Abstract
Because of great challenges and workload in deleting genes on a large scale, the functions of most genes in pathogenic fungi are still unclear. In this study, we developed a high-throughput gene knockout system using a novel yeast-Escherichia-Agrobacterium shuttle vector, pKO1B, in the rice blast fungus Magnaporthe oryzae. Using this method, we deleted 104 fungal-specific Zn2Cys6 transcription factor (TF) genes in M. oryzae. We then analyzed the phenotypes of these mutants with regard to growth, asexual and infection-related development, pathogenesis, and 9 abiotic stresses. The resulting data provide new insights into how this rice pathogen of global significance regulates important traits in the infection cycle through Zn2Cys6TF genes. A large variation in biological functions of Zn2Cys6TF genes was observed under the conditions tested. Sixty-one of 104 Zn2Cys6 TF genes were found to be required for fungal development. In-depth analysis of TF genes revealed that TF genes involved in pathogenicity frequently tend to function in multiple development stages, and disclosed many highly conserved but unidentified functional TF genes of importance in the fungal kingdom. We further found that the virulence-required TF genes GPF1 and CNF2 have similar regulation mechanisms in the gene expression involved in pathogenicity. These experimental validations clearly demonstrated the value of a high-throughput gene knockout system in understanding the biological functions of genes on a genome scale in fungi, and provided a solid foundation for elucidating the gene expression network that regulates the development and pathogenicity of M. oryzae.
Author Summary
Magnaporthe oryzae is not only the fungus causing the rice blast disease, which leads to 20–30% losses in rice production, but also a primary model pathosystem for understanding host-pathogen interactions. However, there is no high-throughput gene knockout system constructed, and little is known about most of the genes in this fungus. We developed a high-throughput gene knockout system, and using this system, we obtained null mutants of 104 fungal-specific Zn2Cys6 transcription factor (TF) genes by screening 8741 primary transformants in M. oryzae. We analyzed the functions of these TF genes in development, pathogenesis, and stress responses under 9 conditions. We found that 61 Zn2Cys6 TF genes play indispensable and diversified roles in fungal development and pathogenicity. CNF1 is the first reported TF gene that strongly and negatively regulates asexual development in the rice blast fungus, and CCA1, CNF1, CNF2, CONx1, GPF1, GTA1, MoCOD1 and PCF1 are required for pathogenicity. We further found via RNA-seq that GPF1 and CNF2 have similar mechanisms in gene expression regulation related to pathogenicity. The resulting data provide new insights into how Zn2Cys6 TF genes regulate important traits during the infection cycle of this rice blast pathogen.
Introduction
Magnaporthe oryzae is the best-studied phytopathogenic fungus, which was voted first in the top 10 list of fungal plant pathogens by an international community of molecular plant pathologists [1]. The importance of this filamentous ascomycete fungus is not only owing to the fact that the rice blast disease caused by the fungus is the most destructive disease of rice throughout the world, which typically leads to 20–30% losses and even complete loss in grain production during regional epidemics [1], [2], but also to its being a primary model in the study of host–fungal pathogen interactions [3]. The rice blast fungus has a complicated life cycle including hyphal growth, conidiogenesis, conidial germination, appressorium formation and plant infection, which provides substantial biological information in eukaryotic development and pathogenesis. The rice blast fungus is highly amenable to molecular genetic manipulation, and the functions of numerous genes are identified by gene knockout or ectopic insertion [4]–[6].
Transcription factors (TFs) are proteins that bind to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA. In the rice blast fungus, more than 522 putative TF proteins have been identified from 12,991 M. oryzae proteins (www.ftfd.snu.ac.kr; www.broadinstitute.org). Therefore, nearly 4.02% of genes in the genome code for TFs, which makes this family the single largest family of M. oryzae proteins. Recent functional analyses of single or several TF genes revealed their critical biological roles in fungal development, pathogenesis and response to the environment, for instance, in hyphal growth (MNH6, MSTU1, MoCRZ1 and MoSWI6) [7]–[10], conidiogenesis (COM1, CON7, COS1, MNH6, and MoHOX2/HTF1) [7], [11]–[16], conidial germination (TRA1) [17], appressorium formation (MoLDB1, MoSOM1 and MoCDTF1) [18], [19], plant infection (COM1, MNH6, MIG1, MST12/MoHOX8, and MoSfl1) [7], [11], [16], [20]–[22], and response to oxidative stress(MoATF1 and MoAP1) [23], [24] or light (MgWC-1) [25]. However, the biological functions of most TFs have not been revealed, mainly because it is difficult to delete genes on a large scale and because a high-throughput gene knockout method has not been previously established in M. oryzae.
Several loss-of-function techniques, such as homologous recombination [4], insertional mutagenesis [6] and RNA interference [26], have been used to investigate gene functions in fungi. Although insertional mutagenesis and RNA interference have been effectively used to inactivate gene functions, they all have limitations that prevent them from becoming high-throughput gene knockout techniques, including 1) inability to cover the genome and the heavy workload to clone genes in the insertional mutagenesis method [6], and 2) the silencing of potential unintended targets by the RNA interference method [26]. Gene knockout through DNA homologous recombination is a primary technique used to inactivate gene functions. More than 650 putative TF genes were disrupted in Fusarium graminearum using homologous recombination [27]. In that study, the double-joint PCR method [28] was used to generate gene knockout constructs, which were then transformed into fungal protoplasts. In Neurospora crassa, homologous recombination was developed into a high-throughput gene knockout technique [29], in which gene knockout constructs generated by making use of a yeast recombinational cloning method were transformed into conidia by electroporation. Yeast recombinational cloning is a more suitable method than double-joint PCR for high-throughput gene knockout study. However, protoplast transformation is an elaborate, time-consuming and inefficient method, and electroporation of germinating conidia has not been built in the rice blast fungus and many other filamentous fungi. The frequency of homologous recombination after transformation is often quite low in filamentous fungi, typically <10% in M. oryzae [30]. In addition, mutant screening is a tedious and inefficient step in high-throughput gene knockout studies. The implementation of KU70/KU80 (mus-51/mus-52) mutations as gene knockout background strains greatly increases the frequency of homologous recombination in N. crassa, Aspergilli and other filamentous fungi, and reduces the workload in screening [29], [31]–[34]. However, the null mutants obtained from KU70/KU80 deletion background strains need to be complemented with native KU70/KU80 genes before analyzing their mutant phenotypes [29]. These shortcomings limit the application of the above methods in research in fungal functional genomics. Until now, there is still a surge in interest in functional genomics research through the systematic mutagenesis of identified genes sequenced in the genomes of a large number of fungi (http://www.ncbi.nlm.nih.gov/genome/).
To learn the biological functions of TFs at the genome level, we constructed a high-throughput gene knockout method that enables the rapid knockout of large numbers of genes in the rice blast fungus. In this method, gene knockout vectors are built by a yeast recombinational cloning method using a high-throughput way, DNA transformations are performed by Agrobacterium tumefaciens-mediated transformation (ATMT), and the null mutants are identified by the negative (GFP)/positive (a resistant gene) screening system through a novel yeast-Escherichia-Agrobacterium shuttle vector, pKO1B. With this system, we deleted 104 putative fungal-specific Zn2Cys6 TF genes in M. oryzae. The null mutants were then examined for their phenotypes in development, pathogenicity and responses to stress conditions. In particular, many Zn2Cys6 TF genes required for fungal growth, asexual development, conidial germination and appressorium formation, pathogenicity, and response to stress were identified. We further identified the genes regulated by GPF1 and CNF2 via RNA-sequencing (RNA-seq) and found that GPF1 and CNF2 have similar mechanisms in the regulation of gene expression related to fungal pathogenicity. Our findings will provide new insights into the transcriptional regulation of fungal development and pathogenicity, and a way to study the biological functions of genes in fungi at the genome level.
Results
Strategy of the high-throughput gene knockout system via pKO1B vector in M. oryzae
Three major challenges in high-throughput gene knockout of fungi are 1) how to build gene-deletion cassettes quickly, 2) how to transfer DNA into fungal cells easily, and 3) how to identify null mutants from the large number of transformants efficiently. To solve these problems, we developed a high-throughput gene knockout system using a newly designed yeast-Escherichia-Agrobacterium shuttle vector, pKO1B, (Figure 1A). Through this plasmid, three techniques suitable for high-throughput manipulation (yeast recombinational cloning to build gene-deletion cassettes, ATMT to transfer DNA into fungal cells and dual selection system to identify null mutants) were combined into the high-throughput gene knockout system.
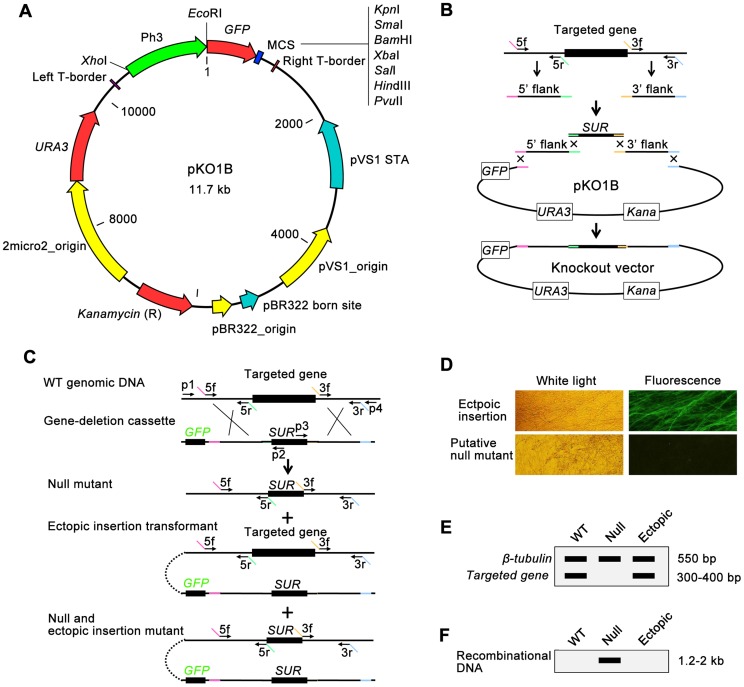
Figure 1. Overview of the high-throughput gene knockout system in fungus.
(A) Features of a new binary yeast-Escherichia-Agrobacterium shuttle vector, pKO1B. (B) Building of gene-deletion cassettes in pKO1B by yeast recombinational cloning. The 5′ and 3′ flanking fragments of the targeted genes were separately amplified from genomic DNA with primers 5f/5r and 3f/3r. Primers 5r and 3f have 5′ tails homologous to the SUR cassette, whereas those for 5f and 3r are homologous to the vector. The two flanks were cotransformed into yeast along with the SUR cassette and gapped pKO1B. Homologous recombination created the circular knockout vector, and the final knockout vector was subsequently transformed into A. tumefaciens. (C) Deletion of the targeted gene. The gene-deletion cassette was transformed into the fungal cells via ATMT. Homologous recombination created three types of transformants: null mutants, ectopic insertion transformants, and null and ectopic insertion mutants. The GFP gene was discarded in the null mutants. Primers p1/p2 or p3/p4 were used to identify the unique recombinational DNA fragment, indicating a knockout event. (D) The transformants are screened for GFP fluorescence under a microscope. Putative null mutants do have not GFP fluorescence, but ectopic transformants do. (E) The transformants are screened by double PCR for the targeted gene using the β-tubulin gene as a positive control. The wild-type strain or ectopic transformants produced a characteristic band, indicating the targeted gene, while the null mutants did not. (F) The transformants are screened by PCR for a unique recombinational DNA fragment marked as a knockout event. The null mutants have a 1.2–2.0 kb band on an electrophoretic gel, while the wild-type strain and the ectopic transformants do not.
pKO1B is a vector built on the framework of the binary vector pCAMBIA1300 (www.cambia.org). It contains the URA3-2micro2_origin sequence from a yeast plasmid pYES2 (Invitrogen, USA) and eGFP gene under the control of a strong promoter of M. oryzae H3 histone gene [35] (Figure 1A). pKO1B has a new characteristic: the ability to be replicated in three organisms, Saccharomyces cerevisiae, Escherichia coli and A. tumefaciens. Through the gapped vector pKO1B, three DNA fragments (5′ and 3′ flanking fragments of the targeted gene and a resistant gene fragment) could be merged into a gene-deletion cassette in one step by yeast recombinational cloning (Figure 1B). The knockout vectors obtained were transformed into A. tumefaciens without transferring the gene-deletion cassettes from a yeast plasmid to another binary Agrobacterium plasmid. The gene-deletion cassettes in pKO1B were transformed into fungal cells using the ATMT method. When the gene-deletion cassettes were ectopically integrated into fungal genomic DNA, GFP was activated in ectopic insertional transformants and was easily observed under a fluorescence microscope as a negative selective marker, but not in null mutants (Figure 1C and 1D).
To screen null mutants efficiently, the genomic DNAs of the transformants without green fluorescence were isolated using an improved CTAB method in a high-throughput way (shown in Materials and Methods) and then were detected for the targeted gene and β-tubulin gene by double PCR. If the targeted gene was deleted in a mutant, only one band for β-tubulin appeared as a positive control on an electrophoretic gel; otherwise, there were two bands with one for the targeted gene and the other for β-tubulin in ectopic insertion transformants (Figure 1C and 1E). For those null mutants identified in the above negative screening PCR, we then continued to search for the unique recombinational DNA fragment that indicated a knockout event and only appeared in the null mutants by PCR. In this PCR, one primer was limited in the genomic DNA outside of the 5′ or 3′ flanking fragment of the targeted gene, and the other primer was limited in the resistant gene of the gene-deletion cassettes (primers p1+p2 or p3+p4 in Figure 1C). There was one band 1.2–2.0 kb in length on the electrophoretic gel that appeared in null mutants. Otherwise, there was no band appearing in the ectopic transformants and wild-type strain (Figure 1F). Gene knockout and ectopic insertion maybe happened coincidentally (Figure 1C). To identify those mutants that were undetected in previous screening steps, copies of transformed gene deletion cassettes in null mutants were confirmed by qPCR after comparison with the wild-type strain using β-tubulin as a control. The null mutants had one copy of the gene deletion cassette, while the null+ectopic transformants had more than two copies and the wild-type strain none. Finally, the mutants containing a single copy of the gene deletion cassette were considered null mutants.
