. 3B). In view of these features, we anticipated that an electrostatic contribution would be important in the interaction between OsRac1(GMPPNP) and OsRbohB(138–313).
FIGURE 3.
Mode of binding between OsRac1 and OsRbohB(138–313). Shown is the electrostatic potential of the molecular surface of OsRac1(GMPPNP) (A) and OsRbohB(138–313) (PDB code 3A8R) (B). The amino acid residues labeled with single-letter code are putative key residues involved in the OsRac1-OsRbohB(138–313) interaction. Positively and negatively charged sites are colored blue and red, respectively. The surface potential was generated using PyMOL with APBS tools (±3 kT). C, GST pulldown binding assay with full-length CA-OsRac1 and OsRbohB(138–313). Alanine substitutions were introduced in place of Asp38, Tyr39, and Asp45 (in Switch I); Glu69 and Asp70 (in Switch II); and Asp135 (in the insert region) of GST-fused full-length CA-OsRac1. Each mutant was overexpressed in E. coli and purified, and equal amounts of each GST-fused protein were used as bait for the OsRbohB(138–313) pulldown. The amount of OsRbohB(138–313) pulled down was quantitated by densitometric analysis. Numerical values below the panel indicate the ratio of OsRbohB(138–313) binding to OsRac1 mutants compared with WT OsRac1. D, two-dimensional 1H-15N HSQC spectra of wild-type CA-OsRac1 (left), CA-OsRac1 Y39A (center), and CA-OsRac1 D45A (right).
To test the importance of the charge interaction, we performed in vitro pulldown assays using OsRbohB(138–313) and CA-OsRac1 as prey and bait proteins, respectively (Fig. 3C). The acidic residues located on the molecular surface of OsRac1 were replaced with alanine. Among the mutants generated, Switch I mutants Y39A and D45A showed markedly attenuated binding with OsRbohB(138–313) (Fig. 3C). To determine whether the tertiary structures of the Y39A and D45A mutants are disrupted, two-dimensional 1H-15N HSQC NMR spectra were measured. The 1H-15N HSQC spectra of the Y39A and D45A mutants were well resolved (Fig. 3D), and no marked chemical shift changes were observed in comparison with the wild-type protein (11). These NMR data indicate that the tertiary structures of the Y39A and D45A mutants are folded and are not substantially altered by the mutations. All of these data indicate that the Switch I Tyr39 and Asp45 residues of OsRac1 are critical for the direct interaction with OsRbohB(138–313).
Disruption of the Interaction between OsRac1 and OsRbohB Compromises ROS Production in Rice Cells
To establish whether the interaction of OsRac1 Tyr39 and Asp45 residues with OsRbohB affects the activation of NADPH oxidase, we examined ROS production in transgenic rice suspension cells overexpressing CA-OsRac1 Y39A and CA-OsRac1 D45A under the control of the maize ubiquitin promoter. RT-PCR analysis confirmed that the OsRac1 mutants were highly expressed in transgenic cells (Fig. 4A). In these experiments, we used L-012, a luminol derivative, to detect ROS (42). When suspension cells were treated with 0.5 mm L-012 and the chemifluorescence was examined using an LAS-4000 image analyzer, all CA-OsRac1 lines generated a large amount of ROS (Fig. 4, B and C), as reported by Kawasaki et al. (7). In contrast, the ROS levels in all lines of both CA-OsRac1 Y39A and CA-OsRac1 D45A were markedly lower than those in CA-OsRac1 suspension cells (Fig. 4, B and C). OsRac1 Tyr39 and Asp45 residues therefore play an important role in the activation of NADPH oxidase in rice suspension cells.
FIGURE 4.
