Background: ESCRT-III can enhance Vta1 stimulation of Vps4 ATPase activity via the Vps4 stimulatory element (VSE) of Vta1.
Results: α7 and α9 of the Vps4 small AAA domain mediate VSE stimulation, contributing to Vps4 function in vivo.
Conclusion: Vta1 contacts Vps4 α7 and α9 during ESCRT-III-enhanced stimulation of Vps4.
Significance: These studies identify a novel mechanism of Vps4 stimulation.
Keywords: ATPases Associated with Diverse Cellular Activities (AAA), Endosomal Sorting Complexes Required for Transport (ESCRT), Endosome, Enzyme Mechanism, Lysosome, Membrane Trafficking, Vps4, Vta1
Abstract
The endosomal sorting complexes required for transport (ESCRTs) function in a variety of membrane remodeling processes including multivesicular body sorting, abscission during cytokinesis, budding of enveloped viruses, and repair of the plasma membrane. Vps4 ATPase activity modulates ESCRT function and is itself modulated by its cofactor Vta1 and its substrate ESCRT-III. The carboxyl-terminal Vta1/SBP-1/Lip5 (VSL) domain of Vta1 binds to the Vps4 β-domain to promote Vps4 oligomerization-dependent ATP hydrolysis. Additionally, the Vps4 stimulatory element (VSE) of Vta1 contributes to enhancing Vps4 oligomer ATP hydrolysis. The VSE is also required for Vta1-dependent stimulation of Vps4 by ESCRT-III subunits. However, the manner by which the Vta1 VSE contributes to Vps4 activation is unknown. Existing structural data were used to generate a model of the Vta1 VSE in complex with Vps4. This model implicated residues within the small ATPase associated with various activities (AAA) domain, specifically α-helices 7 and 9, as relevant contact sites. Rational generation of Vps4 mutants defective for VSE-mediated stimulation, as well as intergenic compensatory mutations, support the validity of this model. These findings have uncovered the Vps4 surface responsible for coordinating ESCRT-III-stimulated Vta1 input during ESCRT function and identified a novel mechanism of Vps4 stimulation.
Introduction
The endosomal sorting complexes required for transport (ESCRTs)2 participate in multivesicular body (MVB) sorting in the endocytic pathway, abscission of the cellular bridge during cytokinesis, the budding of enveloped viruses, and the repair of small wounds in the plasma membrane (1–5). Most of these processes are similar in that cytosolic machinery (i.e. ESCRTs) effects membrane scission from within the neck of a vesicle budding away from the cytoplasm (e.g. MVB sorting, viral budding) or from within a membrane tubule (e.g. abscission), and this class of processes have been termed “reverse topology” membrane fission events (6). The sorting of cargo into the MVB pathway is the best understood of these ESCRT-dependent phenomena. Cargoes destined for inclusion into the MVB pathway are covalently modified with ubiquitin (7–9). These modified cargoes are recognized by ubiquitin-binding domains in the early ESCRTs (ESCRT-0, -I, and -II) and sequestered into endosomal microdomains that bud into the endosome as intralumenal vesicles (10, 11). Intralumenal vesicle formation results from the activity of ESCRT-III and associated factors (e.g. Bro1·Doa4 and Vps4·Vta1 complexes) that interact on the endosomal membrane (12, 13). ESCRT-III and associated factors are responsible for coordinating intralumenal vesicle formation with recycling of ubiquitin and disassembly of ESCRT-III (14–16). ESCRTs function on the cytoplasmic face of the endosome to drive membrane vesiculation into the endosomal lumen.
Coordination of ESCRT-III polymerization and disassembly is central in this MVB sorting process. The ESCRT-III subunits (Vps20/CHMP6, Snf7/CHMP4, Vps24/CHMP3, Vps2/CHMP2, Did2/Fti1/CHMP1, Ist1/hIst1, and Vps60/CHMP5) are structurally similar metastable molecules that exist as soluble monomers in the cytoplasm and assemble into a membrane-associated polymer (17–20). Polymerization of ESCRT-III into a spiral fibril has been implicated in the deformation and scission of membranes (21–24). This polymerization is thought to lead to the exposure of carboxyl-terminal motifs (e.g. microtubule interacting and trafficking (MIT)-interacting motifs (MIMs)) responsible for recruiting ESCRT-III associated factors such as the Vps4·Vta1 hetero-oligomer (25–32). Vps4 (Vacuolar protein sorting 4) is the ATPase associated with various activities (AAA)-ATPase responsible for disassembling ESCRT-III (33–35), and Vta1 (Vps twenty associated 1) is a cofactor that activates Vps4 ATP hydrolysis and promotes Vps4 function (36–41). Both Vps4 and Vta1 harbor MIT domains in their amino termini (see Fig. 1A) (42–45). MIM-MIT associations can enhance activity of Vps4·Vta1 in addition to facilitating recruitment to ESCRT-III (16, 27–32, 35, 46–48). The presumption is that ESCRT-III associations with Vps4·Vta1 serve to coordinate the timing of ESCRT-III polymerization with ESCRT-III disassembly to permit efficient ESCRT function (14, 49).
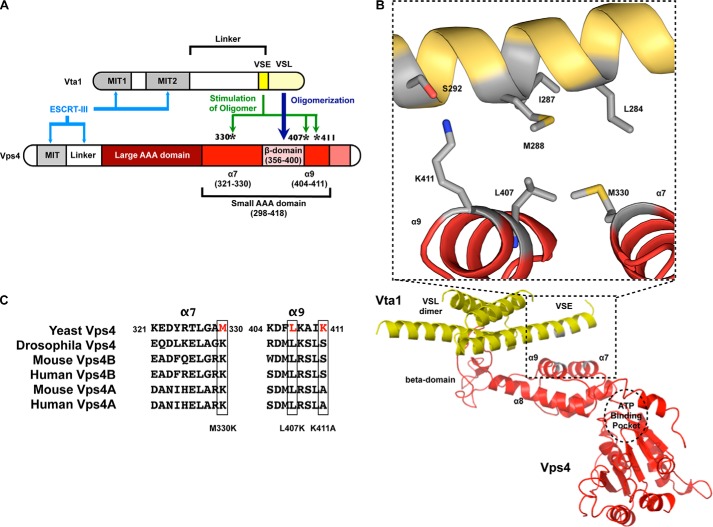
FIGURE 1.
Modeling of the VSE into the structure of the VSL domain in complex with the small AAA and β-domains of Vps4. A, cartoon of Vps4 structural domains. The β-domain is an insert within the small AAA domain. Met-330 is in α7 preceding the β-domain, whereas Leu-407 and Lys-411 are in α9 immediately following the β-domain. α9 is located closer to the β-domain (proximal helix), whereas α7 is more distant (distal helix). B, the structure of the Vta1 VSL dimer bound to the Vps4 β-domain and small AAA domain (Protein Data Bank code 3MHV; MMDB 85335) was used to align structures of the Vps4 AAA domain (residues 119–437) (Protein Data Bank code 2QP9; MMDB 59522; red) and the Vta1 carboxyl terminus (residues 281–330) (Protein Data Bank code 2RKL; MMDB 62002; yellow). The Vta1 VSE falls on a flexible helix preceding the VSL domain, and alignment positioned the VSE neighboring Vps4 α7 and α9. Residues of the Vta1 VSE (Leu-284, Ile-287, and Met-288) and Vps4 small AAA domain (Met-330 of α7 and Leu-407 of α9) that appear to mediate VSE-Vps4 interaction are indicated in gray in the inset. Vta1 Ser-292 is not required for VSE stimulation but appears to interact with Vps4 Lys-411 of α9. These residues are also depicted in gray. C, sequence alignment of α7 and α9 from S. cerevisiae Vps4, D. melanogaster Vps4, M. musculus Vps4A and Vps4B, and H. sapiens Vps4A and Vps4B. Leu-407 is conserved from yeast to man.