TFs containing fungal-specific Zn2Cys6 zinc finger and fungal_TF_MHR Domain are the largest group of TFs in the rice blast fungus. The 163 putative fungal-specific Zn2Cys6 genes, previously annotated at the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr/index.php) [36] or obtained by BLAST searches at NCBI under organism item (rice blast fungus, taxid: 318829) with the Conserved Domain program [37], were selected to generate gene deletion mutants (Table S1 in Text S1). As a result, 104 TF genes were identified to be deleted in M. oryzae (Table S2 and Table S3 in Text S1). The knockout rate of 104 genes was 15.03±12.02% (from 0.68 to 62.07%). The causes for tens of TF genes not deleted in this experiment lie in the following. 1) Knockout vectors of twenty-one genes had not been constructed. 2) For those genes with a knockout rate less than 1%, more transformants need to be screened. 3) Because the SUR gene was selected as a resistance gene, the mutants must be screened on DCM medium. But the mutants of TF genes that are involved with amino acid metabolism cannot grow on DCM medium. 4) Some genes may be essential genes.
Phenotypic analyses of fungal-specific Zn2Cys6 transcription factor deletion mutants at developmental stages
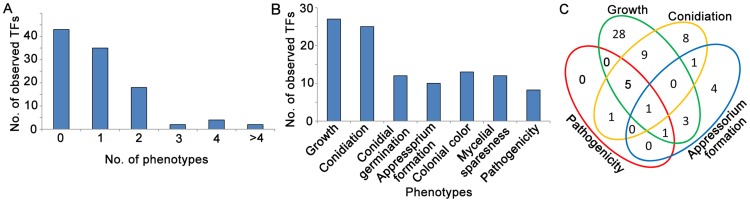
We analyzed the phenotypes of the 104 Zn2Cys6 TF null mutants at multiple developmental stages that M. oryzae likely encounters during its infection cycle in rice. These phenotypes analyzed included: developmental characteristics (mycelial growth, colony color and mycelial shape; conidiation; conidial germination; and appressorium formation) and virulence to plants (barley leaf explants, rice leaf explants and rice seedlings). A substantial fraction of the mutants (58.7%, 61/104) clearly displayed visible phenotypes (Figure 2A). The results (Figure 2B) revealed that 27 of the 104 TF genes studied were involved in mycelial growth (Figure S1), 25 TF genes in conidial production, 12 TF genes in conidial germination and 10 TF genes in appressorium formation, and 5 TF genes were involved in pathogenicity in barley (Figure S2), while 7 TF genes were involved in pathogenicity in rice (Figure S3). Table 1 and Table S4 in Text S1 provide a complete list of the phenotypic analyses. Among these mutants displaying clearly visible mutant phenotype changes, 42.6% (26/61) of mutants exhibited multiple mutant phenotypes and 57.4% (35/61) exhibited a single mutant phenotype (Figure 2A). The Zn2Cys6 TFs regulating asexual reproduction also often controlled vegetative growth (conidial germination, colony growth, pigmentation and mycelial appearance) of the fungus (Figure 2C). Interestingly, 7 TF genes required for pathogenicity had multiple mutant phenotypes (Figure 2C, Table 1), and this phenomenon implied that the fungal pathogenic process is complex, where it is closely linked with many fungal development stages, and that the pathogenicity-related Zn2Cys6 TF genes are also involved in other developmental processes.
Figure 2. Analysis of Zn2Cys6 transcription factor mutant phenotypes in fungal development stages.
(A) Number of TFs showing multiple mutant phenotypes. (B) Number of mutants showing mutant phenotypes in each developmental stage. (C) Venn diagram showing the number of mutant phenotypes. The phenotypes included vegetative growth (conidial germination, colony growth, pigmentation and mycelial appearance), conidiation (asexual reproduction), appressorium formation and pathogenicity to rice and barley.
Table 1. Phenotypic summary of 104 Zn2Cys6 TF gene-deleted mutants.
| Gene locus (MG8) | Gene | Growth | Conidiation | Conidial germination | Appressorium formation | Virulence | Stress |
| MGG_04387 | FZC1 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_01486 | FZC2 | Normal | Normal | Normal | Reduced | Normal | Affected |
| MGG_01518 | MoNIT4 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_01624 | FZC3 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_01734 | FZC4 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_01833 | FZC5 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_02377 | FZC6 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_02595 | FZC7 | Normal | Normal | Normal | Reduced | Normal | Unaffected |
| MGG_02879 | FZC8 | Reduced | Normal | Normal | Normal | Normal | Unaffected |
| MGG_02880 | FZC9 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_03463 | FZC10 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_04108 | TAS1 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_04843 | FZC11 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_05829 | FZC12 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_05891 | FZC13 | Reduced | Reduced | Normal | Normal | Normal | Affected |
| MGG_06355 | FZC14 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_07063 | GCC1 | Reduced | None | Normal | Affected | ||
| MGG_07800 | FZC15 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_08094 | FZC16 | Normal | Normal | Reduced | Normal | Normal | Affected |
| MGG_08130 | FZC17 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_08777 | FZC18 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_10197 | TRA1 | Normal | Normal | Reduced | Normal | Normal | Unaffected |
| MGG_11116 | FZC19 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_12037 | FZC20 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_12424 | FZC21 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_16444 | FZC22 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_16756 | FZC23 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_17060 | FZC24 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_17669 | FZC25 | Normal | Reduced | Normal | Normal | Normal | Unaffected |
| MGG_17821 | FZC26 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_17841 | GPF1 | Reduced | Normal | Reduced | Reduced | None | Affected |
| MGG_08058 | MoACEII | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_00329 | FZC27 | Normal | Normal | Normal | Reduced | Normal | Affected |
| MGG_07681 | FZC28 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_01779 | FZC29 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_01887 | FZC30 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_02866 | FZC31 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_04674 | FZC32 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_05033 | FZC33 | Reduced | Normal | Reduced | Normal | Normal | Affected |
| MGG_05343 | MoCOD1 | Reduced | None | Reduced | Affected | ||
| MGG_06279 | FZC34 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_06312 | FZC35 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_06416 | FZC36 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_06626 | FZC37 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_06778 | FZC38 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_06832 | FZC39 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_06954 | FZC40 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_07149 | GTA1 | Reduced | Reduced | Reduced | Normal | Reduced | Affected |
| MGG_07215 | PIG1 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_07534 | FZC41 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_07549 | FZC42 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_07777 | FZC43 | Normal | Reduced | Reduced | Normal | Normal | Affected |
| MGG_07845 | FZC44 | Increased | Normal | Normal | Normal | Normal | Unaffected |
| MGG_08185 | FZC45 | Increased | Reduced | Normal | Normal | Normal | Affected |
| MGG_08314 | FZC46 | Reduced | Normal | Normal | Normal | Normal | Unaffected |
| MGG_08784 | FZC47 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_14728 | FZC48 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_09027 | FZC49 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_09273 | FZC50 | Reduced | Normal | Reduced | Normal | Normal | Affected |
| MGG_09312 | FZC51 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_09676 | FZC52 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_09829 | FZC53 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_09950 | FZC54 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_11764 | FZC55 | Normal | Normal | Normal | Reduced | Normal | Affected |
| MGG_12776 | FZC56 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_13350 | FZC57 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_13360 | CNF3 | Normal | Increased | Normal | Normal | Normal | Unaffected |
| MGG_13385 | FZC58 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_13629 | FZC59 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_14175 | FZC60 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_14852 | FZC61 | Normal | Reduced | Normal | Normal | Normal | Affected |
| MGG_15023 | CNF2 | Normal | Increased | Normal | Normal | Reduced | Affected |
| MGG_17623 | PCF1 | Normal | Reduced | Reduced | Normal | Reduced | Affected |
| MGG_00021 | FZC62 | Increased | Reduced | Normal | Normal | Normal | Affected |
| MGG_00096 | FZC63 | Increased | Normal | Normal | Normal | Normal | Affected |
| MGG_00320 | FZC64 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_00417 | FZC65 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_00494 | MoPRO1 | Normal | Reduced | Normal | Reduced | Normal | Unaffected |
| MGG_02089 | FZC66 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_02289 | FZC67 | Increased | Normal | Normal | Normal | Normal | Affected |
| MGG_02962 | CNF1 | Normal | Increased | Reduced | Normal | Reduced | Affected |
| MGG_03183 | CNF4 | Normal | Increased | Normal | Normal | Normal | Affected |
| MGG_03669 | FZC68 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_04141 | FZC69 | Reduced | Normal | Normal | Reduced | Normal | Affected |
| MGG_04326 | FZC70 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_04360 | FZC71 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_04933 | FZC72 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_05153 | FZC73 | Reduced | Normal | Normal | Normal | Normal | Affected |
| MGG_05659 | CCA1 | Reduced | Reduced | Reduced | Reduced | Reduced | Affected |
| MGG_05724 | FZC74 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_05845 | FZC75 | Normal | Normal | Normal | Reduced | Normal | Affected |
| MGG_06243 | FZC76 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_06455 | FZC77 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_07458 | FZC78 | Normal | Normal | Normal | Normal | Normal | Unaffected |
| MGG_07636 | FZC79 | Increased | Normal | Normal | Normal | Normal | Affected |
| MGG_08361 | FZC80 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_08974 | FZC81 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_12339 | FZC82 | Normal | Normal | Reduced | Reduced | Normal | Affected |
| MGG_12349 | CONx1 | Normal | Reduced | Reduced | Affected | ||
| MGG_13927 | FZC83 | Increased | Normal | Normal | Normal | Normal | Affected |
| MGG_14816 | FZC84 | Normal | Normal | Reduced | Normal | Normal | Affected |
| MGG_15139 | FZC85 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_17012 | FZC86 | Normal | Normal | Normal | Normal | Normal | Affected |
| MGG_03711 | FZC87 | Reduced | Normal | Normal | Normal | Normal | Affected |
Notes: The phenotypes of the mutants in colony growth, conidiation, conidial germination, appressorium formation, virulence and response to stress were compared with the wild-type strain 70-15. The genes are named as FZC1∼FZC87 (Fungal-specific Zn2 Cys6 transcription factor 1 to 87) if not designated in the succeeding text. The detailed phenotypic information of mutants was shown in Table S4 in Text S1.
We identified the homologs of the 104 M. oryzae Zn2Cys6 TF genes in N. crassa and F. graminearum by Blastp (www.broadinstitute.org), and then compared the phenotypes of the knockout mutants with the homologs studied in N. crassa [29] or F. graminearum [27] (Table S5 in Text S1). Deletion of members of the Zn2Cys6 TF family in F. graminearum and N. crassa resulted in mutant phenotypes at 16% (46/296) and 42% (30/72) of the mutants [27], [29]. However, more mutants (58.7%) in M. oryzae displayed visible mutant phenotypes. But only two mutants showed defects in growth and asexual and/or sexual development in three species simultaneously (Table S5 in Text S1). This comparison of mutant phenotypes between the three fungi suggested that the large majority of Zn2Cys6 TF genes evolved to have a unique, not conserved function in regulating fungal development.
Zn2Cys6 transcription factors involved in fungal growth
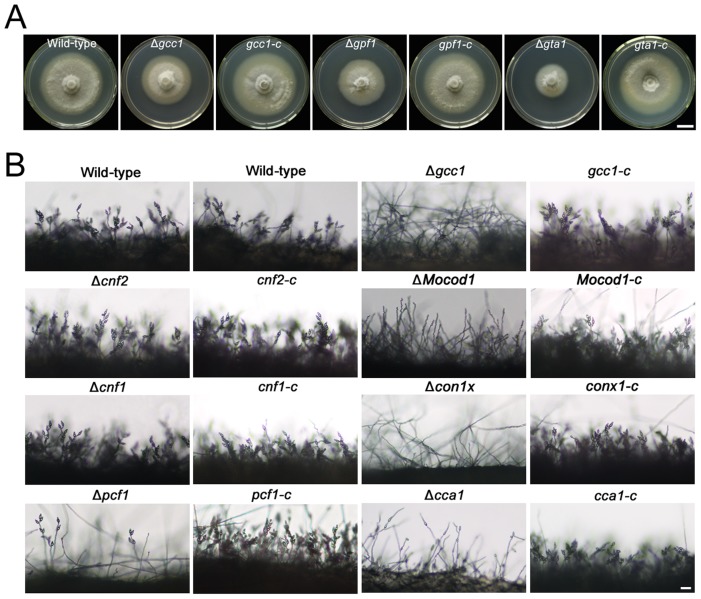
The null mutants of 27 Zn2Cys6 TFs showed significant differences in fungal growth with the wild-type strain (Figure 2B, Table 1). Among these null mutants, three mutants (01C9-1/ΔMGG_07063, 01F1-1/ΔMGG_17841 and 02G4-1/ΔMGG_07149) showed reduced colony growth on CM medium at 24.7, 26.6 and 43.5%, respectively (Table S4 in Text S1). The TF genes deleted in these mutants were designated GCC1 (growth, conidiation and cell wall regulatory factor 1, MGG_07063), GPF1 (growth and pathogenicity regulatory factor 1, MGG_17841) and GTA1 (growth and tolerance to acidic stress regulatory factor 1, MGG_07149), respectively. To confirm the defects of the mutants in growth were caused by the knockout of the TF genes, we complemented the three mutants Δgcc1, Δgpf1 and Δgta1 with their respective native copies from wild-type strain 70-15 (Figure S4C). The phenotypic analyses showed the GCC1-rescued strain (gcc1-c) and GTA1-rescued strain (gta1-c) all recovered from their defects in growth, and GPF1-rescued strain (gpf1-c) also recovered most defects in growth when compared to the mutants and the wild-type strain (Table 2, Figure 3A). These results implied that Zn2Cys6 TFs GCC1, GPF1 and GTA1 were required for the regulation of fungal growth in the rice blast fungus.