Biological significance of Tyr39 and Asp45 in ROS production in rice cells. The CA-OsRac1 Y39A and CA-OsRac1 D45A mutants were overexpressed in rice cells, and their effect on OsRbohB NADPH oxidase activity was evaluated by quantifying ROS production. A, the expression of endogenous WT OsRac1, exogenous CA-OsRac1 and its Y39A and D45A mutants, hygromycin phosphotransferase (HPT), and actin-1 (ACT1) in these transgenic rice cells was confirmed by RT-PCR. B, example of ROS detection in wild-type OsRac1, CA-OsRac1-5, CA-OsRac1 Y39A-5, and CA-OsRac1 D45A-2 rice suspension cells using the L-012 reagent. C, quantification of ROS production in the wild-type OsRac1, CA-OsRac1, CA-OsRac1 Y39A, and CA-OsRac1 D45A lines. The luminescence intensity recorded from each well was measured by ImageJ. Data are mean ± S.E. of four biological replicates.
Important Residues in Switch I for OsRbohB Binding
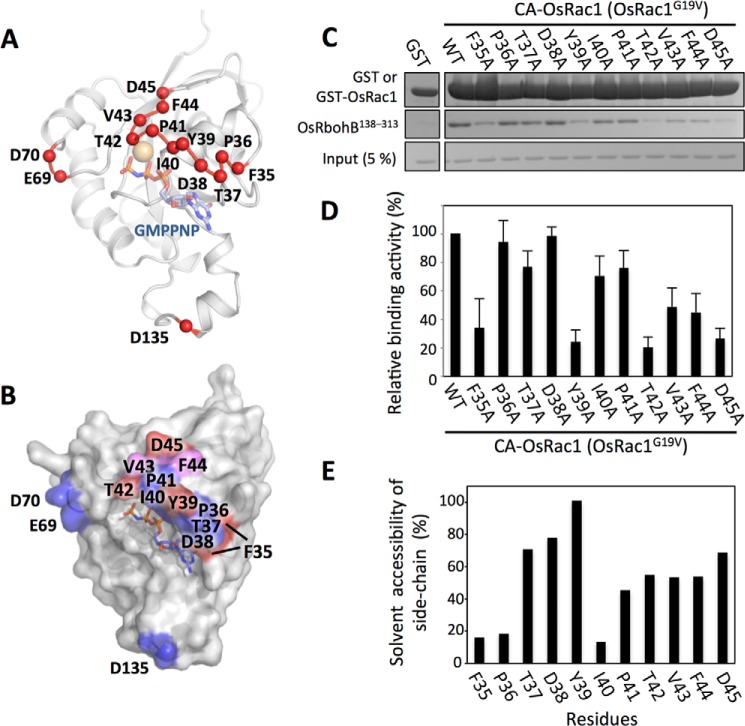
OsRac1 Switch I Tyr39 and Asp45 residues are necessary for the direct interaction of OsRac1 with OsRbohB(138–313), which leads to NADPH oxidase activation and ROS production. To identify other residues of Switch I that may also be important, each of the 11 residues in OsRac1 Switch I was replaced with alanine, and the mutants were subjected to in vitro pulldown assays (Fig. 5). The F35A, Y39A, T42A, and D45A mutants showed markedly reduced binding with OsRbohB(138–313), whereas the V43A and F44A mutants showed moderately reduced binding (Fig. 5, C and D). The solvent accessibility of all residues located in Switch I was calculated using the structure of OsRac1(GMPPNP) (Fig. 5E). The solvent accessibility values for Phe35, Tyr39, Thr42, Val43, Phe44, and Asp45 were 16, 100, 55, 54, 54, and 69%, respectively. The side chains of Phe35 and Thr42 interacted with GMPPNP (supplemental Fig. S5), and substitution with alanine destabilized binding to GMPPNP. Tyr39, Val43, Phe44, and Asp45 were exposed to the solvent and were not involved in direct binding to GMPPNP or coordination of the Mg2+ ion (supplemental Fig. S5B). Accordingly, we concluded that the Switch I Tyr39, Val43, Phe44, and Asp45 residues are involved in the OsRbohB(138–313)-binding site (Fig. 5B).