Regulation of Vps4 ATPase activity is key during this coordination. Vps4 ATPase activity is regulated in a number of manners. First, oligomerization is one mechanism regulating Vps4 ATPase activity (34, 35, 50, 51). Second, ESCRT-III interactions with Vps4 enhance ATP hydrolysis by relieving autoinhibition of the AAA domain; this occurs via MIM-MIT interactions and ESCRT-III acidic region interactions with the Vps4 linker region (31, 51, 52). Third, Vta1 activation of Vps4 occurs through both promoting Vps4 oligomerization and enhancing ATP hydrolysis by the Vps4 oligomer (40, 53). The Vta1 carboxyl-terminal Vta1/SBP1/Lip5 (VSL) domain is responsible for dimerization of Vta1 and interacts with the Vps4 β-domain (an insert within the AAA domain) to promote Vps4 oligomerization (39, 40, 43, 54, 55) (see Fig. 1A). The Vps4 stimulatory element (VSE), an element just upstream from the Vta1 VSL domain, mediates Vta1 enhancement of Vps4 oligomer ATP hydrolysis (53). Moreover, binding of ESCRT-III subunits Did2 or Vps60 to the amino-terminal Vta1 MIT domains relieve autoinhibition within Vta1 to further enhance stimulation of Vps4 ATP hydrolysis via the Vta1 VSE (31, 43, 46, 47, 53). These observations indicate a complex orchestration of inputs from ESCRT-III and Vta1 that regulate Vps4 AAA domain ATP hydrolysis via the Vps4 MIT domain, linker region, and β-domain.
The Vta1 VSE has been implicated in mediating ESCRT-III-enhanced Vta1 stimulation of Vps4 (53), but the surface of Vps4 that is contacted by the VSE to effect increased ATP hydrolysis is unknown. Structures of the Vta1 carboxyl terminus dimer, including both the VSL and VSE (43), and the VSL dimer in complex with a Vps4 fragment containing the β-domain and part of the small AAA domain (55) have been determined. This information was used to model VSE interaction with Vps4 to understand Vta1 VSE stimulation (see Fig. 1B). Three residues (located on α7 and α9) of the small AAA domain were identified as a potential interaction surface: methionine 330, leucine 407, and lysine 411. Models of the Vps4 oligomer (49, 50) place these residues on the outer surface of the proposed oligomer where they would be accessible to the Vta1 VSE. Vps4 mutants with these residues altered exhibited concentration-dependent ATPase activity comparable with that of WT Vps4 and were responsive to direct stimulation by ESCRT-III, indicating that these mutants do not globally compromise Vps4 structure and function. However, these Vps4 mutants exhibited defects in VSE-mediated activation by Vta1. Vps4(L407K) exhibited the most severe defects in stimulation by the Vta1 VSE, but stimulation could be restored with compensatory mutation of the Vta1 VSE. Vps4(L407K) also exhibited deficits in the maturation of the MVB cargo CPS and recycling of ESCRT-III subunit Snf7 in vivo. These observations support a model in which the Vta1 VSE contacts the Vps4 small AAA domain via α7 and α9 to mediate ESCRT-III enhancement of Vps4·Vta1, thereby promoting ATP hydrolysis and coordinating ESCRT function.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
pET28 Vps4 was generated by PCR-amplifying the VPS4 ORF (along with 252 bp of sequence 3′ to the STOP codon) with NdeI (5′) and SalI (3′) restriction sites and subcloning the resulting fragment into the NdeI and SalI sites of pET28b (Novagen). Mutagenesis of Vps4 was performed using the Gene Tailor site-directed mutagenesis system (Invitrogen). The NcoI to SalI fragments of these pET28 Vps4 mutants were then subcloned into the pMB28 (VPS4 in pRS416) (52) after shuttling through pRS415. All cloned PCR products and mutant plasmids were sequenced to exclude unexpected mutations. The generation of pGST-Vta1, pET28-Vta1, Vta1 fragments, Vta1(VSEΔ) mutant, and pET28-Ist1 (WT and L168A,Y172A) expression plasmids have been previously described (26, 40, 53). The BY4742 vps4Δ::NEO strain was obtained from Open Biosystems.
Protein Expression and Purification
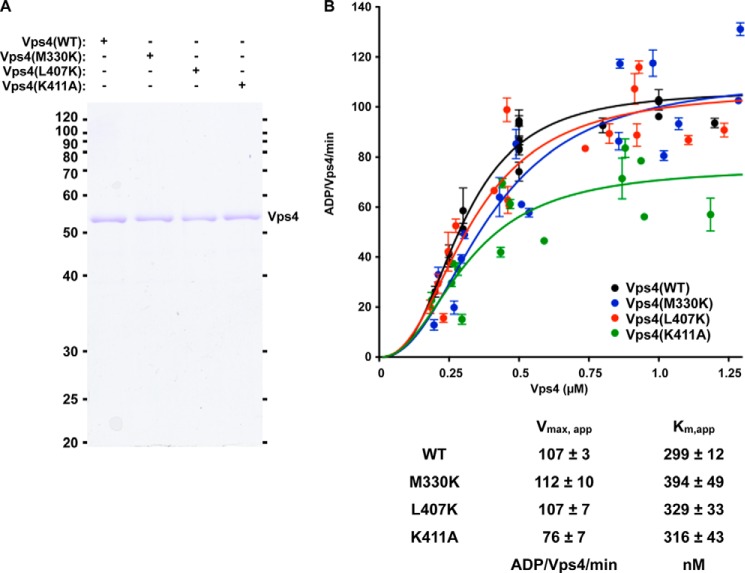
Protein expression was performed in the BL21-DE3 bacterial strain at 16–20 °C for 14–20 h with 0.5 mm isopropyl β-d-thiogalactopyranoside. His6-Vps4 WT and mutant fusion proteins were purified by Ni2+ affinity chromatography (5 ml HiTrap Chelating FF), treated with thrombin, incubated with ATP to dissociate chaperones, and subjected to anion exchange (Bioscale Q2) chromatography with (A: 20 mm Hepes, pH 7.5; B: 20 mm Hepes, 1 m KCl, pH 7.5). WT Vta1 and Vta1(VSEΔ) were purified by the same procedure. His6-Vta1(1–330), (275–330), and (290–330) were purified by Ni2+ affinity chromatography (5 ml HiTrap Chelating FF), incubated with ATP to dissociate chaperones, and subjected to size exclusion (Superdex 75 HiLoad 16/60) chromatography (20 mm Hepes, 150 mm KCl, pH 7.5). Ist1 was purified by Ni2+ affinity chromatography (5 ml HiTrap Chelating FF), treated with thrombin, incubated with ATP to dissociate chaperones, and subjected to size exclusion (Superdex 200 HiLoad 16/60) chromatography (20 mm Hepes, 200 mm KCl, pH 7.5). For Vta1 and Ist1 proteins, fractions from the gel filtration and anion exchange chromatography were checked for ATPase activity to avoid including contaminating ATPases. For Vta1 and Vps4, pooled fractions were concentrated; adjusted to 20 mm Hepes, 150 mm KCl, 10% glycerol, 2 mm DTT, pH 7.5; and stored at −80 °C. Ist1 was concentrated and stored at −80 °C in 20 mm Hepes, 200 mm KCl, pH 7.5. Purity of the Vps4 proteins is indicated in Fig. 2A. Vps4 regulators (Ist1 and Vta1 proteins) were of similar purity (see Figs. 3, inset, and 5A).
FIGURE 2.
Vps4 ATPase activity and Ist1 inhibition are not disrupted by mutations in Vps4 α7 and α9. A, Coomassie staining of equivalent amounts of WT and mutant Vps4 proteins. B, titration (250 nm to 1.25 μm) of WT (black) and Vps4 mutants (M330K, blue; L407K, red; K411A, green) was performed in the presence of 4 mm ATP. Rates are indicated as ADP generated/Vps4/min. The concentrations yielding half-maximal activity (Km, app) and the apparent Vmax are indicated in the accompanying table.
FIGURE 3.