Table 2. Developmental characteristics of M. oryzae strains.
| Strain | Growth (mm) | Conidiation (×103/cm2) | Conidial germination (%) | Appressorium formation (%) | Strain | Growth (mm) | Conidiation (×103/cm2) | Conidial germination (%) | Appressorium formation (%) |
| Wild-type | 48.7±0.6A* | 62.5±10.0C | 95.2±1.8A | 95.9±1.6A | |||||
| Δgcc1 | 38.7±0.6C | 0.0±0.0E | gcc1-c | 48.7±0.6A | 61.0±8.3C | ||||
| Δgpf1 | 38.0±0.1C | 76.4±6.4C | 86.5±1.1C | gpf1-c | 47.0±1.0B | 94.0±1.5A | 97.3±0.7A | ||
| Δgta1 | 27.3±0.6D | 84.5±0.5B | 97.8±0.3A | gta1-c | 49.7±0.6A | 92.5±2.6A | 97.8±1.6A | ||
| Δcnf1 | 1229.7±101.1A | 86.7±2.7B | 89.9±3.5B | cnf1-c | 66.4±8.8C | 91.8±2.4A | 96.2±0.3A | ||
| Δcnf2 | 188.9±7.6B | cnf2-c | 71.0±5.2C | ||||||
| ΔMonit4 | 20.2±4.6D | Monit4-c | 59.3±15.6C | ||||||
| Δpcf1 | 28.5±6.1D | 86.4±2.4B | 87.4±2.5B | pcf1-c | 58.6±8.0C | 94.4±0.9A | 97.1±2.1A | ||
| Δcca1 | 7.3±1.7E | 3.6±0.6E | 10.2±0.8E | cca1-c | 65.9±6.4C | 94.4±1.7A | 96.8±0.7A | ||
| Δconx1 | 0.1±0.0E | conx1-c | 65.7±4.4C | ||||||
| ΔMocod1 | 0.0±0.0E | Mocod1-c | 60.8±10.3C | ||||||
| Δfzc16 | 84.6±3.0B | 97.1±1.2A | fzc16-c | 95.1±1.8A | 98.5±0.8A |
Notes: The strains (5-mm mycelial blocks) were grown on CM medium for 8 days, and the diameter of colonies was then measured (growth) or conidia were collected and counted (conidiation). 40-µl (1×105 conidia/ml) conidial suspensions were incubated on plastic slides for 4 hpi (conidial germination assay) and 24 hpi (appressorium formation assay).
*Same capital letters indicate non-significant difference estimated by Duncan's test in each developmental item (P≤0.05).
The experimental strains were the wild-type strain, the mutants (Δcca1, Δconx1, Δcnf1, Δcnf2, Δfzc16, Δgcc1, Δgpf1, Δgta1, ΔMocod1, ΔMonit4 and Δpcf1) and complemented strains (cca1-c, conx1-c, cnf1-c, cnf2-c, fzc16-c, gcc1-c, gpf1-c, gta1-c, Mocod1-c, Monit4-c and pcf1-c).
Figure 3. Colony growth and conidiophores of M. oryzae strains.
(A) Colony growth of the wild-type strain 70-15, mutants (Δgcc1, Δgpf1 and Δgta1), and complemented strains (gcc1-c, gta1-c, gta1-c) on CM medium for 8 days. Bar = 1 cm. (B) Conidiophores of the wild-type, mutants (Δcnf1, Δcnf2, Δpcf1, Δcca1, Δconx1, Δgcc1 and ΔMocod1), and complemented strains (cnf1-c, cnf2-c, pcf1-c, cca1-c, conx1-c, gcc1-c and Mocod1-c). Bar = 20 µm.
Zn2Cys6 transcription factors involved in fungal asexual development
Among the 104 Zn2Cys6 TFs, the null mutants of 25 genes displayed distinct changes in asexual development (Figure 2B, Table 1). Of these mutants, 21 mutants produced less conidia or even no conidia, but the other 4 mutants produced more conidia (Table 1, Table S4 in Text S1). We selected 2 mutants without conidiation, 4 mutants with less conidiation and 2 mutants with more conidiation to be complemented by their native genes to show the relationship between mutant's phenotype and TF gene deleted (Figure S4C). The genes deleted in these null mutants were CNF1 (conidial production negative regulatory factor 1, MGG_02962 in mutant 06B5-7), CNF2 (conidial production negative regulatory factor 2, MGG_15023 in mutant 03C7-2), PCF1 (pathogenicity and conidiation regulatory factor 1, MGG_17623 in mutant 03C11-4), MoNIT4 (a homolog of the nit-4 gene, which is a pathway-specific regulatory gene that mediates nitrate induction in N. crassa [38], MGG_01518 in mutant 01A7-1), CCA1 (conidiation, conidial germination and appressorium formation required transcription factor 1, MGG_05659 in mutant 06C2-14), CONx1 (conidiation required transcription factor x1, MGG_12349 in mutant 06D5-21), GCC1 (MGG_07063 in mutant 01C9-1) and Mocod1 (homolog of amyR from Aspergillus nidulans [3], [39], [40], MGG_05343 in mutant 02F2-6). The mutants Δcnf1 and Δcnf2 produced more conidia than did the wild-type; mutants Δpcf1, Δcca1 and Δconx1 produced less conidia; and mutants Δgcc1 and ΔMocod1 did not produce any conidia. The complementation experiments displayed that the defects of mutants in conidiation were caused by the deletion of the TF genes, since the complemented strains of these mutants all recovered from their abnormal asexual developmental phenomena (Table 2, Figure 3B).
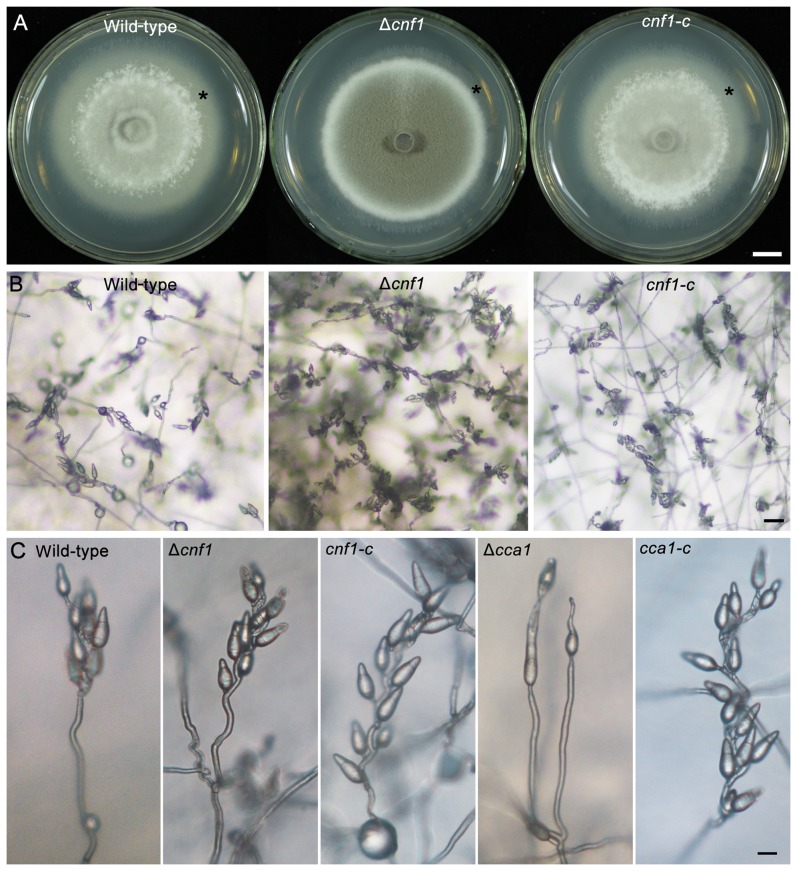
CNF1 is a strong conidial production negative regulatory factor in rice blast fungus, and its deletion may result in greatly increased conidial production (usually 20- to 40-fold of the wild-type strain). Moreover, the mutant Δcnf1 had significant changes in mycelial appearance, such as deep dark colony color from the gray-white of the wild-type strain, short spore-bearing aerial hyphae from long aerial hyphae of the wild-type strain, and spore-bearing aerial hyphae formed at the rim of mycelial colony earlier than in the wild-type strain (Figure 4A). Although the conidiophore development of Δcnf1 seemed similar to that of the wild-type strain and CNF1-rescued mutant cnf1-c (Figure 3B, Figure 4C), more aerial hyphae differentiated into conidiaophores in Δcnf1 (Figure 4B). The increase in conidial production of Δcnf1 was possibly partly due to the spore-bearing hyphae differentiating earlier and more from the aerial hyphae than in the wild-type strain.
Figure 4. Mycelial appearance and spore-bearing aerial hyphae of the M. oryzae strains.
(A) The mycelial appearance of the wild-type strain, Δcnf1 and its complemented strain cnf1-c. Stars indicate non-sporulating hyphae. Bar = 1 cm. (B) Spore-bearing aerial hyphae of the wild-type strain, Δcnf1 and its complemented strain cnf1-c on CM medium. Bar = 20 µm. (C) Conidial development on conidiophores of the M. oryzae strains Δcnf1 and Δcca1. The conidiophore pictures of the wild-type, mutants (Δcnf1 and Δcca1), and their complemented strains (cnf1-c and cca1-c) were taken on the strains grown on CM medium on microscope slides for 1 day. Bar = 40 µm.
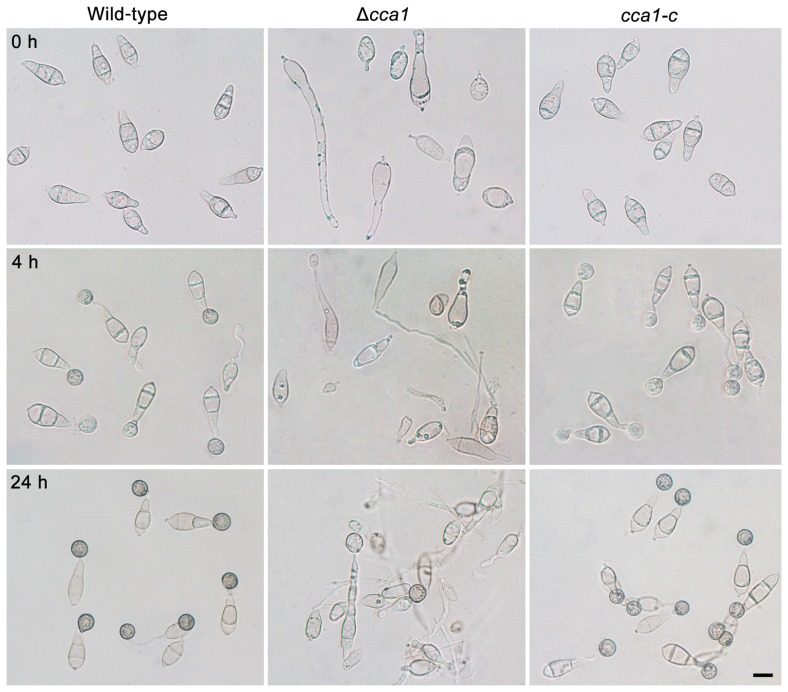
Δcca1 produced few and odd, long and vacuolated conidia (Table 2, Figure 5). Single or two tandem spores differentiated from the apex of conidiophores in the mutant Δcca1, and the CCA1-rescued mutant cca1-c produced normal conidiophores and conidia similar as the wild-type strain (Figure 4C, Figure 5). These results suggested that CCA1 is required for the differentiation of conidiophores and the formation of conidia.
Figure 5. Conidia, conidial germination and appressorium formation of the M. oryzae strain Δcca1.
The conidia (0 h) of the wild-type strain, mutant Δcca1, and its complemented strain cca1-c were grown in H2O on plastic cover slides for 4 h (conidial germination) and 24 h (appressorium formation). Bar = 10 µm.
Zn2Cys6 transcription factors involved in conidial germination and appressorium formation
When the null mutants of 101 Zn2Cys6 TFs were assayed for their conidial germination rate at 4 hpi (hour post inoculation) and their appressorium formation rate at 24 hpi, 12 mutants and 10 mutants displayed significant differences in conidial germination and appressorium formation, respectively (Figure 2B, Table 1). We assayed conidial germination and appressorium formation in the complemented strains of 6 null mutants (Δfzc16, Δgpf1, Δgta1, Δpcf1, Δcnf1 and Δcca1) (Figure S4C) and found that these complemented strains all recovered from their defects in conidial germination or in appressorium formation (Table 2). These experiments implied that Zn2Cys6 TFs GPF1, PCF1, CNF1, and CCA1 are required for conidial germination and appressorium formation and that FZC16 and GTA1 are required for conidial germination in the rice blast fungus.
Among these 6 mutants, the mutant Δcca1 showed marked defects not only in conidial germination and appressorium formation, but also in conidiation (Table 2). The conidial germination rate of the mutant Δcca1 was 3.6%, and appressorium formation rate was 10.2% (Table 2). The structure and shape of conidium, germinated conidium and appressorium of the mutant Δcca1 displayed notable differences with the wild-type strain (Figure 5). Therefore, the mutant Δcca1 produced few multifarious and vacuolated conidia, which germinated and formed not fully melaninized appressoria at a low ratio.
Zn2Cys6 transcription factors required for virulence
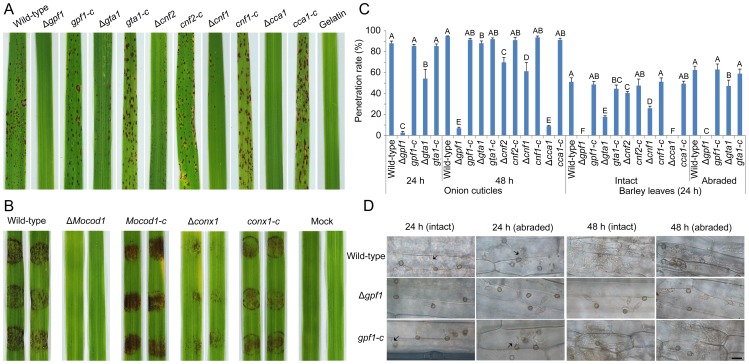
The pathogenicity of 104 null mutants was tested first on intact barley leaves by placing agar plugs containing mycelia on them. Mutant Δgpf1 (MGG_17841) was found to lose pathogenicity to barley, and 4 mutants Δcnf1 (MGG_02962), Δcnf2 (MGG_15023), ΔMocod1 (MGG_05343) and Δpcf1 (MGG_17623) showed reduced pathogenicity (Table S4 in Text S1, Figure S2). We then determined the pathogenicity of 101 mutants on rice seedlings (CO-39) by spraying with conidial suspension (1×105 spores/ml). The results showed that it was decreased in 3 mutants Δcnf1 (MGG_02962), Δcnf2 (MGG_15023) and Δgta1 (MGG_07149) and that 2 mutants Δgpf1 (MGG_17841) and Δcca1 (MGG_05659) lost their virulence to rice (Table S4 in Text S1, Figure S3A). Impressively, Δcca1 caused lesions on barley leaves when inoculated with mycelial plugs, but lost virulence on rice seedlings when spayed as a conidial suspension. The other 3 mutants Δconx1 (MGG_12349), Δgcc1 (MGG_07063) and ΔMocod1 (MGG_05343) with little or no conidiation were tested on intact rice leaves with agar plugs containing mycelia. Δconx1 and ΔMocod1 were found to have lower virulence (Table S4 in Text S1, Figure S3B).