FIGURE 5.
Effects of mutations in OsRac1 Switch I residues on OsRbohB(138–313) binding. A, the alanine-substituted residues in CA-OsRac1 are mapped on the OsRac1 crystal structure as red spheres. B, surface representation of the OsRbohB(138–313)-binding interface on OsRac1. OsRac1 mutations that markedly affected binding (red) or had no effect (blue) are indicated. Mutation of residues colored pink was shown to moderately affect OsRbohB(138–313) binding. C, pulldown assay using OsRac1 mutants. Single alanine substitutions in Switch I were introduced in the full-length CA-OsRac1 background of the GST fusion construct. Equal amounts of each GST-fused OsRac1 protein were used as bait for OsRbohB(138–313) (middle). D, quantification by densitometric analysis of the amount of OsRbohB(138–313) pulled down in the in vitro pulldown assay using CA-OsRac1 mutants and OsRbohB(138–313). The relative binding activities were assessed by comparison with the value of GST-CA-OsRac1 (WT) binding activity to OsRbohB(138–313), which was set to 100%. The binding level of each mutant was categorized by the value as markedly reduced (below 40%), moderately reduced (between 40 and 60%), and weak or no effect on binding (above 60%). Average and error values were determined from three independent experiments. E, analysis of solvent accessibility for each amino acid in Switch I. The calculation was performed using Naccess.
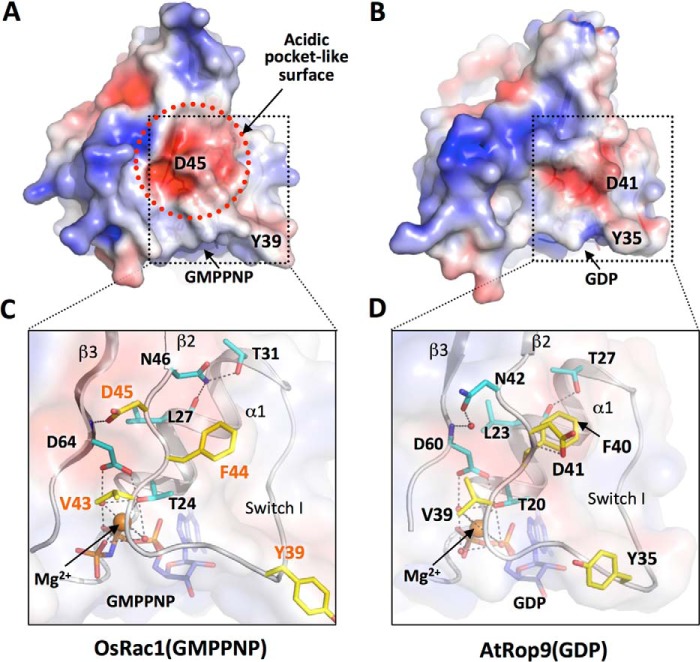
GTP-bound OsRac1 binds much more strongly than GDP-bound OsRac1 to OsRbohB (11). To elucidate the differences between GTP- and GDP-bound OsRac1, we used AtRop9, which shows the highest sequence similarity to OsRac1 in the PDB, as a model of GDP-bound OsRac1. A structural comparison of OsRac1 (GTP-bound form) with AtRop9 (GDP-bound form) showed a difference in their NADPH oxidase-binding sites. In OsRac1, the OsRbohB-binding site formed an acidic pocket-like surface centered on Asp45 (Fig. 6A), but this feature was not observed in AtRop9 (Fig. 6B). The acidic surface in OsRac1 was well stabilized by a hydrogen bond network among GMPPNP-Mg2+ and residues in α1, β2, β3, and the C-terminal portion of Switch I. The main chain of the Mg2+ ion-coordinating residue Asp64 in β3 contacted the side chain of the Switch I residue Asp45, and the side chain of the neighboring residue Asn46 was held by hydrogen bonds from Leu27 to Thr31 in α1 (Fig. 6C). In the AtRop9(GDP) complex, these hydrogen bonds did not form, and the position of the Asp41 side chain was largely different from that of Asp45 in OsRac1 (6 Å apart at the Cγ atom positions in superposed structures) so that the acidic pocket-like surface was covered (Fig. 6, B and D). We noted that all of these important residues in OsRac1 (Leu27, Thr31, Asp45, Asn46, and Asp64) are conserved in AtRop9. The GDP-to-GTP exchange in OsRac1 may induce a conformational change and increase the stability of the C-terminal acidic region of Switch I so that OsRac1 associates more strongly with the basic surface of OsRbohB.