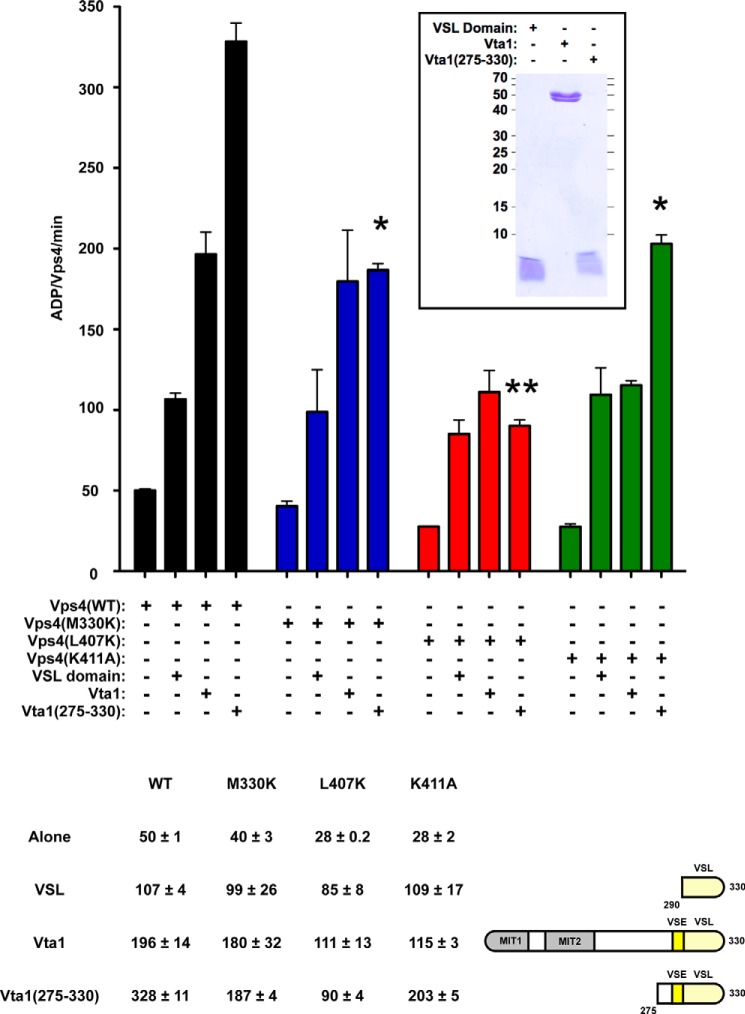
Mutations in Vps4 α7 and α9 disrupt aspects of VSE stimulation. WT (black) and mutant Vps4 (M330K, blue; L407K, red; K411A, green) ATPase activity alone (250 nm) or in the presence of 8 μm VSL domain (His6-Vta1(290–330)), WT Vta1 (His6-Vta1(1–330)), and Vta1(275–330) (His6-Vta1(275–330)). The rates are indicated as ADP generated/Vps4/min; the rates are also indicated in the table. WT and mutant Vps4 proteins exhibit comparable ATPase activity in the presence of VSL domain, and these rates are enhanced from the activities of the Vps4 proteins alone. Vps4(L407K) and Vps4(K411A) in the presence of Vta1 exhibit reduced activity compared with WT Vps4 and Vps4(M330K) in the presence of Vta1 (p value < 0.05). Vps4(M330K) and Vps4(K411A) in the presence of Vta1(275–330) (ESCRT-III-activated VSE) exhibit reduced activity compared with WT Vps4 in the presence of Vta1(275–330) (p value < 0.05; indicated by *), and activity of Vps4(L407K) with Vta1(275–330) is further reduced (p value < 0.01; indicated by **). Coomassie staining of equivalent amounts of VSL, Vta1, and Vta1(275–330) is indicated in the inset.
FIGURE 5.
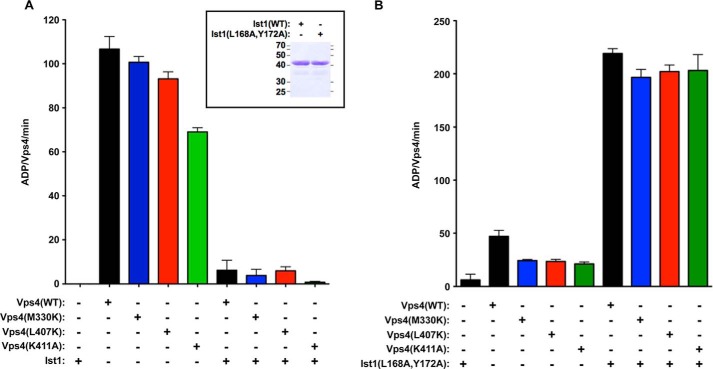
Direct ESCRT-III stimulation and Ist1 inhibition are not disrupted by mutations in Vps4 α7 and α9. A, 500 nm WT (black) and mutant Vps4 (M330K, blue; L407K, red; K411A, green) ATPase activity alone or in the presence of 10 μm WT Ist1. WT Ist1 inhibits WT Vps4 and the Vps4 mutants similarly. The rates are indicated as ADP generated/Vps4/min. Coomassie staining of equivalent amounts of Ist1 and Ist1(L168A,Y172A) is indicated in the inset. B, 250 nm WT (black) and mutant Vps4 (M330K, blue; L407K, red; K411A, green) ATPase activity alone or in the presence of 1 μm Ist1(L168A,Y172A). Ist1(L168A,Y172A) stimulates WT Vps4 and the Vps4 mutants similarly.
ATPase Assay
Measurement of Vps4 ATPase activity was performed as previously described (34, 40). Vps4 protein concentration was assessed using a Bio-Rad protein assay, and the stock was diluted to 1 or 2 μm in ATPase reaction buffer (0.1 m KOAc, 20 mm Hepes, and 5 mm MgOAc, pH 7.5). Portions of 1 or 2 μm dilutions were checked by SDS-PAGE and Coomassie staining to normalize for differences between WT and mutant Vps4 proteins during rate calculations. Titration of Vps4 (0.2–1.5 μm) was performed by diluting appropriate amounts of 2 μm Vps4 in ATPase buffer for a total of 18 μl. Reactions were initiated by the addition of ATP/[α-32P]ATP to 4 mm. 1-μl samples were removed at 4, 8, 12, and 16 min after ATP/[α-32P]ATP addition and resolved by thin layer chromatography using precoated PEI Cellulose TLC glass plates (Merck) and developing buffer (0.75 m KPO4, pH 3.5). The plates were dried, exposed to phosphorimaging screens for 12–16 h, processed using the Typhoon FLA 7000 system (GE Life Sciences), and ADP and ATP signal was quantitated using ImageQuant software package (GE Life Sciences). The percentage of ATP hydrolysis was used to calculate ADP generated per Vps4 molecule (using corrected Vps4 concentration) per min. Vps4 (0.25 μm) activity in the presence of Vta1 proteins (8 μm), a concentration previously demonstrated to be saturating (53), or Ist1 (10 μm, with 0.5 μm Vps4) were determined similarly with the additional step that 18-μl reactions were preincubated on ice for at least 30 min prior to initiation at 30°. Reactions were performed in duplicate or triplicate within experiments, and rates presented are from experiments performed on at least 2 separate days. The error bars indicate standard error from the mean. The significances of differences in rates were assessed by T tests using Prism5 (GraphPad). An example of this analysis including images of TLC plates, and determination of rates has been presented in Norgan et al. (53).
GST Pulldown Assays
GST and GST-Vta1 were coupled to glutathione-Sepharose 4B beads (GE Healthcare) in PBS with 0.05% Tween 20 (PBST). Beads were washed with PBST and with ATPase reaction buffer supplemented with 0.05% Tween 20 and 1 mm ATP. GST- or GST-Vta1-coupled beads were dispensed into 0.8-ml microspin columns (Pierce) along WT or mutant Vps4 proteins at a final concentration of 10 ng/μl. Incubations were performed with 1 mm ATP at 4 °C for 1 h. Samples were then spun briefly and washed three times with ATPase reaction buffer containing 0.05% Tween 20 and 1 mm ATP. Samples were eluted with 5× Laemmli sample buffer, resolved by SDS-PAGE, and analyzed by staining with Coomassie Blue or Western blotting with Vps4 antisera (33).