After complemented with native genes (Figure S4C), we retested the pathogenicity of the 7 mutants with their complemented strains on rice seedlings by spraying with conidial suspensions (Δgpf1, Δcnf1, Δcnf2, Δgta1, Δcca1 and gpf1-c, cnf1-c, cnf2-c, gta1-c, and cca1-c) (Figure 6A) or on rice leaf explants by inoculating with mycelial plugs (ΔMocod1, Δconx1 and Mocod1-c, conx1-c) (Figure 6B). The facts that the complemented strains recovered from their defects in virulence on rice implied that the loss or weakening of virulence in the mutants was caused by the deletion of the TF genes GPF1, CNF1, CNF2, GTA1, CCA1, CONx1 and MoCOD1, respectively. To assay the roles of these TF genes in plant penetration, onion cuticle and barley leaves were inoculated with a conidial suspension of the mutants and their complemented strains. Compared with the wild-type strain, the ability of Δgpf1, Δgta1, Δcnf2, Δcnf1 and Δcca1 to penetrate the onion cuticle and barley leaf cuticle was weakened to some extent (Figure 6C). Notably, the mutants Δgpf1 and Δcca1 were unable to penetrate barley cuticle, even after barley cuticle was abraded (for Δgpf1), while the wild-type strain penetrated into the epidermal cells of barley leaves and grew invasively after 24 hpi (Figure 6C, Figure 6D). These results suggested that GPF1 is required by the rice blast fungus for penetration into the plant cuticle and possibly for invasive growth. Although the conidia of Δcca1 were unable to penetrate barley cuticle and could not cause lesions in rice, the mycelia of Δcca1 could cause lesions in barley and rice. The difference in virulence between conidia and mycelia may lie in the conidial defects of Δcca1 in germination, appressorium formation and host penetration ability.
Figure 6. Pathogenicity assay of the M. oryzae strains.
(A) Pathogenicity assay of 5 mutants and their complementation strains on rice. These 5 mutants are Δgpf1, Δgta1, Δcnf2, Δcnf1, Δcca1 and their complementation strains gpf1-c, gta1-c, cnf2-c, cnf1-c, and cca1-c. The rice seedlings were sprayed with a conidial suspension of M. oryzae strains and cultured for 7 days. (B) Pathogenicity assay of the mutants on rice leaf explants. The mycelial agar plugs of the mutants ΔMocod1 and Δconx1, their complementation strains Mocod1-c and conx1-c, and the wild-type strain 70-15 were placed on intact rice leaves for 4 days. (C) Penetration assay. 10-or 20-µl (5×104 conidia/ml) conidial suspensions were inoculated on onion cuticle or barley leaves (intact or abraded) and incubated for 24 or 48 h. The experimental strains were the wild-type strain 70-15, mutants (Δgpf1, Δgta1, Δcnf2, Δcnf1 and Δcca1) and complemented strains (gpf1-c, gta1-c, cnf2-c, cnf1-c and cca1-c). Same capital letters in same treatment item indicate non-significant difference estimated by Duncan's test (P≤0.05). (D) Penetration of Δgpf1. 20 µl (5×104 conidia/ml) conidial suspensions of Δgpf1, complemented strain gpf1-c or wild-type strain were inoculated on barley leaf explants (intact or abraded) and incubated for 24 or 48 h. Arrows indicate the in planta hyphae invaded. Bar = 25 µm.
Phenotypic analyses of Zn2Cys6 transcription factor deletion mutants under stress conditions
There are various stress conditions encountered by M. oryzae when it completes its infection cycle in rice. It is possible to predict the biological functions of these deleted Zn2Cys6 TF genes in cell activity regulation by assaying the resistance ability to stress conditions of the mutants. We screened and assayed in a rapid way the mycelial growth of the mutants under 9 abiotic stress conditions (0.5 mM H2O2, 0.005% SDS, 0.3 M CaCl2, 0.5 M NaCl, 1 M sorbitol, olive oil as a single organic carbon source, pH 5.0 and pH 9.0, and minimal medium). Four mycelial inoculation blocks were placed in a 9-cm plate by needle point inoculation and the diameter of mycelial colonies were measured after 6 days culture. The results displayed that many mutants had significant visible phenotypes under those stress conditions when compared with the wild-type strain (Table S4 in Text S1): 14 mutants showed mutant phenotypes in MM medium (nitrogen metabolism), 16 mutants in CM-C medium containing olive oil (carbon metabolism), 22 mutants in CM medium with SDS (cell wall stress), 17 mutants or 19 mutants in CM medium with high concentration of Ca2+ or Na+ (ionic regulation), 18 mutants in CM medium with 1 M sorbitol (hypertonic pressure), 30 mutants in CM medium with 0.5 mM H2O2 (oxidative stress), and 30 mutants or 14 mutants in CM medium at pH 5.0 or pH 9.0 (ambient pH regulation).
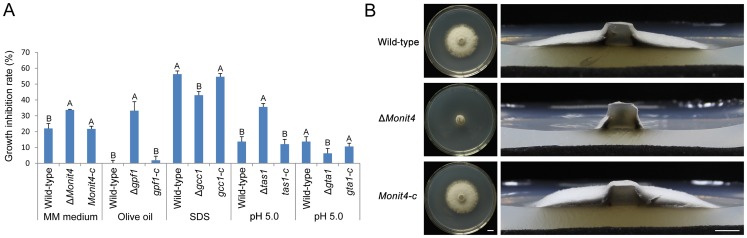
After comparison of the relative growth rate of mutants on CM medium, we selected 5 mutants (01A7-1, 01B11-1, 01C9-1, 01F1-1 and 02G4-1) that had significant changes in mycelial growth under stress conditions to be complemented with native genes to see if these phenotypes were caused by the deletion of TF genes (Figure S4). The TF genes in these 5 mutants were: MoNIT4 in mutant 01A7-1 (MGG_01518), GPF1 in mutant 01F1-1 (MGG_17841), GCC1 in mutant 01C9-1 (MGG_07063), TAS1 (tolerance to acidic stress regulatory factor 1) in mutant 01B11-1 (MGG_04108) and GTA1 in mutant 02G4-1 (MGG_07149). The mycelial growth of ΔMonit4 in minimal medium, Δgpf1 in CM-C medium containing olive oil, and Δtas1 at pH 5.0 was reduced (Figure 7A). However, the mycelial growth of Δgcc1 in CM medium containing SDS and Δgta1 at pH 5.0 was increased. When rescued with native genes, these mutants all recovered their growth under stress conditions (Figure 7A). Interestingly, the mutant ΔMonit4 grew normally as the wild-type strain on CM medium (Table S4 in Text S1, Figure S1), but grew slowly as a very sparse and thin mycelium layer on MM medium (Figure 7B). As previously reported, MoNIT4 is necessary for the regulation of nitrogen metabolism [38], [41]. These data also suggested that TF gene GPF1 is necessary for gene expression regulation relative to fat metabolism, that GCC1 is necessary for gene expression regulation relative to cell wall synthesis, and that TAS1 and GTA1 are necessary for gene expression regulation in response to acidic stress.
Figure 7. Growth of the M. oryzae strains under stress conditions.
(A) Growth inhibition rate. 5-mm mycelial blocks of M. oryzae strains were inoculated on CM medium and medium with stress conditions for 8 days, and the diameter of colonies was then measured to calculate the growth inhibition rate. The experimental strains were the wild-type strain, mutants (ΔMonit4, Δgpf1, Δgcc1, Δtas1 and Δgta1), and complemented strains (Monit4-c, gpf1-c, gcc1-c, tas1-c and gta1-c). Same capital letters in same stress item indicate non-significant difference estimated by Duncan's test (P≤0.05). (B) Mycelial growth of ΔMonit4 on MM medium. The wild-type strain, ΔMonit4, and complemented strain Monit4-c were grown on MM medium at 25°C for 8 days. Bar = 5 mm.
Genes regulated by two virulence-required genes GPF1 and CNF2
Eight Zn2Cys6 TF genes (CCA1, CNF1, CNF2, CONx1, GPF1, GTA1, MoCOD1 and PCF1) are required for fungal pathogenicity to rice or barley. Δgpf1 and Δcnf2 are two mutants that have similar phenotypes on virulence with relatively limited differences in other fields, because ΔMocod1 and Δconx1 do not produce conidia, Δcca1 and Δgta1 have virulence when inoculated on rice leaves with mycelial plugs, Δpcf1 shows reduced virulence on barley inoculated with mycelial plugs, but normal when sprayed on rice, and Δcnf1 produces greatly increased conidia. To understand how these TF genes affect fungal pathogenicity, the two genes GPF1 and CNF2, whose mutants lose (Δgpf1) or have reduced (Δcnf2) virulence to both barley and rice, were selected to analyze their functions in gene expression regulation. Since Δgpf1 could not penetrate rice cuticle, we could not obtain its mRNA during infection. The rice blast fungus experiences temporal nitrogen starvation when it infects the plant, and the gene expression pattern of the mycelia under starvation is similar to that at plant infection [42]–[45], and the hyphae enduring starvation are similar to those inoculated as mycelial plugs in plant infection. We therefore analyzed the genome-wide gene expression of the mycelia after 4 h starvation of the wild-type strain, Δgpf1 and Δcnf2 by RNA-seq. The RNA-seq experiments were performed in biologic triplicate for each strain.
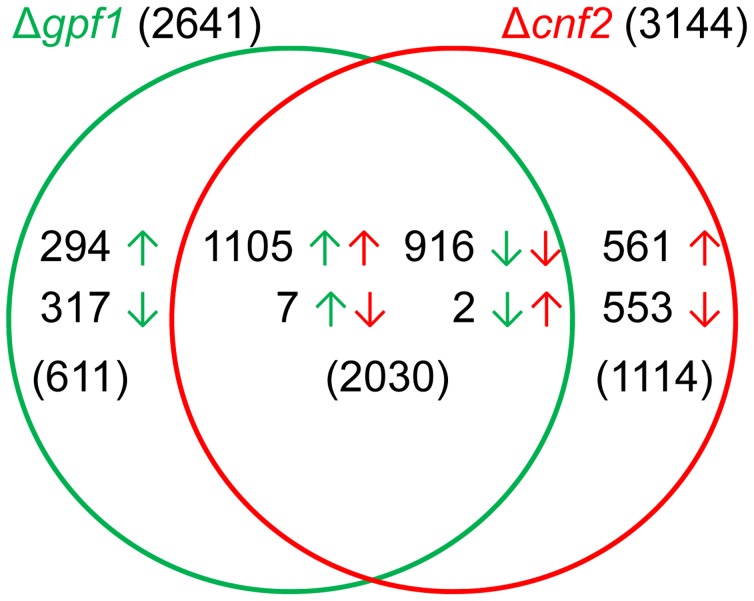
The transcripts of 10,864, 11,048 and 10,681 genes were identified in the starved mycelia of the wild-type strain, Δgpf1 and Δcnf2, respectively. Compared with the wild-type strain, 2641 genes were differentially expressed significantly in Δgpf1 (FDR<0.05), with 1406 genes up-regulated and 1235 down-regulated (Table S6 in Text S1), and 3144 genes in Δcnf2 (FDR<0.05), with 1668 genes up-regulated and 1476 down-regulated (Table S7 in Text S1). The similarities and differences between genes regulated by GPF1 and CNF2 were analyzed in detail by comparing the DEGs in Δgpf1 or Δcnf2. The comparison assays showed that the expression of 611 genes or 1114 genes were independently regulated by GPF1 or CNF2, while 2030 genes were regulated by both GPF1 and CNF2 together (Figure 8). Among the 2030 genes commonly regulated by GPF1 and CNF2, 916 genes were down-regulated and 1105 genes up-regulated simultaneously in both Δgpf1 and Δcnf2, while only 9 genes were regulated in the opposite direction in both mutants (Figure 8, Table S8 in Text S1). It is very surprising that the expression of so many DEGs were regulated in the same direction in Δgpf1 and Δcnf2 (Figure 8). The correlation coefficient between DEGs of Δgpf1 and Δcnf2 was determined by the linear trend model, and the result showed R2 = 0.88 with p<0.0001. These similar patterns were not mainly caused by the starvation, as we also assayed the mutants of five TF genes containing non-Zn2Cys6 domains by RNA-seq with the same treatment at the same time, but the mutants displayed specific DEG patterns different from Δgpf1 and Δcnf2. Therefore, these similar patterns in gene expression regulation in two mutants were possibly caused by the fact that TFs Gpf1 and Cnf2 had the same DNA-binding domain (Zn2Cys6 domain), and Δgpf1 or Δcnf2 had similar phenotypes in pathogenicity. However, GPF1 and CNF2 still showed great differences in gene regulation since 1152 DEGs appeared in Δcnf2 when compared with Δgpf1 (Table S9 in Text S1).
Figure 8. Comparison of differentially expressed genes between Δgpf1 and Δcnf2.
Green or red arrows are the directions of gene expression regulation in Δgpf1 or Δcnf2.
To see how GPF1 and CNF2 affect the pathogenicity to plants, we reviewed the functions of the DEGs which were studied in previous reports. Fifty DEGs in Δgpf1 and 68 DEGs in Δcnf2 (total of 80 DEGs in both were studied by knockout. Of the 80 genes, the mutants of 57 genes (36 DEGs in Δgpf1 and 49 DEGs in Δcnf2) displayed defects in virulence (Table 3) but the mutants of 23 genes (14 DEGs in Δgpf1 and 19 DEGs in Δcnf2) were dispensable for fungal pathogenicity (Table S10 in Text S1). Interestingly, of these reported pathogenicity-required genes, the number of down-regulated genes (45 DEGs) was much higher than that of up-regulated genes (12 DEGs) in the two mutants Δgpf1 and Δcnf2 (Table 3). These pathogenicity-required genes are mainly related to transcription (such as CON7, MoLDB1, MoMIG1, MoPAC2, MSTU1, MST12 and PTH12) [8], [12], [18], [20], [21], [46], [47], G-protein signaling (such as MAGB, MoRGS1 and MGB1) [48]–[51], MAPK pathway (such as MST7, MoMCK1 and MoMPS1) [52]–[54], autophagy (such as MoATG1, MoATG5, MoATG9 and SNX41) [55]–[58], regulation of reactive oxygen species (such as MoHYR1 and GTR1) [59], [60], and amino acid, lipid and carbohydrate metabolism (such as ILV2, MoSNF1, ICL1 and MoPLAA) [61]–[64]. We also checked the relationships between Zn2Cys6 TFs and Gpf1 or Cnf2 in transcription regulation. Surprisingly, 60 or 66 Zn2Cys6 TF genes were listed as DEGS of Δgpf1 or Δcnf2 (total of 79 genes in two mutants) (Table S11 in Text S1). The Zn2Cys6 TF genes are also the important targeted genes regulated by two Zn2Cys6 TF genes GPF1 and CNF2. These facts implied that the well-balanced and sophisticated expression regulation of TF genes and pathogenicity-required genes is necessary for the maintenance of pathogenicity in M. oryzae.