FIGURE 6.
Surface charge difference between OsRac1 (GTP-bound form) and AtRop9 (GDP-bound form). The surface electrostatic potential (±3 kT) and the hydrogen bond network around the C-terminal portions of Switch I of OsRac1(GMPPNP) (A and C) and AtRop9-GDP (B and D) are shown. AtRop9 shows the highest sequence similarity to OsRac1 in the PDB. The surface and ribbon representations are rotated by 50° about the horizontal axis compared with the depiction in Fig. 3A. The acidic pocket-like structure of OsRac1 is demarcated by the red dashed circle (A). Critical residues in OsRac1 for OsRbohB(138–313) binding are shown in stick representation (Tyr39, Val43, Asp45, and Phe44; colored yellow with red labels). Other residues that form hydrogen bonds with these critical residues and with GMPPNP/GDP and the Mg2+ ions are colored cyan (C and D).
Interaction Site of HsRac1 with p67phox, an Essential Component of the NADPH Oxidase Complex
As described above, OsRac1 Switch I Tyr39, Val43, Phe44 and Asp45 residues interact with OsRbohB(138–313), with Tyr39 and Asp45 playing a key role in the activation of NADPH oxidase in rice cells (Fig. 7). Switch I of HsRac1/HsRac2 has been reported to be important for the activation of NADPH oxidase (43, 44). For example, substitutions of Phe28, Thr35, Val36, and Asp38 of HsRac2, which correspond to Phe35, Thr42, Val43, and Asp45 of OsRac1, respectively, resulted in a significant reduction in activation ability (43), and alanine substitutions at Thr35 and Asp38 in HsRac2 abolished it completely. In animals, phagocytic NOX2 is the best characterized NADPH oxidase, and its activation process is relatively well understood. Unlike NADPH oxidase of plants (Rboh), NOX2 does not interact directly with HsRac1/HsRac2 (44), and NOX2 activation requires the formation of a multicomponent complex (45, 46). One essential component is the direct interaction of p67phox with NOX2 and HsRac1/HsRac2 (45, 47). The crystal structure of the HsRac1-p67phox complex shows that HsRac1 binds to p67phox via α1, Switch I, and the loop connecting β6 to α5 (Fig. 7 and supplemental Fig. S6) (48). Although the Switch I region is therefore critical for the activation of NADPH oxidase in both plants and animals, the molecular details of the interacting regions differ between plants and animals (Fig. 7 and supplemental Fig. S6).
FIGURE 7.