Biochemical Analyses
Analysis of CPS transport to the vacuole by pulse-chase immunoprecipitation was performed as previously described (17). The significances of differences in maturation rates were assessed by two-way analysis of variance tests using Prism5 (GraphPad). Subcellular fractionation was performed as previously described (17, 53). Briefly, 5 OD equivalents of yeast spheroplasts were resuspended at 10 OD/ml in lysis buffer (50 mm Tris, pH 7.5, 200 mm sorbitol, 2 mm EDTA with protease inhibitors), lysed by 15 strokes in a Dounce homogenizer, and subjected to a 10 min, 13,000 × g spin at 4 °C to separate the S13 and P13 fractions. Samples (0.04 OD equivalents) were resolved by SDS-PAGE and Western blotted for Snf7 (polyclonal antibody, 1:5,000), phosphoglycerate kinase (mAb, 1:2000) (Invitrogen), and Pep12 (mAb, 1:2000) (Invitrogen). Western blots were developed using both film and the UVP Autochemi System (Upland, CA), and quantitation was performed using ImageQuantTL software (GE Life Sciences). The error bars indicate standard error from the mean. The significances of differences in rates were assessed by t tests using Prism5 (GraphPad). Vps4 protein levels in yeast were assessed by harvesting 5 OD equivalents of rapidly growing yeast, precipitating proteins with addition of TCA to 10%, and lysing samples in 5× Laemmli sample buffer with glass beads. Samples (0.5 OD equivalents) were resolved by SDS-PAGE and Western blotted for Vps4 (polyclonal antibody, 1:1,000) and phosphoglycerate kinase (mAb, 1:10,000) (Invitrogen).
Molecular Modeling
The PyMOL molecular graphics system (Schrödinger) was used to model Vta1 VSE interaction with the Vps4 AAA domain. The structure of the Vta1 VSL in complex with Vps4 small AAA and β-domains (Protein Data Bank code 3MHV; MMDB 85335) (55) was used to align the extended Vta1 carboxyl terminus dimer structure (Protein Data Bank code 2RKL, includes VSE; MMDB 62002) (43) with the extended Vps4 AAA domain (Protein Data Bank code 2QP9; MMDB 59522) (56).
RESULTS
Residues within the Vps4 Small ATPase Domain Mediate Vta1 VSE Activation
ESCRT-III can stimulate Vps4 ATPase activity directly (31, 51, 52) or indirectly through binding Vta1 and derepressing activity of the Vta1 VSE (31, 53). However, the mechanisms by which these inputs are transmitted to the Vps4 AAA domain to alter ATP hydrolysis are unclear. We sought to understand this phenomenon by addressing the Vps4 AAA domain surface contacted by the Vta1 VSE. The structure of the Vta1 VSL domain dimer in complex with the small AAA and β-domains of Vps4 (55) was utilized to constrain a region of Vps4 that may be contacted by VSL-proximal Vta1 VSE residues (Fig. 1A highlights these domains). The Vps4-VSL cocrystal structure does not include the flexible helix containing the VSE, but this structure (Ref. 5; Protein Data Bank code 3MHV; MMDB 85335) can serve as a guide to align the previously determined structure of the Vta1 carboxyl terminus containing the flexible helix that includes the VSE (Ref. 43; Protein Data Bank code 2RKL; MMDB 62002) with the Vps4 AAA domain (Ref. 56; Protein Data Bank code 2QP9; MMDB 59522) (Fig. 1B). In this model, Vta1 residues implicated for the stimulatory activity of the VSE (Leu-284, Ile-287, and Met-288) (53) are proximal to Vps4 residues Met-330 and Leu-407 within the small AAA domain. Additionally, a third residue within the small AAA domain of Vps4 (Lys-411) appeared to interact with Ser-292 of Vta1, although mutation of this Vta1 residue did not compromise Vta1 stimulation of Vps4 (53). These three Vps4 residues fall onto two helices, α7 (Met-330) and α9 (Leu-407 and Lys-411), with α9 more proximal to the β-domain. Examination of yeast, fly, and mammalian sequence conservation of Vps4 α7 and α9 revealed that Leu-407 is the most conserved of the three residues across the Vps4 family (Fig. 1C). To test the model that the Vta1 VSE stimulates Vps4 via these residues, mutant forms of Vps4 were generated and analyzed for basal ATPase activity (Fig. 2), activity upon stimulation with Vta1 and ESCRT-III-activated Vta1 (Fig. 3), and direct regulation by ESCRT-III (Fig. 5).
Mutation of α7 and α9 Does Not Eliminate Vps4 ATPase Activity
WT and mutant forms of Vps4 were expressed in bacteria, purified (Fig. 2A), and characterized for inherent ATPase activity (i.e. ATPase activity in the absence of other factors). Vps4 exhibits concentration-dependent increases in specific activity (ADP generated/Vps4/min) in the low micromolar range (34, 40). This behavior is attributed to the role of oligomerization in forming the active enzyme. All forms of Vps4 displayed similar concentration-dependent ATPase activities with half-maximal activities (apparent Km, or Km, app) between 299 nm for WT and 394 nm for Vps4(M330K) (Fig. 2B). The apparent maximal specific activities (Vmax, app) of WT, Vps4(M330K), and Vps4(L407K) were similar (107, 112, and 107 ADP/Vps4/min, respectively), whereas Vps4(K411A) exhibited a reduced Vmax, app (76 ADP/Vps4/min). This analysis indicated that these mutant Vps4 proteins assemble into active oligomers in a manner comparable with WT Vps4, but a mutation within this portion of the small ATPase domain (K411A) subtly alters inherent ATP hydrolysis by the Vps4 oligomer.
Mutation of the Vps4 Small AAA Domain Impacts Vta1 Stimulation
Vta1 effects Vps4 stimulation through both the VSL domain and the VSE to promote Vps4 oligomerization and maximal activity of the Vps4 oligomer (40, 53). The Vta1 VSL domain primarily impacts Vps4 oligomerization (40), whereas the Vta1 VSE impacts additional stimulation of the oligomer (53). ESCRT-III binding to Vta1 enhances stimulation via the VSE, and Vta1(275–330) recapitulates this ESCRT-III-activated VSE stimulation (53). We therefore examined Vta1 stimulation of Vps4 in three contexts: 1) stimulation by the VSL domain; 2) stimulation by full-length Vta1, in which the VSE is partially repressed (i.e. basal VSE stimulation); and 3) stimulation by Vta1(275–330) to examine ESCRT-III-activated VSE stimulation. For these experiments examining VSL- or VSE-mediated stimulation, a concentration of Vps4 less than the Km, app was used (250 nm) to facilitate examination of stimulatory effects; the activities of WT and mutant Vps4 proteins alone at this concentration range from 28 to 50 ADP/Vps4/min (Fig. 3, consistent with Fig. 2B). The addition of VSL domain (8 μm) stimulated Vps4 activity to 107 ADP/Vps4/min (Fig. 3), comparable with the Vps4 intrinsic Vmax, app (Fig. 2B). The addition of Vta1 (8 μm) stimulated Vps4 activity to 196 ADP/Vps4/min (i.e. basal VSE stimulation), and Vta1(275–330) (8 μm) further stimulated activity to 328 ADP/Vps4/min (Fig. 3). This profile of stimulation is consistent with previous analyses indicating that Vta1 stimulates Vps4 more robustly than the VSL domain and that Vta1(275–330) exhibits additional VSE stimulation reflective of ESCRT-III (40, 53).
ESCRT-III-activated VSE stimulation (i.e. Vta1(275–330) stimulation) of Vps4(M330K), Vps4(L407K), and Vps4(K411A) were all significantly reduced compared with WT Vps4; however, these three mutants exhibited distinct profiles of stimulatory responses. VSL domain stimulated the mutants to activities comparable with their intrinsic Vmax, app values (99, 85, and 109 ADP/Vps4/min, respectively; Figs. 3 and 2B). This observation suggested that these mutations do not compromise Vta1 stimulation via the Vps4 β-domain, as predicted (because of intact interactions between the VSL and β-domain that contribute to oligomerization). However, Vta1 or Vta1(275–330) was unable to further stimulate Vps4(L407K) above the VSL domain stimulation (Fig. 3). This observation indicated that Vps4(L407K) is responsive to VSL domain stimulation via the β-domain but is defective for both basal and ESCRT-III-activated VSE-mediated stimulation.