Table 3. GPF1- or CNF2-dependent genes encoding known virulence factors.
| Gene locus (MG8) | Gene | Δgpf1 a | Δcnf2 a | Virulence on plants/Function | Reference |
| MGG_00365 | MAGB | - | −1.58 | Reduced virulence/G-protein signaling | [48] |
| MGG_00466 | MoCDC42 | −1.52 | −2.00 | Reduced virulence/CDC42 | [74] |
| MGG_00501 | TGD2 | −1.67 | −2.68 | Reduced virulence/Transcription factor | [15], [72], [83] |
| MGG_00692 | MSTU1 | −7.80 | −11.85 | Reduced virulence/Transcription factor | [8] |
| MGG_00800 | MST7 | −2.00 | −1.67 | No virulence/MAPK pathway | [52] |
| MGG_00803 | MoSNF1 | −1.43 | −1.79 | Reduced virulence/Glucose derepression pathway | [62] |
| MGG_00883 | MoMCK1 | −1.39 | −1.52 | No virulence/MAPK pathway | [53] |
| MGG_01057 | MoLDB1 | −1.63 | −2.18 | Reduced virulence/Transcription factor | [18] |
| MGG_01204 | MoMIG1 | - | −1.95 | No virulence/Transcription factor | [20] |
| MGG_01230 | MoSSADH | −1.54 | - | Reduced virulence/γ-aminobutyric acid degradation | [23] |
| MGG_01282 | 1.46 | 2.26 | No virulence/polyubiquitin | [84] | |
| MGG_01558 | SET3 | - | −1.53 | No virulence/Set3 complex | [85] |
| MGG_01662 | MoAAT | - | −1.56 | Reduced virulence/4-aminobutyrate aminotransferase | [23] |
| MGG_01663 | HOS2 | −1.46 | −1.51 | No virulence/Set3 complex | [85] |
| MGG_02531 | MoVPR | −3.04 | −3.00 | Reduced virulence | [23] |
| MGG_02731 | MoRAC1 | - | −1.45 | Reduced virulence/Small GTPase | [86] |
| MGG_02848 | MC69 | −1.57 | - | Reduced virulence/Secreted proteins | [87] |
| MGG_02897 | Mossk1 | - | −1.81 | Reduced virulence/Response regulator | [79] |
| MGG_03065 | MoVMA11 | −1.53 | - | No virulence/ATPase | [46] |
| MGG_03694 | MoATG6 | −1.39 | - | No virulence/Autophagy | [88] |
| MGG_04050 | MoSEC22 | −2.81 | −3.80 | No virulence/R-Snare | [89] |
| MGG_04895 | ICL1 | −1.72 | −1.59 | Reduced virulence/Glyoxylate cycle | [63] |
| MGG_04943 | MoMPS1 | - | −1.75 | No virulence/MAPK pathway | [54] |
| MGG_05133 | MoCRZ1 | - | −1.42 | Reduced virulence/Ca2+ signaling | [9], [90] |
| MGG_05201 | MGB1 | −1.67 | −1.80 | No virulence/G-protein signaling | [51] |
| MGG_05287 | CON7 | - | −1.93 | No virulence/Transcription factor | [12] |
| MGG_05428 | MoVAM7 | - | −1.89 | No virulence/Snare | [91] |
| MGG_05664 | MoPDEH | - | −1.77 | Reduced virulence/cAMP signaling | [92] |
| MGG_05755 | MoMON1 | 1.38 | - | No virulence/Vacuole and vesicle fusion | [93] |
| MGG_06033 | MoMSB2 | - | −1.78 | Reduced virulence/Signaling | [94] |
| MGG_06249 | MoTCTP | - | 1.43 | Reduced virulence/Mitochondrial protein | [80] |
| MGG_06393 | MoATG1 | −1.36 | −1.51 | No virulence/Autophagy | [55] |
| MGG_06439 | MoTEA4 | −1.88 | −2.26 | Reduced virulence | [95] |
| MGG_06564 | MoPAC2 | - | −1.67 | Reduced virulence/Transcription factor | [96] |
| MGG_06868 | ILV2 | 4.24 | 4.03 | Reduced virulence/Amino acid metabolic pathway | [61] |
| MGG_06971 | MoSFL1 | - | −1.73 | Reduced virulence/Transcription factor | [22] |
| MGG_07460 | MoHYR1 | 1.73 | 2.32 | Reduced virulence/Reactive oxygen species | [59] |
| MGG_08212 | MoATF1 | 1.42 | - | Reduced virulence/Transcription factor | [24] |
| MGG_08850 | MoGTI1 | −1.58 | −2.55 | Reduced virulence/Stress | [96] |
| MGG_08895 | GTR1 | 2.06 | 1.82 | Reduced virulence/Reactive oxygen species | [60] |
| MGG_09125 | MoSHO1 | - | −1.80 | Reduced virulence/Signaling | [94] |
| MGG_09262 | MoATG5 | 7.64 | 8.88 | No virulence/Autophagy | [56] |
| MGG_09559 | MoATG9 | −1.41 | −1.42 | No virulence/Autophagy | [57] |
| MGG_09565 | PMK1 | −1.49 | −1.93 | No virulence/MAPK pathway | [52] |
| MGG_09912 | MoCMK1 | −1.75 | −1.46 | Reduced virulence/Ca2+ signaling | [97] |
| MGG_10315 | MPG1 | −12.49 | −15.83 | Reduced virulence/Hydrophobin | [75] |
| MGG_10323 | MoRHO3 | −1.46 | −1.69 | No virulence/Small GTPase | [98] |
| MGG_11346 | MoCDTF1 | - | −1.72 | No virulence/Transcription factor | [19] |
| MGG_12828 | MoATG15 | 1.48 | - | No virulence/Autophagy | [88] |
| MGG_12832 | SNX41 | −1.36 | - | No virulence/Autophagy | [58] |
| MGG_12865 | PTH12 | 2.97 | 2.09 | No virulence/Transcription factor | [16], [47] |
| MGG_12868 | ECH1 | - | 1.47 | Reduced virulence/Enoyl-CoA hydratase | [99] |
| MGG_12958 | MST12 | - | −1.71 | No virulence/Transcription factor | [21] |
| MGG_13624 | MoABC1 | - | −1.59 | No virulence/ABC transporter | [100] |
| MGG_14014 | MoPLAA | 2.12 | 2.09 | Reduced virulence/Lipid metabolism | [64] |
| MGG_14517 | MoFLBA | - | −1.69 | Reduced virulence/G-protein signaling | [49], [50], [101] |
| MGG_14847 | MST11 | - | −1.33 | No virulence/MAPK pathway | [52] |
Note:
) The expression of genes increased or decreased in Δgpf1 and Δcnf2.
The number means the fold change, and the symbol “−” means no significant change (FDR<0.05) afterwards compared with the wild-type strain.
Discussion
Since the genomes of a large number of fungi have been sequenced and are being sequenced, there is a surge of interest in functional genomics research through the systematic mutagenesis of identified genes. The construction of a genome-wide gene deletion mutant set of fungi based on the homologous recombinational gene knockout procedure is a valuable resource for the analysis of fungal development, pathogenicity, and many aspects of cell biology and biochemistry, such as those done in the budding yeast S. cerevisiae [65], the fission yeast Schizosaccharomyces pombe [66], and the saprobe filamentous fungus N. crassa [29]. However, except for N. crassa, F. graminearum and Aspergilli which are highly efficient in gene deletion experiments, no efficient system has been available to perform high-throughput gene knockout in filamentous fungi. In this study, we present an approach to knockout large numbers of genes that utilizes several methods suitable to high-throughput manipulation. Several time-consuming steps of the gene knockout procedure could be performed in a high-throughput way. In this procedure, the gene-deletion cassettes were built using a yeast homologous recombination method [29] with a yeast-Escherichia-Agrobacterium shuttle vector pKO1B, which is the first reported artificial plasmid that could be replicated in yeast, E. coli and Agrobacterium cells. The gene-deletion cassettes obtained in a binary vector pKO1B could be directly used to transform fungal cells through the ATMT method [67]. This advantage avoids the tedious and inefficient work to transfer the gene-deletion cassettes from a yeast plasmid to another binary Agrobacterium plasmid. The use of GFP fluorescence as a negative marker to eliminate most ectopic insertion transformants reduced the number of transformants one has to identify by PCR or Southern blot. The employment of the negative (for the targeted gene)/positive (for the unique recombinational DNA) identification PCR and qPCR to identify null mutants instead of Southern blot, which is not high throughput, makes the identification of null mutants a high-throughput procedure. The copies of the gene-deletion cassette in mutant genomic DNA quantified by qPCR has been widely performed in animals and plants [68], [69]. The reliability and stability of PCR or qPCR were guaranteed by the extraction of high quality genomic DNA with CTAB [70] performed in a high-throughput way in this study. The result of the gene knockout events (Figure S4A) were also reconfirmed by Southern blot at the DNA level in 10 mutants (Figure S4B) and at the transcript level in 16 mutants (Figure S4C). As a result, two researchers were able to complete one cycle of the gene knockout experiment for 96 genes in a month, including preparing knockout vectors, performing ATMT transformation, and identifying null mutants, which is a major improvement over the individual gene knockout protocol. More importantly, this procedure can be adapted for knocking out genes in other fungi without any modification or after the substitution of organism-specific promoter to drive the GFP reporter. Besides the GFP gene, a second drug resistance gene (HPH/hygromycin B phosphotransferase gene, NEO/neomycin phosphotransferase II gene or BAR/glufosinate resistance gene, etc.) or a herpes simplex virus thymidine kinase (HSVtk) gene, which converts 5-fluoro-2′-deoxyuridine to a toxic compound [71], could be used as an alternative negative selection marker against ectopic transformants. As an alternative vector, pKO1B-HPH (shown in Materials and Methods) is another yeast-Escherichia-Agrobacterium shuttle which could be used in this high-throughput gene knockout system besides pKO1B. To increase knockout efficiency, M. oryzae KU80 null mutants, which are defective for non-homologous end joining (NHEJ) DNA repair [31], could be alternative strains used in targeted gene replacement instead of the wild-type strain.
In this study, we generated 104 fungal-specific Zn2Cys6 TF gene-deleted mutants in M. oryzae by a high-throughput gene knockout procedure and analyzed the phenotypes of individual TF mutants. The deletion of Zn2Cys6 TF genes resulted in phenotype changes in fungal development and pathogenicity in 58.7% mutants compared to the wild-type strain, while 26 mutants were defective in pleiotropic phenotype. Colony growth and asexual reproduction (conidiation) were the two phenotypic categories most observed in mutants, and most mutants defective in conidiation were often defective in vegetative growth (conidial germination, colony growth, pigmentation and mycelial appearance). Seven Zn2Cys6 TF genes functional for pathogenicity in M. oryzae are also especially required for vegetative growth, conidiation or appressorium formation. Comparison of phenotypes between M. oryzae, F. graminearum and N. crassa Zn2Cys6 TF gene orthologs seemed to agree with the previously reported viewpoint that Zn2Cys6 TFs evolved divergently in how to regulate fungal growth and asexual development, rather than keeping the same function in different fungi [27]. These divergences in functions may be due to the differences in the life cycles between the three fungi. N. crassa is an obligate saprophyte which lives on dead organic material and cannot attack a living host, while M. oryzae and F. graminearum are facultative saprophytic plant pathogens that additionally need specialized structures (such as an appressorium) and functions to infect and obtain nutrients from living plants.
Until now, 6 Zn2Cys6 TF genes (MoCOD1, MoCOD2, MoNIT4, PIG1, TRA1 and XLR1) have been identified in the rice blast fungus [17], [39], [41], [72], [73], and 4 of them were also in our Zn2Cys6 TF gene knockout mutant set. The expression of MoNIT4 and MoCOD1 were up-regulated during conidiation [41], and their mutants ΔMonit4 and ΔMocod1 had reduced conidiation [39], [41], and ΔMocod1 also had lower pathogenicity [39]. In our study, the mutants ΔMonit4 and ΔMocod1 were also defective in conidiation or pathogenicity; furthermore, our mutant ΔMocod1 did not produce any conidia on CM medium. Similar to a previous report [74], our ΔTra1 mutant also showed defects in conidial germination. PIG1 was identified to be involved in melanin biosynthesis, but not confirmed by knockout [73]; however, the Δpig1 mutant did not show notable phenotypes in fungal development in our study. These small discrepancies in mutant phenotypes with previously reported data may be due to the differences in wild-type strains and experimental conditions.
TFs Mnh6, Moatf1, Mocrz1 and Mstu1 were reported to regulate hyphal growth in the rice blast fungus, and the colony growth of the mutants Δmnh6, ΔMoatf1, ΔMocrz1 and ΔMstu1 was about 70–90% of the wild-type strain [7]–[9], [24]. We found 27 Zn2Cys6 TF genes involved in fungal growth, and 3 genes GCC1, GPF1 and GTA1 were required for normal colony growth. Interestingly, the colony growth of Δgta1 was only about 56% of that of wild-type strain, and GTA1 is a gene which affects colony growth the most among known TF genes in the rice blast fungus. GTA1 also functions in conidiation and pathogenicity to plants.
The change in asexual reproduction in a mutant is a phenotype that mostly happens when TF genes or other genes are deleted in the rice blast fungus. The TF gene-deleted mutants (Δcom1, Δcon7, Δcos1, Δmnh6, ΔMohox2) showed reduced conidiation [7], [11]–[13], [15], [16]. In our study, 25 Zn2Cys6 TF genes were identified as being involved in fungal conidiation. Of them, CCA1, GCC1, MoCOD1 and CONx1 are necessary for the differentiation of conidiophores. Unexpectedly, the deletion of four Zn2Cys6 TF genes (CNF1, CNF2, CNF3 and CNF4) led to increased conidial production. In particular, Cnf1 is the strongest negative regulatory factor of conidial production identified in the rice blast fungus until now. The deletion of CNF1 led to earlier and more differentiation of spore-bearing hyphae than in the wild-type strain. Impressively, the mutants Δcnf1 and Δcnf2 produced more conidia, but their conidia had reduced virulence to barley and rice. Conidia are the main way to spread the rice blast disease. It is clear that a balance exists between the ability to produce conidia and the pathogenicity to plant in the phytopathogenic fungus.