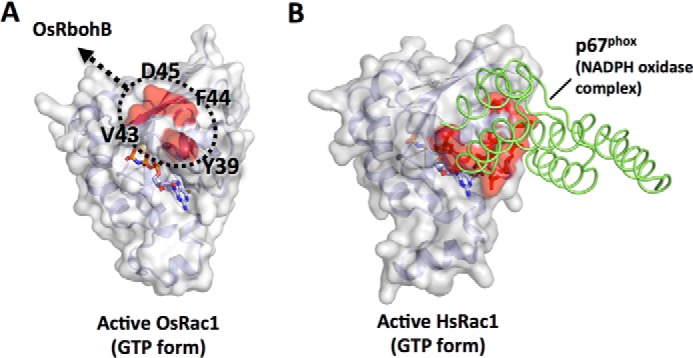
Modes of binding of plant and animal Rac/Rop proteins to NADPH oxidase. A, surface representation of OsRac1(GMPPNP). Four residues that are critical for the recognition and activation of the plant NADPH oxidase OsRbohB(138–313), revealed in this study, are shown in red. B, x-ray structure of the HsRac1-p67phox complex (PDB code 1E96). HsRac1 and p67phox are shown in surface (gray) and ribbon (green) representations, respectively. The p67phox-binding interface on HsRac1 is colored red. Activated OsRac1 interacts directly with the N-terminal domain of OsRbohB through Tyr39 and Asp45 and induces ROS production by OsRbohB, leading to an immune response in plant cells. Although Switch I is critical for the activation of NADPH oxidase in both plant and animal Rac/Rop proteins, details of their binding regions are different.
Conclusions
OsRac1 Switch I was identified as the region that binds OsRbohB(138–313), and four residues in this region (Tyr39, Val43, Phe44, and Asp45), particularly Tyr39 and Asp45, were found to be critical for the interaction and for ROS production in rice cells. Previous studies have shown that ROS production by NADPH oxidase is regulated by Rac/Rop proteins (11, 16, 49). Combined with our previous results (11, 13, 29), our data assist in delineating the mechanism of rice immunity. In particular, OsCERK1, the receptor for pathogen-associated molecular patterns, such as chitin, phosphorylates and activates OsRacGEF1, and activated OsRacGEF1 subsequently activates OsRac1 by the guanine nucleotide exchange process. Finally, OsRac1 activates ROS production via direct interaction with the NADPH oxidase OsRbohB (Fig. 7 and supplemental Fig. S6). Our results have also contributed toward delineation of the molecular mechanisms associated with various plant cellular responses, such as pollen tube growth and root hair development, that are influenced by Rac/Rop proteins.
Supplementary Material
Acknowledgments
We thank Hiroko Kinoshita, Akiko Yasuba, and Momoko Yoneyama for help with sample preparations; Rie Kurata and Yuki Nishigaya for help with mass spectrometry; Kohei Takeshita and Yumiko Hara for help with crystallization; and Kokoro Hayashi, Tomoyuki Mori, and Kentaro Ihara for helpful comments. We also thank the beamline staff of BL41XU and BL44XU at SPring-8 (Proposals 2013A1502, 2012B1544, 2012B6708, 2012A1248, and 2012A6708) and of NW12 at the Photon Factory (Proposal 2012G154) for help with the data collection and Academia Sinica and the National Synchrotron Radiation Research Center (Taiwan, Republic of China) for use of the MX225HE detector at beamline BL44XU.
This work was supported in part by Grants-in-aid for science research (to I. O., Y. K., C. K., and K. S.), the Targeted Proteins Research Program (to C. K. and K. S.), a Ministry of Agriculture, Forestry and Fisheries Genomics for Agricultural Innovation grant (to K. S.), Grants-in-aid for JSPS Fellows (to K. K. and M. N.), and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The atomic coordinates and structure factors (code 4U5X) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Figs. S1–S6.