The two other Vps4 mutants display more complex responses (Fig. 3). Vps4(M330K) exhibited enhanced Vta1 stimulation compared with the VSL domain (180 and 99 ADP/Vps4/min, respectively), but Vta1(275–330) was unable to further stimulate Vps4(M330K) (187 ADP/Vps4/min). This observation suggested that Met-330 does not participate in basal VSE stimulation, whereas this residue is critical for ESCRT-III-activated VSE-mediated stimulation. By contrast, Vps4(K411A) exhibited Vta1 stimulation comparable with VSL domain stimulation (115 and 109 ADP/Vps4/min, respectively), suggesting that basal VSE stimulation requires Lys-411. However, Vta1(275–330) enhanced Vps4(K411A) activity (203 ADP/Vps4/min) above levels observed with Vta1 or the VSL domain, but this activity was reduced from Vta1(275–330) stimulation of WT Vps4 (328 ADP/Vps4/min). This observation suggested that Vps4(K411A) remains responsive to the ESCRT-III-activated VSE stimulation mediated by Met-330, although Lys-411 participates in this stimulation. Lys-411 is present on α9, the helix proximal to the β-domain, whereas Met-330 is present on α7, the helix more distal to the β-domain. These results suggested that: 1) basal VSE stimulation is mediated by contacting α9, 2) ESCRT-III-activated VSE stimulation is mediated by additionally contacting α7, and 3) L407K disrupts stimulation via α7 and α9 to prevent both modes of VSE-mediated stimulation, suggesting that this represents a relevant contact site for both modes.
These ATPase assays suggested that Vps4 α7 and α9 contribute to Vta1 interaction with Vps4. To examine this model, in vitro association was examined by GST pulldown experiments. GST-Vta1 was able to isolate WT and Vps4 mutants more effectively than GST alone under conditions analogous to the ATPase reaction (Fig. 4). However, variation of isolation was observed among the Vps4 mutants: Vps4(K411A) exhibited isolation comparable with that of Vps4(WT), but Vps4(M330K) and Vps4(L407K) were less effectively isolated. This result indicated that the VSE contributes to Vta1 interaction with Vps4 in addition to VSL-β-domain interaction.
FIGURE 4.
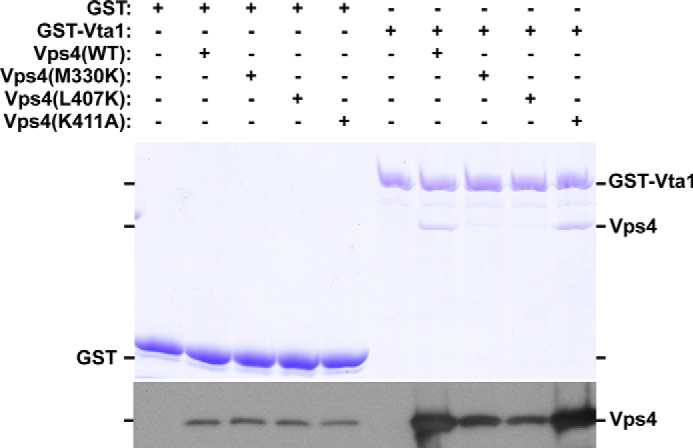
Vps4 M330K and L407K partially disrupt in vitro association with GST-Vta1. GST or GST-Vta1 pulldown experiments were performed with purified WT and Vps4 mutants. Eluted material was analyzed by Coomassie staining and Western blotting for Vps4. WT and Vps4 mutants were isolated with GST-Vta1 more effectively than with GST alone. Vps4(WT) and Vps4(K411A) exhibited similar isolation with GST-Vta1, but isolation of Vps4(M330K) and Vps4(L407K) was reduced. The data presented are representative of experiments performed more than three times.
Mutation of α7 and α9 Does Not Impact Direct ESCRT-III Regulation of Vps4
In addition to stimulating Vps4 via Vta1, ESCRT-III subunits can stimulate Vps4 directly (30, 31, 51, 52). We have implicated the α7-α9 surface of the small AAA domain in mediating ESCRT-III-activated Vta1 stimulation (i.e. Vta1(275–330) stimulation) (Fig. 3). Therefore, the contribution of this α7-α9 surface to direct ESCRT-III regulation of Vps4 was also examined.
The ESCRT-III subunit Ist1 was used for this analysis because recombinant Ist1 is more soluble than other ESCRT-III subunits. Although the Ist1 carboxyl terminus stimulates Vps4 ATPase activity, full-length Ist1 inhibits Vps4 ATPase activity (57). However, an Ist1 mutant defective for Did2 MIM1 binding (L168A,Y172A) (26) stimulates Vps4 in a manner dependent on both the Ist1 MIM1 and the Vps4 MIT domain (Fig. 5B and data not shown). Vps4 activities in the presence of WT Ist1 or Ist1(L168A,Y172A) were assessed to examine direct ESCRT-III regulation of Vps4. WT Vps4 alone (500 nm, rather than the 250 nm concentration used in Fig. 3) exhibited activity of 107 ADP/Vps4/min, and the addition of 10 μm WT Ist1 reduced ATPase activity to 6 ADP/Vps4/min (Fig. 5A). Similarly, the activities of the Vps4 small AAA domain mutants were inhibited by Ist1 (101, 93, and 69 ADP/Vps4/min for M330K, L407K, and K411A alone were reduced to 4, 6, and 0.7 ADP/Vps4/min with Ist1 addition). These observations indicated that these mutations within the small AAA domain do not disrupt Ist1 inhibition of Vps4 ATPase activity.
Direct stimulation of Vps4 by ESCRT-III was examined using the model protein Ist1(L168A,Y172A). WT Vps4 (250 nm, as used in Fig. 3 for the purpose of observing stimulation) generated 219 ADP/Vps4/min in the presence of 1 μm Ist1(L168A,Y172A) (Fig. 5B). Similar stimulation was observed with Ist1(L168A,Y172A) addition to Vps4(M330K), Vps4(L407K), and Vps4(K411A) (197, 202, and 203 ADP/Vps4/min, respectively). These similarities indicated that Ist1(L168A,Y172A) stimulation of Vps4 is not dependent on Met-330, Leu-407, or Lys-411. Stimulation of Vps4 mutants was also observed with addition of GST-Vps2 coupled to glutathione beads (data not shown). These observations indicate that direct ESCRT-III stimulation of Vps4 is not dependent on α7 and α9. In total, these in vitro biochemical characterizations demonstrate that these three residues of the Vps4 small AAA domain (Met-330, Leu-407, and Lys-411) are not required for intrinsic Vps4 ATPase activity, direct ESCRT-III stimulation of Vps4, or Ist1 inhibition of Vps4, but these residues contribute to Vta1 VSE stimulation of Vps4 in a specific manner.
VSE Stimulation via the Small AAA Domain Promotes Vps4 Function in Vivo
We next sought to address the significance of this VSE contact with α7 and α9 in vivo. Deletion of Vta1 only partially disrupts MVB sorting (40), and ESCRT-III activation of Vta1 only partially contributes to this defect as suggested by two observations: 1) deletion of the Vta1 MIT domains disrupts Vta1-ESCRT-III associations, but expression of this mutant in vta1Δ yeast exhibited only weak defects in MVB sorting (31); and 2) mutation of the Vta1 VSE (L284E, I287E, and M288E; VSEΔ) compromised MVB sorting and ESCRT-III recycling in vivo, although the extents of the defects were less than vta1Δ effects (53). These observations predicted that mutation of the Vps4 residues mediating Vta1 VSE function would also only weakly disrupt MVB sorting and ESCRT-III recycling.