We identified 2641 genes regulated by GPF1 and 3144 genes by CNF2 through RNA-seq. Interestingly, 2021 genes were regulated in the same direction in Δgpf1 and Δcnf2. This fact suggested GPF1 and CNF2 have similar mechanisms in the regulation of fungal pathogenicity. Nearly sixty DEGs in Δgpf1 and Δcnf2 were confirmed to be required for fungal pathogenicity (Table 3). In particular, the expression of two pathogenicity-required genes, an APSES TF gene MSTU1 [8] and a hydrophobin gene MPG1 [75], were greatly down-regulated in Δgpf1 (7.80- and 12.49- fold, respectively) and Δcnf2 (11.85- and 15.83-fold, respectively). About half of Zn2Cys6 TF genes and many other DNA-binding domain TF genes were regulated by GPF1 and CNF2. These data primarily revealed the gene expression network in the regulation of pathogenicity controlled by GPF1 and CNF2. It is necessary to keep the well-balanced expression of TF genes and pathogenicity-required genes for maintenance of pathogenicity in M. oryzae.
The high-throughput analysis of mutant phenotypes showed that some TF gene mutants shared highly overlapping phenotypes, such as mycelial growth in Δgcc1, Δgpf1 and Δgta1, and pathogenicity in Δgpf1, Δgta1, Δcnf1 and Δcnf2. On the other hand, several mutant phenotypes were shared by one TF gene mutant. Also, it is fascinating to know how to regulate similar phenotypes by different TF genes. In general, every TF gene regulates the expression of many downstream genes and the expression of every gene is regulated by many TF genes, and therefore each mutant phenotype is controlled by the changes in expression of several genes. When two TF genes show similar mutant phenotype, there are at least 3 types of phenotype regulation mechanisms between them. First, TF gene A regulates another TF gene B, and TF gene B continues to regulate a group of downstream genes. Second, TF genes A and B commonly regulate a group of downstream genes. Third, TF genes A and B regulate two different groups of downstream genes independently, but they lead to similar phenotypes. However, two or three of these gene regulation types often occur simultaneously. For example, GPF1 and other TF genes were down-regulated in Δcnf2, while CNF2 and other TF genes were also down-regulated in Δgpf1 (Table S11 in Text S1). A common group of genes (Figure 8) were regulated both by CNF1 and GPF1 which led to similar pathogenicity phenotypes in mutants, while two other different groups of genes (Figure 8) were regulated by CNF1 and GPF1 independently which led to different phenotypes in conidiation in mutants.
In conclusion, this study represents a major advance in a high-throughput gene knockout system suitable for filamentous fungi, provides the functional characterization of 104 Zn2Cys6 TF genes in the rice blast fungus, and reveals gene expression patterns of two virulence-required TF genes GPF1 and CNF2. These studies will help us build more fungal gene-deletion mutant libraries in an economical way and uncover the transcriptional network in fungi and fungal pathogenic mechanisms.
Materials and Methods
Strains
M. oryzae strain 70-15 and its mutants, S. cerevisiae strain FY834, and E. coli strain DH5α and A. tumefaciens strain AGL1 were used in this study.
Building of a yeast-Escherichi-Agrobacterium shuttle vector pKO1B
A 1184-bp promoter fragment of M. oryzae H3 histone gene amplified from pKD5 [35] with primers H3sF and H3SR (Table S1 in Text S1) and 720-bp eGFP CDS fragment amplified from pEGFP (Clontech, USA) with primers KoGFPF and KoGFPR (Table S1 in Text S1) were inserted into the XhoI/EcoR1 sites and the EcoR1/SacI sites of the binary vector pCAMBIA1300 (Cambia, USA) to produce the vector pKO1. pKO1B was then produced by inserting a 2.9-kb URA3-2micro2_origin fragment amplified from pYES2 (Invitrogen, USA) with primers uraf and urar (Table S1 in Text S1) into the SacII site of pKO1 by a yeast recombinational cloning method using S. c. EasyComp Transformation Kit (Invitrogen, USA).
Primer design and construction of gene-deletion cassettes
A list of 2-kb regions on both sides of each ORF (12775 ORF totally) was retrieved from the rice blast fungus database MG8 (www.broadinstitute.org) and saved in Excel type files by a program written by Mr. Tan Cheng. 23-nt primers (Table S1 in Text S1) of each gene-specific flank (1000 bp–1500 bp in length and 1200 bp in optimum length) were designed by the BatchPrimer3 program [76]. For each gene, primers were designed and synthesized with the following common 33-nt 5′ regions:
5f:GCTGTACAAGTAAGAGCTCGGTACCCGGGGATC… (Homologous to pKO1B)
5r:CCGGGAGATGTGGGGCACTGTGGCGTTGGCACA…(Homologous to SUR gene)
3f:TTGATTATTGCACGGGAATTGCATGCTCTCACA… (Homologous to SUR gene)
3r:TTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCA… (Homologous to pKO1B)
The flank fragments were produced from genomic DNA of M. oryzae strain 70-15 using Primer Star or EX Taq (TaKaRa, China) in 96-well PCR plates. The SUR cassette fragment was generated by PCR with primers surf and surr (Table S1 in Text S1) from pBS-SUR [35]. All PCR products were verified by agarose gel electrophoresis analysis and then used in subsequent steps without further purification. Among the 163 TF genes, both flanking fragments were successfully amplified for 142 genes. However, one or sometimes both flanking fragments failed to be amplified for 21 genes even after trying several reaction conditions.
The yeast transformation procedure was conducted following a small-scale yeast transformation protocol in the pYES2 user manual (Invitrogen, USA) in 96-well deep well plates as described by Colot et al [29]. The cocktail mixture was made by adding 1.8 ml competent yeast cells, 100 µl linearized pKO1B by XbaI and HindIII (100 ng/µl), 210 µl PCR production of SUR cassette, 60 µl denatured salmon sperm DNA (Sangon, China) and 2.6 ml DMSO to 20.7 ml freshly prepared 1×LiAc/40%PEG-3350/1×TE solution (100 mM lithium acetate, pH 7.5; 50% PEG-3350; 10 mM Tris-HCl, pH 7.5; 1 mM EDTA). Next, 200 µl of the mixture, followed by 4 µl of 5′ and 3′ flank PCR fragments of the targeted gene, were pipetted into each well of a 96-deep-well plate with a multichannel pipette. The plates were sealed and processed further following the user manual.
After 3 days, the yeast cells cultured in SC-Ura liquid medium were collected and disrupted in Fastprep-24 homogenizer (MP, USA) at 4.0 m/s for 2 min. The plasmids were extracted with TIANprep yeast plasmid DNA kit (Tiangen Biotech, China) following the user manual. Next, the yeast plasmids were transformed into the competent cells of E. coli strain DH5α prepared following the Inoue method for “ultra-competent” cells [77]. Four bacterial colonies on each LB plate were placed in the wells of 96-deep-well plates, with each well containing 1 ml of LB liquid medium with 50 µg/ml kanamycin. The plates were shaken at 220 rpm overnight at 37°C.
The correctness of homologous recombinational cloning for gene-deleted cassettes was confirmed by bacterial double PCR. Common primers were designed to amplify the 5′ and 3′ flanking fragment of each target gene-deleted cassette. Forward primer KO1Bf1 for the 5′ flanking fragment and reverse primer KO1Br2 for the 3′ flanking fragment were located inside pKO1B, and reverse primer SURr1 for the 5′ flanking fragment and forward primer SURf2 for the 3′ flanking fragment were located in SUR cassette (Table S1 in Text S1). The resulting PCR products in knockout plasmids constructed correctly would be 1.0–1.5 kb for 5′ flanking fragment and 1.5–2.0 kb for 3′ flanking fragment. The gene deletion cassettes were further confirmed by sequencing with the primers SURf2 and SURr1. PCR and DNA sequencing results showed a success rate of 100% for the recombinant plasmids (Table S2 in Text S1). We found that it was enough to confirm the recombinant plasmids by PCR and not necessary to confirm by sequencing plasmids. The correctly built knockout plasmids were extracted from the bacterial cultures using the AxyPrep-96 plasmid purification kit (Axygen, China) following the protocol in the user manual.
Agrobacterium tumefaciens-mediated transformation with knockout vectors
The knockout plasmids were transformed into the competent cells of A. tumefaciens strain AGL1 using the freeze/thaw shock transformation method following the procedure described elsewhere [67]. The plasmids in Agrobacterium cell were confirmed by culture PCR with primer set SurP1 and SurP2 (Table S1 in Text S1) for a 368-bp SUR gene fragment.
M. oryzae strain 70-15 was grown on CM medium for 12 days at 25°C under constant fluorescent light, and the conidia were harvested and transformed with the knockout plasmids mediated by A. tumefaciens according to a previously reported procedure [67], whereas performed in groups, usually 24 or 48 genes in a batch each time. The nitrocellulose membrane strips containing the conidia co-cultivated with A. tumefaciens were placed on AIM medium in the dark at 22°C for 2 days and then transferred onto the selection defined complex medium (DCM; 0.17% yeast nitrogen base without amino acids, 0.2% asparagine, 0.1% ammonium nitrate and 1% glucose, pH 6.0 with Na2HPO4) plates containing 100 µg/ml sulfonylurea, 50 µg/ml kanamycin, 400 µg/ml cefotaxime and 60 µg/ml streptomycin. Sulfonylurea-resistant transformants grown on selection medium were individually transferred onto new selection DCM plates using sterile toothpicks. A total of 8741 primary transformants corresponding to 133 Zn2Cys6 TF genes were selected on the basis of sulfonylurea resistance (Table S2 in Text S1). No transformant was obtained for the other nine genes.
Identification of gene-deleted mutants by GFP fluorescence, double PCR and qPCR
After culture for 2 days, a little mycelium of each transformant was picked out and placed onto a glass slide. We usually placed the mycelia of six transformants on a slide in proper order. The green fluorescence emitted by transformants was then observed one by one under a fluorescence microscope. Ectopic transformants emitted green fluorescence and null mutants did not when excited under the fluorescence microscope (Figure 1D). The transformants without green fluorescence were picked out and inoculated on a new selective plate and grown for 3 days. In total, 3191 transformants without green fluorescence were screened from the primary transformants.
Extraction of genomic DNA was performed following the CTAB protocol of Rogers and Bendich [70] with modifications. A small piece (>9 mm2) of mycelium along with some medium in the selective plate was transferred to a 2-ml round-bottom tube, and 400 µl ddH2O were added along with 0.1 g porcelain beads. The mycelial cells were disrupted in a Fastprep-24 homogenizer at 4.0 m/s for 2 min; 400 µl 4×CATB buffer (4% CTAB, 100 mM Trisma base, 20 mM EDTA, 1.4 M NaCl) were then added to each tube and the tubes incubated at 65°C for 30 min. Next, genomic DNA was extracted following the normal CTAB protocol [70]. In total, genomic DNA of the 3191 transformants was extracted with the CTAB method in a high-throughput way.
The gene-deleted mutants from the transformants without green fluorescence were identified by negative screening double PCR. Double PCR was performed using primers CKF and CKR internal to the targeted gene and primers Tbl-gF and Tbl-gR for the β-tubulin gene (Table S1 in Text S1). PCR was performed in 25-µl reaction mixtures in 96-well PCR plates: 0.15 µl primers Tbl-gF and Tbl-gR (20 µM), 0.5 µl primers CKF and CKR (20 µM), 2.5 µl 10×PCR buffer, 0.4 µl dNTP mix (25 µM), 0.3 µl Taq (5 U/µl), 19.5 µl ddH2O and 1 µl genomic DNA. The PCR program was: 94°C 3 min followed by 35 cycles of 94°C for 30 s, 57°C for 30 s and 72°C for 30 s, and a final extension at 72°C for 10 min. The PCR products were detected by 1.0% agarose gel electrophoresis. If the targeted gene was deleted in a transformant, there was only one band for β-tubulin with 554 bp in length, appearing as a positive control; otherwise, in ectopic transformants and the wild-type strain there were two bands with one of 300–400 bp (the targeted gene) and another of 554 bp (β-tubulin) (Figure 1E). After checking 2280 genomic DNA samples, we identified 477 transformants with deletions in a total of 104 TF genes (Table S2 in Text S1, Figure S4A). No null mutants were found for the other 29 genes after screening all DNA samples.
For the transformants identified as null mutants in the negative screening double PCR, a second PCR was performed to verify the gene deletion event. One primer p1 or p4 (Table S1 in Text S1) was limited in the genomic DNA outside the 5′ or 3′ flanking fragment in gene-deletion cassettes, and another primer p2 or p3 (Table S1 in Text S1) was limited in the SUR gene in gene-deletion cassettes. If the targeted gene was deleted, there was a band with 1.2–2.0 kb in length appearing in the gel. Otherwise, there was no band for the ectopic transformant and wild-type strain 70-15 (Figure 1F). A total of 477 transformants of 104 genes identified by the preceding negative screening PCR were also identified as null mutants in this positive screening PCR (Table S2 in Text S1, Figure S4A).
The copies of transformed gene-deletion cassettes in null mutants were identified by qPCR. The concentration of DNA samples of null mutants was standardized to 25 ng/µl by DNA fluorometry. The primers for the genomic DNA of SUR gene are qSurF and qSurR (Table S1 in Text S1). The genomic DNA fragment coding for β-tubulin gene (one copy in the genome) was selected as a control. The primers of β-tubulin gene are qtblF and qtblR (Table S1 in Text S1). PCR mixture of 25 µl was prepared: 12.5 µl 2×RT-PCR buffer (SYBR Green, premix Ex Taq, TaKaRa), 0.5 µl forward primer and 0.5 µl reverse primer (20 µM), 2 µl genomic DNA, and 9.5 µl ddH2O. Real-time PCR was performed in a Mastercycler (Eppendorf, USA) with the following program: 95°C for 2 min, 40 cycles (95°C for 10 s, 60°C for 20 s), and ending with a melting curve step. Each sample was repeated three times. The copies of transformed gene-deletion cassettes in null mutants were calculated by comparing with the data of the β-tubulin gene and wild-type strain. The exogenous gene was inserted into the mutant's genomic DNA in an integral multiple number. If the copy number of the selective marker gene was 1.0±0.2 times the β-tubulin gene in genomic DNA, the mutant was regarded as containing a single insertion of the selective marker gene. As a result, 477 null mutants of 104 genes were identified as single insertion null mutants (Table S2 and Table S3 in Text S1).
The gene deletion events in mutants of ten randomly selected TF genes were reconfirmed by Southern blot (Figure S4B), which was performed according to a previously reported procedure [7].