- ROS
- reactive oxygen species
- GEF
- guanine nucleotide exchange factor
- CA
- constitutively activated
- DN
- dominant-negative
- GMPPNP
- guanosine 5′-(β,γ-imido)triphosphate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank
- HSQC
- heteronuclear single quantum coherence
- GTPγS
- guanosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell 14, suppl., S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng Z. L., Yang Z. (2000) The Rop GTPase: an emerging signaling switch in plants. Plant Mol. Biol. 44, 1–9 [DOI] [PubMed] [Google Scholar]
- 3. Nibau C., Wu H. M., Cheung A. Y. (2006) RAC/ROP GTPases: 'hubs' for signal integration and diversification in plants. Trends Plant Sci. 11, 309–315 [DOI] [PubMed] [Google Scholar]
- 4. Yang Z., Fu Y. (2007) ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10, 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu T., Wen M., Nagawa S., Fu Y., Chen J. G., Wu M. J., Perrot-Rechenmann C., Friml J., Jones A. M., Yang Z. (2010) Cell surface- and Rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oda Y., Fukuda H. (2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337, 1333–1336 [DOI] [PubMed] [Google Scholar]
- 7. Kawasaki T., Henmi K., Ono E., Hatakeyama S., Iwano M., Satoh H., Shimamoto K. (1999) The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. U.S.A. 96, 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ono E., Wong H. L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A. 98, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suharsono U., Fujisawa Y., Kawasaki T., Iwasaki Y., Satoh H., Shimamoto K. (2002) The heterotrimeric G protein α acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A. 99, 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujiwara M., Umemura K., Kawasaki T., Shimamoto K. (2006) Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol. 140, 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong H. L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., Kojima C., Yoshioka H., Iba K., Kawasaki T., Shimamoto K. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H. L., Kawasaki T., Shimamoto K. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7, 362–375 [DOI] [PubMed] [Google Scholar]
- 13. Akamatsu A., Wong H. L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., Shimamoto K. (2013) An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476 [DOI] [PubMed] [Google Scholar]
- 14. Kawano Y., Chen L., Shimamoto K. (2010) The function of Rac small GTPase and associated proteins in rice innate immunity. Rice 3, 112–121 [Google Scholar]
- 15. Kawano Y., Shimamoto K. (2013) Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 16, 496–504 [DOI] [PubMed] [Google Scholar]
- 16. Potocký M., Pejchar P., Gutkowska M., Jiménez-Quesada M. J., Potocká A., Alché Jde D., Kost B., Žárský V. (2012) NADPH oxidase activity in pollen tubes is affected by calcium ions, signaling phospholipids and Rac/Rop GTPases. J. Plant Physiol. 169, 1654–1663 [DOI] [PubMed] [Google Scholar]
- 17. Craddock C., Lavagi I., Yang Z. (2012) New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 22, 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X., Naramoto S., Robert S., Tejos R., Löfke C., Lin D., Yang Z., Friml J. (2012) ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr. Biol. 22, 1326–1332 [DOI] [PubMed] [Google Scholar]
- 19. Lin D., Nagawa S., Chen J., Cao L., Chen X., Xu T., Li H., Dhonukshe P., Yamamuro C., Friml J., Scheres B., Fu Y., Yang Z. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y., Yang Z. (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10, e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng Z. L., Nafisi M., Tam A., Li H., Crowell D. N., Chary S. N., Schroeder J. I., Shen J., Yang Z. (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14, 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Li Z., Gao X., Chinnusamy V., Bressan R., Wang Z. X., Zhu J. K., Wu J. W., Liu D. (2012) ROP11 GTPase negatively regulates ABA signaling by protecting ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9 in Arabidopsis. J. Integr. Plant. Biol. 54, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nibau C., Tao L., Levasseur K., Wu H. M., Cheung A. Y. (2013) The Arabidopsis small GTPase AtRAC7/ROP9 is a modulator of auxin and abscisic acid signaling. J. Exp. Bot. 64, 3425–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sørmo C. G., Leiros I., Brembu T., Winge P., Os V., Bones A. M. (2006) The crystal structure of Arabidopsis thaliana RAC7/ROP9: the first RAS superfamily GTPase from the plant kingdom. Phytochemistry 67, 2332–2340 [DOI] [PubMed] [Google Scholar]