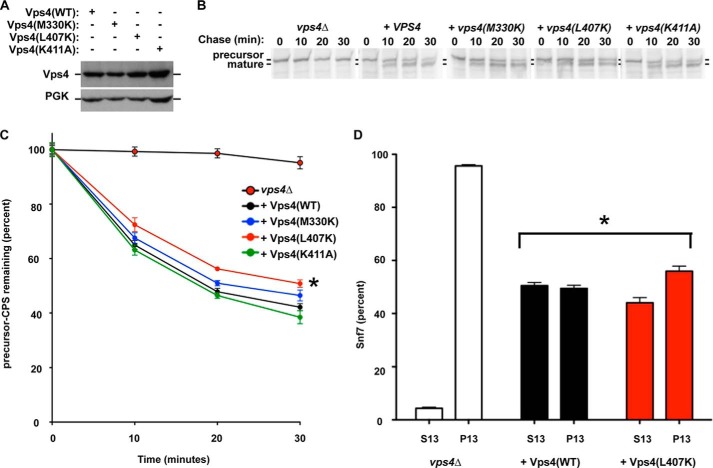
To test this prediction, M330K, L407K, and K411A forms of Vps4 were expressed in vps4Δ cells (Fig. 6A), and the maturation kinetics of the MVB cargo CPS were evaluated as an indicator of MVB sorting pathway function (Fig. 6, B and C). CPS is synthesized as an inactive precursor (pCPS). Sorting of pCPS into the MVB pathway and subsequent delivery to the vacuolar lumen permits proteolytic activation of CPS (mCPS). The kinetic analysis of the conversion of pCPS to mCPS serves as an indicator of MVB pathway function (31, 53). Loss of Vps4 (vps4Δ) disrupted MVB sorting, resulting in the persistence of pCPS (Fig. 6, B and C, and Ref. 17). Reintroduction of WT Vps4 restored CPS maturation with only 42% of pCPS remaining at 30 min (black). Reintroduction of Vps4(K411A) yielded CPS maturation kinetics (green) indistinguishable from WT Vps4. Reintroduction of Vps4(M330K) resulted in a subtle delay in CPS maturation kinetics (blue), but this effect was not significantly different from the CPS maturation kinetics with WT Vps4. By contrast, the CPS maturation kinetics with reintroduction of Vps4(L407K) were delayed from the CPS maturation kinetics with WT Vps4 (two-way analysis of variance, p value < 0.01; red). This partial defect for Vps4(L407K) is similar to partial defects in CPS maturation observed with Vta1(MITΔ) and Vta1(VSEΔ) (31, 53). Importantly, Vps4(M330K), Vps4(L407K), and Vps4(K411A) are all expressed similarly to WT Vps4 (Fig. 6A). These results support the conclusion that Leu-407 contributes to Vps4 function in the MVB sorting pathway by permitting Vta1 stimulation via the VSE.
FIGURE 6.
Leu-407 contributes to Vps4 function in vivo. A, Western blotting analysis of Vps4 levels in vps4Δ yeast expressing WT and Vps4 mutants. Western blotting for phosphoglycerate kinase was performed as a loading control. B and C, pulse-chase immunoprecipitation was performed on endogenous CPS in vps4Δ cells transformed with vector or plasmids expressing WT Vps4 or Vps4 point mutants. CPS maturation was quantitated using a phosphoimager and plotted relative to time zero. CPS maturation kinetics in cells expressing Vps4(L407K) was significantly different (*, p values < 0.01) from cells lacking Vps4 or cells expressing WT Vps4. D, subcellular fractionation of the ESCRT-III subunit Snf7 was performed on vps4Δ cells transformed with vector or plasmids expressing WT Vps4 or Vps4(L407K). Snf7 fractionation in cells expressing Vps4(L407K) was significantly different (*, p values < 0.01) from cells lacking Vps4 or cells expressing WT Vps4.
Vps4 disassembles ESCRT-III, and defects in Vps4 function lead to enhanced ESCRT-III membrane association (17, 34). To more directly examine Vps4(L407K) function in ESCRT-III disassembly, the membrane association of the ESCRT-III subunit Snf7 was examined in vps4Δ yeast expressing Vps4(L407K) (Fig. 6D). Loss of Vps4 (vps4Δ) resulted in 95% of Snf7 in the 13,000 × g pellet, indicating increased membrane association of ESCRT-III. Reintroduction of WT Vps4 reduced Snf7 association with the 13,000 × g pellet to 48%. By contrast, reintroduction of Vps4(L407K) reduced Snf7 association with the 13,000 × g pellet to 56%. These observations indicate a small but significant reduction in Snf7 recycling with reintroduction of Vps4(L407K) (p value < 0.01), consistent with both predictions of the model (only a partial defect) and the Vps4(L407K) partial defect in CPS maturation. This observation supports the conclusion that L407K contributes to Vps4 recycling of ESCRT-III to enable efficient MVB sorting.
Compensatory Mutation of the VSE Restores Stimulation of Vps4(L407K)
Although these biochemical and in vivo studies support the model that the Vta1 VSE contacts Vps4 α7 and α9 to stimulate ATP hydrolysis, one possibility is that these effects are a secondary consequence of altered small AAA domain conformation(s) rather than directly implicating these residues in mediating Vta1 VSE-Vps4 interactions. To address this possibility, we examined whether disrupted activity could be restored by compensatory mutations in Vta1. Characterization of the Vta1 VSE involved mutating hydrophobic residues to acidic residues (L284E, I287E, and M288E) (53), whereas Vps4 Leu-407 was mutated to a basic residue. The ability of the Vta1 L284E, I287E, and M288E triple mutant (Vta1(VSEΔ)) to compensate for the Vps4 L407K mutation was examined (Fig. 7). Vta1(VSEΔ) stimulated WT Vps4 less effectively than WT Vta1 (122 versus 177 ADP/Vps4/min; p value < 0.01), consistent with previous analysis (53). By contrast, Vta1(VSEΔ) stimulated Vps4(L407K) more effectively than WT Vta1 or VSL (149 versus 80 and 78 ADP/Vps4/min; p values < 0.05). These observations indicated that mutation of the Vta1 VSE residues to acidic residues (L284E, I287E, and M288E) compensates for mutation of Vps4 Leu-407 to a basic residue (L407K) to restore Vta1 VSE stimulation of Vps4 via the small AAA domain. This result supports the model that the Vta1 VSE contacts the proximal (α9: Leu-407 and Lys-411) and distal (α7: Met-330) helices of the small AAA domain to enhance Vps4 ATP hydrolysis.
FIGURE 7.
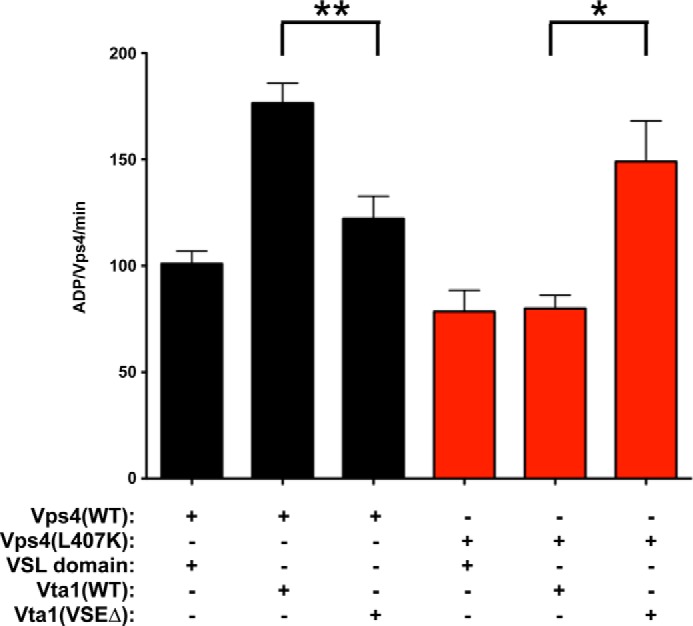
Compensatory mutation of the VSE restores activation of Vps4(L407K). ATPase activity of 250 nm WT Vps4 (black) and Vps4(L407K) (red) in the presence of 8 μm VSL domain, WT Vta1, and Vta1(VSEΔ). The rates are indicated as ADP generated/Vps4/min. The activity of WT Vps4 in the presence of Vta1(VSEΔ) is reduced compared with activity with Vps4 and WT Vta1 (p value < 0.01; **). By contrast, the activity of Vps4(L407K) in the presence of Vta1(VSEΔ) is increased compared with activity with Vps4(L407K) with WT Vta1 (p value < 0.05; *).