Complementation of null mutants with native genes
The mutants were complemented with native gene copies of the wild-type strain 70-15. First, the pKO1B-HPH was built by the replacement of Ph3-GFP cassette with a HPH gene from pCB1003 [78] in pKO1B. The copies of the complementation genes were then cloned from the genomic DNA of the wild-type strain with the primers listed in Table S1 in Text S1 and were inserted into the XbaI/HindIII sites of pKO1B-HPH by the yeast recombinational cloning method. The constructed complementation plasmids were transformed into the mutants using the ATMT method, and the transformants were screened on selective medium containing 200 µg/ml hygromycin B. The gene-rescued transformants were identified by RT-PCR at the mRNA level (Figure S4C) with primers specific for the targeted genes (Table S1 in Text S1).
Phenotypic screening analyses of knockout mutants at development stages
The phenotypic screening analyses were performed by testing 20–30 knockout mutants and one control (the wild-type strain) once in a batch according to previously reported protocols [7], [79], [80]. Mutant phenotypes were assayed in triplicate with five replicates for each strain. The values of colony growth, conidiation, conidial germination and appressorium formation of mutants in the different experimental groups were compared after normalization with the wild-type strain 70-15 in the same group.
Colony growth, morphology and conidiation
Mycelial blocks of 5 mm from 9-day-old wild-type strain 70-15 or mutants were inoculated in the center of CM solid medium in 6-cm plates followed by culture at 25°C under constant fluorescent light. For colony growth, the diameter of the mycelial colony was recorded and colony images were captured at 6 days post inoculation (dpi). For conidiation, the whole conidia of a 7-day-old CM-grown culture in 6-cm plates were harvested and counted using a hemacytometer.
Conidial germination and appressorium formation
Conidia were harvested from a 10-day-old colony growing on a CM plate by adding 0.5 ml 25 ppm Tween-20 solution to the surface of the culture. The suspension was pipetted up and down once more to ensure enough conidia for the assay. A 40-µl aliquot of spore suspension was dropped onto a sterilized plastic coverslip, which was incubated in a moist chamber at 25°C for 4 and 24 h under dark conditions. A total of 300–500 conidia were counted for conidial germination at 4 hpi and for appressorium formation at 24 hpi.
Phenotypic screening analyses of knockout mutants under stress conditions
The phenotypes of the mutants were also screened under different stress conditions (Table S4 in Text S1) with 20–30 knockout mutants once in a batch and the wild-type strain in an experiment. Four pinpoint-like mycelia of 9-day-old strains were inoculated in the solid medium of different stress conditions in a 9-cm plate with a space interval between each other and then incubated at 25°C. Colony images were captured and the diameters of the mycelial colonies were recorded at 6 dpi. The growth rates under stress conditions were compared between strains after normalization with the wild-type strain.
The phenotypes of the mutants of 5 TF genes and their complementation strains under stress conditions were reassessed. Mycelial blocks of 5 mm from 9-day-old strains were inoculated in the center of solid medium of different stress conditions in 6-cm plates followed by culture at 25°C under constant fluorescent light, along with the strains grown on CM medium as controls. Colony images were captured and the diameters of the mycelial colonies were recorded at 8 dpi. The experiments were performed in triplicate with five replicates for each strain. Growth inhibition rate of each mutant was calculated as the growth inhibition rate = (colony diameter on CM medium−colony diameter under stress condition)÷colony diameter on CM medium×100.
Pathogenicity tests on barley and rice
The virulence tests on barley (Hordeum vulgare) and rice (Oryza sativa cv CO39) were performed following previously reported protocols [7]. For assays with leaf explants of barley or rice, 5-mm mycelium blocks of mutants (along with the wild-type strain 70-15 and mock) were inoculated on the leaf, followed by incubation in a wet box at 25°C for 4 days. For spraying assay on rice seedlings, 4 ml conidial suspension (1×105 conidia/ml) containing 0.2% (w/v) gelatin were sprayed onto 15–20 rice seedlings between the third and fourth leaf stages using an artist's airbrush. Inoculated plants were placed in a wet box at 25 C for 2 days and then allowed to grow in controlled environment chambers with a photoperiod of 12 h using fluorescent lights for 5 days.
Plant penetration by the wild-type strain or mutant appressoria was assayed on leaf explants of barley or onion cuticles according to previously reported protocols [7]. A droplet of conidia (5×104 conidia/ml) was inoculated on barley leaf cuticles or onion cuticles and incubated at 25°C for 24 or 48 h. The barley leaves were then treated in methanol (overnight, room temperature) to remove chlorophyll, and fixed in alcoholic lactophenol (1 h, 95% alcohol/lactophenol = 2∶1). Appressorium penetration on barley leaf cuticle or onion cuticles was assessed under a microscope.
RNA-sequencing analysis
The wild-type strain, Δgpf1 and Δcnf2 were grown in liquid CM medium at 25°C with shaking at 180 rpm for 2 days. The cultures were then collected and incubated in H2O at 25°C for 4 h. The treatments were repeated in triplicate for each strain. Total RNA was extracted from ground mycelia in liquid nitrogen with the RNeasy Plant Mini Kit (QIAGEN), and mRNA was isolated using AMPure XP beads (Beckman). RNA-seq libraries were constructed using NEBNext RNA sample preparation kit (NEB) in accordance with the standard low-throughput protocol. Samples were sequenced in a 1×100 nt way on an Illumina Hiseq2500 instrument using the TruSeq PE Cluster Kit v3 - cBot - HS (Illumina) and TruSeq SBS Kit v3-HS (Illumina). The clean reads were generated by removing adaptor sequences, tags with>10% “N”, and low quality tags by FastQC (http://www.bioinformatics.babraham.ac.uk), and were mapped to the M. oryzae genome database (MG8) (www.broadinstitute.org) using Tophat software [81]. The data from triple biological replicates were then analyzed using Cufflinks software and resulted in quantified genome-wide transcript levels of genes (expressed in fragments per kilobase of exon model per million mapped fragments – FPKM) [82]. The significant differences in FPKM between different samples were assayed by the Cuffdiff component of the Cufflinks package [82].
List of genes deleted in this study
The functions of 104 Zn2Cys6 TF genes were studied by knockout and phenotypic analysis in this study (Table 1).
Accession numbers
RNA-sequencing data were deposited in NCBI's Gene Expression Omnibus (GEO accession number GSE57146).
Supporting Information
The colonies of M. oryzae strains on CM medium. The mutants of 104 Zn2Cys6 transcription factor genes and the wild-type strain 70-15 were cultured at 25°C for 6 days.
(PDF)
Pathogenicity screening assay of the mutants on barley leaf explants. The mycelial agar plugs of the mutants of 104 Zn2Cys6 transcription factor genes and the wild-type strain 70-15 were placed on intact barley leaves for 4 days.
(PDF)
Pathogenicity screening assay of the mutants of 104 Zn2Cys6 transcription factor genes on rice. (A) The rice seedlings were sprayed with conidial suspension (1×105 spores/ml) of 101 M. oryzae mutants and cultured for 7 days. (B) The mycelial agar plugs of the mutants of 3 TF genes and the wild-type strain were placed on intact rice leaves for 4 days.
(PDF)
Knockout and complementation of Zn2Cys6 transcription factor genes in M. oryzae . (A) Knockout event of null mutants confirmed by PCR. One mutant of each TF gene (which was assayed in mutant phenotype) was selected as a representative to show the PCR identification results. The size of DNA standards are indicated on the right of lanes (M). WT, M. oryzae strain 70-15; a, bands for β-tubulin; b, bands for the targeted genes; c, bands for unique recombinational DNA fragments relative to the target gene-deletion event. (B) Null mutants of ten randomly selected TF genes confirmed by Southern blot. Genomic DNAs were digested with restriction enzymes shown in Figure S4B and separated on 0.7% agarose gels. The DNAs were individually hybridized with the probes (indicated in Figure S4B). Only one band was detected in mutants and its size was different from that in the wild-type strain, indicating that homologous recombination occurred at a single site. (C) Complementation of 16 Zn2Cys6 transcription factor gene-deleted mutants. The mutants were rescued with their native copy of gene in M. oryzae strain 70-15. RT-PCR results were shown after amplification with 35 cycles. RNA was isolated from the mycelia of the wild-type strain, mutants and complemented strains grown on CM medium. a, the targeted genes; b, β-tubulin gene; 1, mutants; 2, complemented strains; and 3, wild-type strain 70-15.
(PDF)
Supplemental tables. Table S1. Primers used in this study. Table S2. Summary of the gene knockout of 163 Zn2Cys6 transcription factor genes in the rice blast fungus. Table S3. List of fungal-specific Zn2Cys6 zinc finger genes being knocked out and identification of copies of SUR gene in the mutants in this study. Table S4. Phenotypic analyses of the null mutants of 104 Zn2Cys6 transcription factor genes. Table S5. Comparison of phenotypes of previously characterized transcription factors between three fungal mutants (M. oryzae, F. graminearum and N. crassa). Table S6. Differentially expressed genes in Δgpf1 when compared with the wild-type strain (FDR<0.05). Table S7. Differentially expressed genes in Δcnf2 when compared with the wild-type strain (FDR<0.05). Table S8. Differentially expressed genes shared by both Δgpf1 and Δcnf2. Table S9. Differentially expressed genes in Δcnf2 when compared with Δgpf1 (FDR<0.05). Table S10. GPF1- or CNF2-dependent genes that are dispensable for pathogenicity reported in previous studies. Table S11. Zn2Cys6 transcription factor genes that were listed as DEGs of Δgpf1 or Δcnf2.
(XLSX)
Acknowledgments
We thank the editorial board and four anonymous reviewers for their constructive comments. We also want to thank Dr. Shuqun Zhang for his worthwhile suggestion in writing the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. And RNA-sequencing data are also available at NCBI's Gene Expression Omnibus (GEO accession number GSE57146).
Funding Statement
This research was supported by the National Basic Research Program of China (Grant No: 2012CB114002) (http://www.973.gov.cn), the Natural Science Foundation of China (Grant No: 31371891) (http://www.nsfc.gov.cn) and the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (http://www.cscse.edu.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, et al. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ou SH (1980) Pathogen variability and host resistance in rice blast disease. Annu Rev Phytopathol 18: 167–187. [Google Scholar]
- 3. Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature 434: 980–986. [DOI] [PubMed] [Google Scholar]
- 4. Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea . Annu Rev Phytopathol 29: 443–467. [DOI] [PubMed] [Google Scholar]
- 5. Talbot NJ (2003) On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea . Annu Rev Microbiol 57: 177–202. [DOI] [PubMed] [Google Scholar]
- 6. Jeon J, Park SY, Chi MH, Choi J, Park J, et al. (2007) Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39: 561–565. [DOI] [PubMed] [Google Scholar]
- 7. Lu JP, Feng XX, Liu XH, Lu Q, Wang HK, et al. (2007) Mnh6, a nonhistone protein, is required for fungal development and pathogenicity of Magnaporthe grisea . Fungal Genet Biol 44: 819–829. [DOI] [PubMed] [Google Scholar]
- 8. Nishimura M, Fukada J, Moriwaki A, Fujikawa T, Ohashi M, et al. (2009) Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea . Biosci Biotechnol Biochem 73: 1779–1786. [DOI] [PubMed] [Google Scholar]
- 9. Choi J, Kim Y, Kim S, Park J, Lee YH (2009) MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae . Fungal Genet Biol 46: 243–254. [DOI] [PubMed] [Google Scholar]
- 10. Qi Z, Wang Q, Dou X, Wang W, Zhao Q, et al. (2012) MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae . Mol Plant Pathol 13: 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang J, Zhao X, Sun J, Kang Z, Ding S, et al. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol Plant Microbe Interact 23: 112–123. [DOI] [PubMed] [Google Scholar]
- 12. Odenbach D, Breth B, Thines E, Weber RW, Anke H, et al. (2007) The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea . Mol Microbiol 64: 293–307. [DOI] [PubMed] [Google Scholar]
- 13. Zhou Z, Li G, Lin C, He C (2009) Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae . Mol Plant Microbe Interact 22: 402–410. [DOI] [PubMed] [Google Scholar]
- 14. Kramer B, Thines E, Foster AJ (2009) MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea . Fungal Genet Biol 46: 667–681. [DOI] [PubMed] [Google Scholar]
- 15. Liu W, Xie S, Zhao X, Chen X, Zheng W, et al. (2010) A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae . Mol Plant Microbe Interact 23: 366–375. [DOI] [PubMed] [Google Scholar]
- 16. Kim S, Park SY, Kim KS, Rho HS, Chi MH, et al. (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae . PLoS Genet 5: e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Battaglia E, Klaubauf S, Vallet J, Ribot C, Lebrun MH, et al. (2013) Xlr1 is involved in the transcriptional control of the pentose catabolic pathway, but not hemi-cellulolytic enzymes in Magnaporthe oryzae . Fungal Genet Biol 57: 76–84. [DOI] [PubMed] [Google Scholar]
- 18. Li Y, Liang S, Yan X, Wang H, Li D, et al. (2010) Characterization of MoLDB1 required for vegetative growth, infection-related morphogenesis, and pathogenicity in the rice blast fungus Magnaporthe oryzae . Mol Plant Microbe Interact 23: 1260–1274. [DOI] [PubMed] [Google Scholar]
- 19. Yan X, Li Y, Yue X, Wang C, Que Y, et al. (2011) Two novel transcriptional regulators are essential for infection-related morphogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathog 7: e1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehrabi R, Ding S, Xu JR (2008) MADS-box transcription factor mig1 is required for infectious growth in Magnaporthe grisea . Eukaryot Cell 7: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park G, Xue C, Zheng L, Lam S, Xu JR (2002) MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea . Mol Plant Microbe Interact 15: 183–192. [DOI] [PubMed] [Google Scholar]
- 22. Li G, Zhou X, Kong L, Wang Y, Zhang H, et al. (2011) MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae . PLoS One 6: e19951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo M, Chen Y, Du Y, Dong Y, Guo W, et al. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS pathogens 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo M, Guo W, Chen Y, Dong S, Zhang X, et al. (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae . Molecular plant-microbe interactions 23: 1053–1068. [DOI] [PubMed] [Google Scholar]
- 25. Lee K, Singh P, Chung WC, Ash J, Kim TS, et al. (2006) Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae . Fungal Genet Biol 43: 694–706. [DOI] [PubMed] [Google Scholar]
- 26. Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, et al. (2008) Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol 68: 1348–1365. [DOI] [PubMed] [Google Scholar]
- 27. Son H, Seo YS, Min K, Park AR, Lee J, et al. (2011) A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. [DOI] [PubMed] [Google Scholar]
- 29. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Talbot NJ, Foster AJ (2001) Genetics and genomics of the rice blast fungus Magnaporthe grisea: developing an experimental model for understanding fungal diseases of cereals. Advances in Botanical Research 34: 263–287. [Google Scholar]