- 25. Thomas C., Fricke I., Scrima A., Berken A., Wittinghofer A. (2007) Structural evidence for a common intermediate in small G protein-GEF reactions. Mol. Cell 25, 141–149 [DOI] [PubMed] [Google Scholar]
- 26. Thomas C., Fricke I., Weyand M., Berken A. (2009) 3D structure of a binary ROP-PRONE complex: the final intermediate for a complete set of molecular snapshots of the RopGEF reaction. Biol. Chem. 390, 427–435 [DOI] [PubMed] [Google Scholar]
- 27. Al-Mulla F., Milner-White E. J., Going J. J., Birnie G. D. (1999) Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J. Pathol. 187, 433–438 [DOI] [PubMed] [Google Scholar]
- 28. Oda T., Hashimoto H., Kuwabara N., Hayashi K., Kojima C., Kawasaki T., Shimamoto K., Sato M., Shimizu T. (2008) Crystallographic characterization of the N-terminal domain of a plant NADPH oxidase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 867–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oda T., Hashimoto H., Kuwabara N., Akashi S., Hayashi K., Kojima C., Wong H. L., Kawasaki T., Shimamoto K., Sato M., Shimizu T. (2010) Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 285, 1435–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kosami K., Ohki I., Hayashi K., Tabata R., Usugi S., Kawasaki T., Fujiwara T., Nakagawa A., Shimamoto K., Kojima C. (2014) Purification, crystallization and preliminary X-ray crystallographic analysis of a rice Rac/Rop GTPase, OsRac1. Acta Crystallogr. F Struct. Biol. Commun. 70, 113–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 36. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: Φ,Ψ and Cβ deviation. Proteins Struct. Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 37. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 38. Hiei Y., Ohta S., Komari T., Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 [DOI] [PubMed] [Google Scholar]
- 39. Longenecker K., Read P., Lin S. K., Somlyo A. P., Nakamoto R. K., Derewenda Z. S. (2003) Structure of a constitutively activated RhoA mutant (Q63L) at 1.55 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 59, 876–880 [DOI] [PubMed] [Google Scholar]
- 40. Hirshberg M., Stockley R. W., Dodson G., Webb M. R. (1997) The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat. Struct. Biol. 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 41. Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273, 9656–9666 [DOI] [PubMed] [Google Scholar]
- 42. Yamaguchi K., Imai K., Akamatsu A., Mihashi M., Hayashi N., Shimamoto K., Kawasaki T. (2012) SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 70, 389–397 [DOI] [PubMed] [Google Scholar]
- 43. Xu X., Barry D. C., Settleman J., Schwartz M. A., Bokoch G. M. (1994) Differing structural requirements for GTPase-activating protein responsiveness and NADPH oxidase activation by Rac. J. Biol. Chem. 269, 23569–23574 [PubMed] [Google Scholar]
- 44. Koga H., Terasawa H., Nunoi H., Takeshige K., Inagaki F., Sumimoto H. (1999) Tetratricopeptide repeat (TPR) motifs of p67phox participate in interaction with the small GTPase Rac and activation of the phagocyte NADPH oxidase. J. Biol. Chem. 274, 25051–25060 [DOI] [PubMed] [Google Scholar]
- 45. Bedard K., Krause K. H. (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
- 46. Han C. H., Freeman J. L., Lee T., Motalebi S. A., Lambeth J. D. (1998) Regulation of the neutrophil respiratory burst oxidase. Identification of an activation domain in p67phox. J. Biol. Chem. 273, 16663–16668 [DOI] [PubMed] [Google Scholar]
- 47. Diekmann D., Abo A., Johnston C., Segal A. W., Hall A. (1994) Interaction of Rac with p67phox and regulation of phagocytic NADPH oxidase activity. Science 265, 531–533 [DOI] [PubMed] [Google Scholar]
- 48. Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. (2000) Structure of the TPR domain of p67phox in complex with Rac-GTP. Mol. Cell 6, 899–907 [DOI] [PubMed] [Google Scholar]
- 49. Jones M. A., Raymond M. J., Yang Z., Smirnoff N. (2007) NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58, 1261–1270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.