DISCUSSION
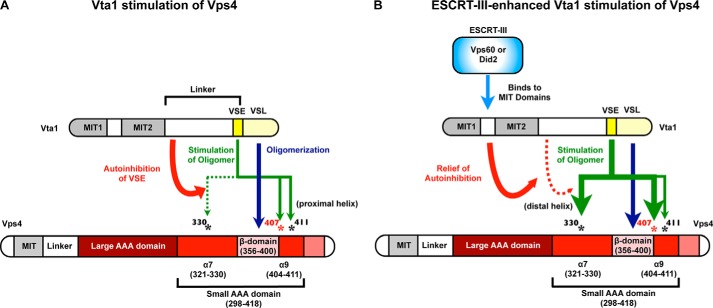
Vta1 promotes Vps4 function to recycle ESCRT-III and permit efficient MVB sorting (36–41). Vta1 stimulation of Vps4 occurs through both the Vta1 VSL domain and the neighboring VSE (31, 53). The VSL domain binds to the Vps4 β-domain to promote oligomerization and stimulate ATP hydrolysis (39, 40, 41, 43, 55). An additional level of Vps4 stimulation is mediated by the VSE, and the VSE is also required for the enhanced stimulation that occurs when ESCRT-III subunits bind to the amino-terminal MIT domains of Vta1 (53). Our results indicate that the Vta1 VSE contacts surface residues in α7 and α9 of the small AAA domain to stimulate Vps4, with Leu-407 playing a key role in this stimulation (Fig. 8). This analysis identifies a novel mechanism by which the AAA domain is regulated to promote Vps4 function.
FIGURE 8.
Vta1 stimulates Vps4 via the β-domain and the small AAA domain α7 and α9. A, the Vta1 VSL domain stimulates Vps4 ATPase activity via the Vps4 β-domain. In the absence of ESCRT-III binding, the VSE contributes to stimulation of Vps4 ATPase activity by contacting the Vps4 α9 residues Leu-407 and Lys-411. However, Vta1 linker region autoinhibition of the VSE curtails the extent of stimulation in the absence of ESCRT-III such that contact with Vps4 α7 (Met-330) is not required. B, ESCRT-III binding Vta1 relieves autoinhibition of the VSE to facilitate contact with Vps4 α7 residue Met-330 to further enhance Vps4 ATPase activity. Contact with α9 also contributes to this ESCRT-III-enhanced stimulation but may not be required. Leu-407 of α9 is key for activation via both α7 and α9 and is conserved from yeast to humans.
Biochemical studies identified the Vta1 VSL domain as necessary to stimulate Vps4 (40). The VSL domain mediates dimerization of Vta1, and the structure of the Vta1 carboxyl terminus (residues 281–330) identified that residues implicated in contacting the Vps4 β-domain decorated two surfaces of the VSL dimer (40, 43). These observations suggested that the VSL domain might stimulate Vps4 by contacting two β-domains from non-neighboring Vps4 subunits within the oligomer to promote oligomer formation. This model linked the importance of VSL dimerization in Vta1 function with the role of the Vta1 VSL domain in promoting Vps4 oligomerization (via the β-domain). However, a structure of the Vta1 VSL domain in complex with the small AAA domain of Vps4 suggested that the orientation of the VSL dimer to the β-domain would not permit additional contacts to other Vps4 subunits within the same oligomer (55). Instead, the VSL dimer was proposed to bridge β-domains across Vps4 oligomers to promote Vps4 function. Another alternative is that VSL dimer contact with a single β-domain facilitates Vps4 oligomerization through subtly altering conformation of the Vps4 monomer (or dimer) rather than inter- or intraoligomeric bridging of Vps4 subunits. Regardless of which mechanism is correct, these models failed to explain how Vta1 could increase the ATPase activity of the Vps4 oligomer above its apparent maximal specific activity.
Further examination of Vta1 stimulation identified an additional element (VSE) that promoted the hydrolysis above the Vps4 inherent maximal rate (maximal specific activity observed with Vps4 alone) (53). The VSE is present in the structure of the Vta1 carboxyl terminus (43) but is not included in the complex of VSL with the Vps4 β- and small AAA domains (55). These structures were combined with the structure of Vps4 AAA domain (56) to generate a model suggesting how the VSE stimulates Vps4 (Fig. 1B). The Vta1 VSE is positioned across from two helices (α7, α9) of the small AAA domain, with α9 proximal to the β-domain. Mutating residues of this proximal helix (L407K and K411A) perturbed stimulation by full-length Vta1 with only stimulation attributed to the VSL domain apparent. These observations suggested that basal Vta1 stimulation occurs through the VSE contacting α9 to stimulate Vps4 ATP hydrolysis beyond the inherent maximal rate (Fig. 8A). ESCRT-III subunits bind to the Vta1 amino-terminal MIT domains to potentiate stimulation via the VSE, and a truncation of Vta1 (residues 275–330) mimics this ESCRT-III activated form (53). Mutation of Vps4 Met-330 disrupted this ESCRT-III-activated VSE stimulation, although basal VSE stimulation was unaffected. These observations suggested that ESCRT-III activation promotes VSE contact with α7 to enhance Vps4 ATP hydrolysis (Fig. 8B). The result that Vps4(K411A) could be stimulated by Vta1(275–330) above the activities observed with full-length Vta1 or the Vta1 VSL domain suggests that VSE stimulation via α7 is not dependent upon stimulation via α9. However, Vps4(L407K) is resistant to VSE stimulation by both full-length Vta1 and Vta1(275–330) (Fig. 3). Although Vps4(L407K) was not stimulated by WT Vta1, mutation of the Vta1 VSE residues from hydrophobic to acidic residues (i.e. Vta1(VSEΔ)) compensated for the mutation of Vps4 Leu-407 to a basic residue. This restoration of stimulation supports the model in which the VSE residues are located in proximity to α7 and α9 to stimulate Vps4 ATPase activity, as illustrated in Fig. 1B. The ATP binding pocket is located in the cleft between the small and large AAA domains, suggesting that perturbations of α7 and α9 are transmitted to the pocket to alter the kinetics of nucleotide binding or the kinetics of nucleophilic attack of the ATP γ-phosphate. This mechanism of contacting α7 and α9 of the small AAA domain to activate Vps4 represents a novel means to enhance AAA-ATPase activity. Further structural studies will be required to resolve the specifics of how contacting α7 and α9 accelerate nucleotide hydrolysis and/or exchange.
The ability of Vta1(VSEΔ) to stimulate Vps4(L407K) highlights the importance of Leu-407 in mediating VSE activation of Vps4. This idea is consistent with the conservation of Leu-407 from Saccharomyces cerevisiae to Drosophila melanogaster, Mus musculus, and Homo sapiens (Vps4A and Vps4B), whereas Met-330 and Lys-411 are not similarly conserved. The importance of Leu-407 is also highlighted by characterization of yeast expressing Vps4(L407K). Expression of Vps4(L407K) in vps4Δ yeast resulted in partial defects in MVB sorting and ESCRT-III disassembly. These phenotypes are similar to the partial defects observed upon mutation of the Vta1 VSE or deletion of the Vta1 MIT domains (31, 53). This similarity is consistent with Leu-407 mediating Vta1 VSE stimulation to contribute to Vps4 function in vivo. These phenotypes are also consistent with other aspects of Vps4 function (i.e. inherent ATPase activity, direct ESCRT-III stimulation, Ist1 inhibition) occurring independent of this α7-α9 surface of the small AAA domain. Although ESCRT-III stimulation via Vta1 is mediated by the VSE contacting α7 and α9, direct ESCRT-III stimulation of Vps4 via the Vps4 MIT domain and the Vps4 linker region occur through a distinct mechanism.
In total, our observations support the model in which α7 and α9 of the small AAA domain are contacted by the Vta1 VSE to stimulate Vps4 ATP activity in addition to VSL association with the β-domain. In the absence of ESCRT-III binding by Vta1, autoinhibition within Vta1 limits the extent of VSE activity such that the VSE primarily contacts α9 (the helix proximal to the β-domain) to stimulate ATPase activity. ESCRT-III binding to the Vta1 MIT domains relieves autoinhibition within Vta1 such that the VSE makes additional contact with α7 (the more distal helix) to further enhance ATPase activity to drive ESCRT-III disassembly. This relief of autoinhibition could occur through subtly reorienting the flexible helix on which the VSE resides or by uncovering or reorganizing the amino-terminal portion of the VSE to permit association with α7 of the Vps4 small AAA domain. The model depicted in Fig. 1B likely represents the ESCRT-III-stimulated Vta1 VSE contacts with Vps4 (including interaction with Met-330), whereas the autoinhibited Vta1 VSE fails to contact α7 of Vps4. Formation of the Vta1 dimer is critical to this model because the VSL dimer creates a four-helix bundle with one chain contacting the β-domain, while the second Vta1 chain supplies the VSE to contact the small AAA domain to enhance ATP hydrolysis (Fig. 1). This model implicates VSL dimer formation in: 1) forming one surface critical for β-domain interaction and 2) permitting simultaneous associations with the β-domain and the small AAA domain by distinct chains of the Vta1 dimer to fully stimulate Vps4.
Acknowledgments
We thank Bruce Horazdovsky, Shirley Dean, and Andreas Schroeder for thoughtful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM 73024 (to D. J. K.), F30 Predoctoral Fellowship 1F30DA26762 (to A. P. N.), and F31 Predoctoral Fellowship GM106693 (to J. A. T.). This work was also supported by funds from the Mayo Graduate School (to A. P. N. and J. A. T.), the Fraternal Order of the Eagles Postdoctoral Fellowship (to B. A. D.), and the Lawrence University LU-R1 Fellowship (to M. E. S.).
- ESCRTs
- endosomal sorting complexes required for transport
- MVB
- multivesicular body
- MIT
- microtubule interacting and trafficking
- MIM
- MIT interacting motif
- AAA
- ATPase associated with various activities
- VSL
- Vta1/SBP1/Lip5
- VSE
- Vps4 stimulatory element
- CPS
- carboxypeptidase S.
REFERENCES
- 1. Jimenez A. J., Maiuri P., Lafaurie-Janvore J., Divoux S., Piel M., Perez F. (2014) ESCRT machinery is required for plasma membrane repair. Science 343, 1247136–1247136 [DOI] [PubMed] [Google Scholar]
- 2. Agromayor M., Martin-Serrano J. (2013) Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 23, 433–441 [DOI] [PubMed] [Google Scholar]
- 3. Weissenhorn W., Poudevigne E., Effantin G., Bassereau P. (2013) How to get out: ssRNA enveloped viruses and membrane fission. Curr. Opin. Virol. 3, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanson P. I., Cashikar A. (2012) Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28, 337–362 [DOI] [PubMed] [Google Scholar]
- 5. Votteler J., Sundquist W. I. (2013) Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCullough J., Colf L. A., Sundquist W. I. (2013) Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 8. Stringer D. K., Piper R. C. (2011) A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192, 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MacDonald C., Buchkovich N. J., Stringer D. K., Emr S. D., Piper R. C. (2012) Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 13, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shields S. B., Piper R. C. (2011) How ubiquitin functions with ESCRTs. Traffic 12, 1306–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurley J. H. (2010) The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45, 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adell M. A., Teis D. (2011) Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 585, 3191–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jouvenet N. (2012) Dynamics of ESCRT proteins. Cell. Mol. Life Sci. 69, 4121–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Babst M., Davies B. A., Katzmann D. J. (2011) Regulation of Vps4 during MVB sorting and cytokinesis. Traffic 12, 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wemmer M., Azmi I., West M., Davies B., Katzmann D., Odorizzi G. (2011) Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adell M. A., Vogel G. F., Pakdel M., Müller M., Lindner H., Hess M. W., Teis D. (2014) Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 205, 33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. (2002) Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271–282 [DOI] [PubMed] [Google Scholar]
- 18. Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teis D., Saksena S., Emr S. D. (2008) Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 20. Shim S., Kimpler L. A., Hanson P. I. (2007) Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 21. Buchkovich N. J., Henne W. M., Tang S., Emr S. D. (2013) Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell 27, 201–214 [DOI] [PubMed] [Google Scholar]
- 22. Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wollert T., Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. (2009) Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16, 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao J., Chen X.-W., Davies B. A., Saltiel A. R., Katzmann D. J., Xu Z. (2009) Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20, 3514–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 28. Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 29. Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. (2008) Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 30. Shim S., Merrill S. A., Hanson P. I. (2008) Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. (2008) ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50–61 [DOI] [PubMed] [Google Scholar]
- 32. Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2010) Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Babst M., Sato T. K., Banta L. M., Emr S. D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Babst M., Wendland B., Estepa E. J., Emr S. D. (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davies B. A., Azmi I. F., Payne J., Shestakova A., Horazdovsky B. F., Babst M., Katzmann D. J. (2010) Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol. Biol. Cell 21, 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yeo S. C., Xu L., Ren J., Boulton V. J., Wagle M. D., Liu C., Ren G., Wong P., Zahn R., Sasajala P., Yang H., Piper R. C., Munn A. L. (2003) Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 116, 3957–3970 [DOI] [PubMed] [Google Scholar]
- 37. Shiflett S. L., Ward D. M., Huynh D., Vaughn M. B., Simmons J. C., Kaplan J. (2004) Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 279, 10982–10990 [DOI] [PubMed] [Google Scholar]
- 38. Ward D. M., Vaughn M. B., Shiflett S. L., White P. L., Pollock A. L., Hill J., Schnegelberger R., Sundquist W. I., Kaplan J. (2005) The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280, 10548–10555 [DOI] [PubMed] [Google Scholar]
- 39. Scott A., Chung H.-Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. (2005) Structural and mechanistic studies of VPS4 proteins. EMBO J. 24, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. (2006) Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. (2006) Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. U.S.A. 103, 6202–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ciccarelli F. D., Proukakis C., Patel H., Cross H., Azam S., Patton M. A., Bork P., Crosby A. H. (2003) The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics 81, 437–441 [DOI] [PubMed] [Google Scholar]
- 43. Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. (2008) Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell 14, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takasu H., Jee J. G., Ohno A., Goda N., Fujiwara K., Tochio H., Shirakawa M., Hiroaki H. (2005) Structural characterization of the MIT domain from human Vps4b. Biochem. Biophys. Res. Commun. 334, 460–465 [DOI] [PubMed] [Google Scholar]
- 45. Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. (2005) Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. U.S.A. 102, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Skalicky J. J., Arii J., Wenzel D. M., Stubblefield W. M., Katsuyama A., Uter N. T., Bajorek M., Myszka D. G., Sundquist W. I. (2012) Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J. Biol. Chem. 287, 43910–43926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Z., Vild C., Ju J., Zhang X., Liu J., Shen J., Zhao B., Lan W., Gong F., Liu M., Cao C., Xu Z. (2012) Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA-ATPase regulator Vta1 in the multivesicular body pathway. J. Biol. Chem. 287, 43899–43908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. (2009) Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hill C. P., Babst M. (2012) Structure and function of the membrane deformation AAA ATPase Vps4. Biochim. Biophys. Acta 1823, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Monroe N., Han H., Gonciarz M. D., Eckert D. M., Karren M. A., Whitby F. G., Sundquist W. I., Hill C. P. (2014) The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 426, 510–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Merrill S. A., Hanson P. I. (2010) Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285, 35428–35438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shestakova A., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2013) The linker region plays a regulatory role in assembly and activity of the Vps4 AAA ATPase. J. Biol. Chem. 288, 26810–26819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Norgan A. P., Davies B. A., Azmi I. F., Schroeder A. S., Payne J. A., Lynch G. M., Xu Z., Katzmann D. J. (2013) Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J. Biol. Chem. 288, 26147–26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vajjhala P. R., Wong J. S., To H.-Y., Munn A. L. (2006) The beta domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 273, 2357–2373 [DOI] [PubMed] [Google Scholar]
- 55. Yang D., Hurley J. H. (2010) Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure 18, 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiao J., Xia H., Yoshino-Koh K., Zhou J., Xu Z. (2007) Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 374, 655–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. (2008) Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]