- 31. Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, et al. (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genetics and Biology 45: 68–75. [DOI] [PubMed] [Google Scholar]
- 32. Takahashi T, Masuda T, Koyama Y (2006) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae . Molecular Genetics and Genomics 275: 460–470. [DOI] [PubMed] [Google Scholar]
- 33. Nayak T, Szewczyk E, Oakley CE, Osmani A, et al. (2006) A Versatile and Efficient Gene-Targeting System for Aspergillus nidulans . Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, et al. (2006) The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus . Eukaryot Cell 5: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li HJ, Lu JP, Liu XH, Zhang LL, Lin FC (2012) Vectors building and usage for gene knockout, protein expression and fluorescent fusion protein in the rice blast fungus. Journal of Agricultural Biotechnology 20: 94–104. [Google Scholar]
- 36. Park J, Jang S, Kim S, Kong S, Choi J, et al. (2008) FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. [DOI] [PubMed] [Google Scholar]
- 37. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. (2010) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan GF, Fu YH, Marzluf GA (1991) nit-4, a pathway-specific regulatory gene of Neurospora crassa, encodes a protein with a putative binuclear zinc DNA-binding domain. Mol Cell Biol 11: 5735–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chung H, Choi J, Park S-Y, Jeon J, Lee YH (2013) Two conidiation-related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genetics and Biology 61: 133–141. [DOI] [PubMed] [Google Scholar]
- 40. Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, et al. (2001) Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans . Curr Genet 39: 10–15. [DOI] [PubMed] [Google Scholar]
- 41. Park SY, Choi J, Lim SE, Lee GW, Park J, et al. (2013) Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog 9: e1003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Donofrio NM, Oh Y, Lundy R, Pan H, Brown DE, et al. (2006) Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea . Fungal Genet Biol 43: 605–617. [DOI] [PubMed] [Google Scholar]
- 43. Talbot NJ, McCafferty HRK, Ma M, Moore K, Hamer JE (1997) Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiological and Molecular Plant Pathology 50: 179–195. [Google Scholar]
- 44. Ebbole DJ, Jin Y, Thon M, Pan H, Bhattarai E, et al. (2004) Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: analysis of expressed sequence tags. Mol Plant Microbe Interact 17: 1337–1347. [DOI] [PubMed] [Google Scholar]
- 45. Wang Y, Wu J, Park ZY, Kim SG, Rakwal R, et al. (2011) Comparative secretome investigation of Magnaporthe oryzae proteins responsive to nitrogen starvation. J Proteome Res 10: 3136–3148. [DOI] [PubMed] [Google Scholar]
- 46. Chen GQ, Liu XH, Zhang LL, Cao HJ, Lu JP, et al. (2013) Involvement of MoVMA11, a putative vacuolar atpase c′ subunit, in vacuolar acidification and infection-related morphogenesis of Magnaporthe oryzae . PLoS ONE 8: e67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sweigard JA, Carroll AM, Farrall L, Chumley FG, Valent B (1998) Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol Plant Microbe Interact 11: 404–412. [DOI] [PubMed] [Google Scholar]
- 48. Liu S, Dean RA (1997) G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea . Mol Plant Microbe Interact 10: 1075–1086. [DOI] [PubMed] [Google Scholar]
- 49. Zhang H, Tang W, Liu K, Huang Q, Zhang X, et al. (2011) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae . PLoS Pathog 7: e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramanujam R, Yishi X, Liu H, Naqvi NI (2012) Structure-function analysis of Rgs1 in Magnaporthe oryzae: role of DEP domains in subcellular targeting. PLoS One 7: e41084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishimura M, Park G, Xu JR (2003) The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea . Mol Microbiol 50: 231–243. [DOI] [PubMed] [Google Scholar]
- 52. Zhao X, Kim Y, Park G, Xu JR (2005) A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea . Plant Cell 17: 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeon J, Goh J, Yoo S, Chi MH, Choi J, et al. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae . Mol Plant Microbe Interact 21: 525–534. [DOI] [PubMed] [Google Scholar]
- 54. Xu JR, Staiger CJ, Hamer JE (1998) Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci U S A 95: 12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu XH, Lu JP, Zhang L, Dong B, Min H, et al. (2007) Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryotic Cell 6: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu JP, Liu XH, Feng XX, Min H, Lin FC (2009) An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae . Curr Genet 55: 461–473. [DOI] [PubMed] [Google Scholar]
- 57. Dong B, Liu XH, Lu JP, Zhang FS, Gao HM, et al. (2009) MgAtg9 trafficking in Magnaporthe oryzae . Autophagy 5: 946–953. [DOI] [PubMed] [Google Scholar]
- 58. Deng YZ, Qu Z, He Y, Naqvi NI (2012) Sorting nexin Snx41 is essential for conidiation and mediates glutathione-based antioxidant defense during invasive growth in Magnaporthe oryzae . Autophagy 8: 1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM (2011) HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog 7: e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fernandez J, Wilson RA (2014) Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS One 9: e87300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Du Y, Zhang H, Hong L, Wang J, Zheng X, et al. (2013) Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae . Mol Plant Pathol 14: 870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yi M, Park JH, Ahn JH, Lee YH (2008) MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae . Fungal Genet Biol 45: 1172–1181. [DOI] [PubMed] [Google Scholar]
- 63. Wang ZY, Thornton CR, Kershaw MJ, Debao L, Talbot NJ (2003) The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea . Mol Microbiol 47: 1601–1612. [DOI] [PubMed] [Google Scholar]
- 64. Liu XH, Zhuang FL, Lu JP, Lin FC (2011) Identification and molecular cloning Moplaa gene, a homologue of Homo sapiens PLAA, in Magnaporthe oryzae . Microbiological research 167: 8–13. [DOI] [PubMed] [Google Scholar]
- 65. Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- 66. Kim DU, Hayles J, Kim D, Wood V, Park HO, et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rho HS, Kang S, Lee YH (2001) Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea . Mol Cells 12: 407–411. [PubMed] [Google Scholar]
- 68. Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30: 503–512. [DOI] [PubMed] [Google Scholar]
- 69. Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31: 132–134, 136–140. [DOI] [PubMed] [Google Scholar]
- 70. Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology 5: 69–76. [DOI] [PubMed] [Google Scholar]
- 71. Khang CH, Park SY, Lee YH, Kang S (2005) A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum . Fungal Genet Biol 42: 483–492. [DOI] [PubMed] [Google Scholar]
- 72. Breth B, Odenbach D, Yemelin A, Schlinck N, Schroder M, et al. (2013) The role of the Tra1p transcription factor of Magnaporthe oryzae in spore adhesion and pathogenic development. Fungal Genet Biol 57: 11–22. [DOI] [PubMed] [Google Scholar]
- 73. Tsuji G, Kenmochi Y, Takano Y, Sweigard J, Farrall L, et al. (2000) Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol Microbiol 38: 940–954. [DOI] [PubMed] [Google Scholar]
- 74. Zheng W, Zhao Z, Chen J, Liu W, Ke H, et al. (2009) A Cdc42 ortholog is required for penetration and virulence of Magnaporthe grisea . Fungal Genet Biol 46: 450–460. [DOI] [PubMed] [Google Scholar]
- 75. Talbot NJ, Ebbole DJ, Hamer JE (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell 5: 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. You FM, Huo N, Gu YQ, Luo MC, Ma Y, et al. (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sambrook J, Russell DW (2001) Molecular cloning, a labortary manual (3nd ed). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. [Google Scholar]
- 78. Carroll AM, Sweigard JA, Valent B (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet Newsl 41. [Google Scholar]
- 79. Motoyama T, Ochiai N, Morita M, Iida Y, Usami R, et al. (2008) Involvement of putative response regulator genes of the rice blast fungus Magnaporthe oryzae in osmotic stress response, fungicide action, and pathogenicity. Curr Genet 54: 185–195. [DOI] [PubMed] [Google Scholar]
- 80. Zhang LL, Cao HJ, Li XD, Lin FC, Feng XX, et al. (2013) MoTCTP, a homolog of translationally controlled tumor protein, is required for fungal growth and conidiation in Magnaporthe oryzae . Chinese J Cell Biol 35: 1141–1154. [Google Scholar]
- 81. Trapnell, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang H, Zhao Q, Guo X, Guo M, Qi Z, et al. (2014) Pleiotropic Function of the Putative Zinc-Finger Protein MoMsn2 in Magnaporthe oryzae . Mol Plant Microbe Interact 27: 446–460. [DOI] [PubMed] [Google Scholar]
- 84. Oh Y, Franck WL, Han S-O, Shows A, Gokce E, et al. (2012) Polyubiquitin Is Required for Growth, Development and Pathogenicity in the Rice Blast Fungus Magnaporthe oryzae . PloS one 7: e42868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ding SL, Liu W, Iliuk A, Ribot C, Vallet J, et al. (2010) The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae . Plant Cell 22: 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen J, Zheng W, Zheng S, Zhang D, Sang W, et al. (2008) Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea . PLoS Pathog 4: e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Saitoh H, Fujisawa S, Mitsuoka C, Ito A, Hirabuchi A, et al. (2012) Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog 8: e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kershaw MJ, Talbot NJ (2009) Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proceedings of the National Academy of Sciences 106: 15967–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Song W, Dou X, Qi Z, Wang Q, Zhang X, et al. (2010) R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae . PLoS One 5: e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang H, Zhao Q, Liu K, Zhang Z, Wang Y, et al. (2009) MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett 293: 160–169. [DOI] [PubMed] [Google Scholar]
- 91. Dou X, Wang Q, Qi Z, Song W, Wang W, et al. (2011) MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae . PloS one 6: e16439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ramanujam R, Naqvi NI (2010) PdeH, a high-affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae . PLoS pathogens 6: e1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gao HM, Liu XG, Shi HB, Lu JP, Yang J, et al. (2013) MoMon1 is required for vacuolar assembly, conidiogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae . Res Microbiol 164: 300–309. [DOI] [PubMed] [Google Scholar]
- 94. Liu W, Zhou X, Li G, Li L, Kong L, et al. (2011) Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog 7: e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Patkar RN, Suresh A, Naqvi NI (2010) MoTea4-mediated polarized growth is essential for proper asexual development and pathogenesis in Magnaporthe oryzae . Eukaryotic cell 9: 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen Y, Zhai S, Zhang H, Zuo R, Wang J, et al. (2013) Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae . Environ Microbiol 16: 788–801. [DOI] [PubMed] [Google Scholar]
- 97. Liu XH, Lu JP, Dong B, Gu Y, Lin FC (2010) Disruption of MoCMK1, encoding a putative calcium/calmodulin-dependent kinase, in Magnaporthe oryzae . Microbiological research 165: 402–410. [DOI] [PubMed] [Google Scholar]
- 98. Zheng W, Chen J, Liu W, Zheng S, Zhou J, et al. (2007) A Rho3 homolog is essential for appressorium development and pathogenicity of Magnaporthe grisea . Eukaryotic cell 6: 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Patkar RN, Ramos-Pamplona M, Gupta AP, Fan Y, Naqvi NI (2012) Mitochondrial beta-oxidation regulates organellar integrity and is necessary for conidial germination and invasive growth in Magnaporthe oryzae . Mol Microbiol 86: 1345–1363. [DOI] [PubMed] [Google Scholar]
- 100. Urban M, Bhargava T, Hamer JE (1999) An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J 18: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, et al. (2007) Rgs1 regulates multiple Gα subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. The EMBO journal 26: 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The colonies of M. oryzae strains on CM medium. The mutants of 104 Zn2Cys6 transcription factor genes and the wild-type strain 70-15 were cultured at 25°C for 6 days.
(PDF)
Pathogenicity screening assay of the mutants on barley leaf explants. The mycelial agar plugs of the mutants of 104 Zn2Cys6 transcription factor genes and the wild-type strain 70-15 were placed on intact barley leaves for 4 days.
(PDF)
Pathogenicity screening assay of the mutants of 104 Zn2Cys6 transcription factor genes on rice. (A) The rice seedlings were sprayed with conidial suspension (1×105 spores/ml) of 101 M. oryzae mutants and cultured for 7 days. (B) The mycelial agar plugs of the mutants of 3 TF genes and the wild-type strain were placed on intact rice leaves for 4 days.
(PDF)
Knockout and complementation of Zn2Cys6 transcription factor genes in M. oryzae . (A) Knockout event of null mutants confirmed by PCR. One mutant of each TF gene (which was assayed in mutant phenotype) was selected as a representative to show the PCR identification results. The size of DNA standards are indicated on the right of lanes (M). WT, M. oryzae strain 70-15; a, bands for β-tubulin; b, bands for the targeted genes; c, bands for unique recombinational DNA fragments relative to the target gene-deletion event. (B) Null mutants of ten randomly selected TF genes confirmed by Southern blot. Genomic DNAs were digested with restriction enzymes shown in Figure S4B and separated on 0.7% agarose gels. The DNAs were individually hybridized with the probes (indicated in Figure S4B). Only one band was detected in mutants and its size was different from that in the wild-type strain, indicating that homologous recombination occurred at a single site. (C) Complementation of 16 Zn2Cys6 transcription factor gene-deleted mutants. The mutants were rescued with their native copy of gene in M. oryzae strain 70-15. RT-PCR results were shown after amplification with 35 cycles. RNA was isolated from the mycelia of the wild-type strain, mutants and complemented strains grown on CM medium. a, the targeted genes; b, β-tubulin gene; 1, mutants; 2, complemented strains; and 3, wild-type strain 70-15.
(PDF)
Supplemental tables. Table S1. Primers used in this study. Table S2. Summary of the gene knockout of 163 Zn2Cys6 transcription factor genes in the rice blast fungus. Table S3. List of fungal-specific Zn2Cys6 zinc finger genes being knocked out and identification of copies of SUR gene in the mutants in this study. Table S4. Phenotypic analyses of the null mutants of 104 Zn2Cys6 transcription factor genes. Table S5. Comparison of phenotypes of previously characterized transcription factors between three fungal mutants (M. oryzae, F. graminearum and N. crassa). Table S6. Differentially expressed genes in Δgpf1 when compared with the wild-type strain (FDR<0.05). Table S7. Differentially expressed genes in Δcnf2 when compared with the wild-type strain (FDR<0.05). Table S8. Differentially expressed genes shared by both Δgpf1 and Δcnf2. Table S9. Differentially expressed genes in Δcnf2 when compared with Δgpf1 (FDR<0.05). Table S10. GPF1- or CNF2-dependent genes that are dispensable for pathogenicity reported in previous studies. Table S11. Zn2Cys6 transcription factor genes that were listed as DEGs of Δgpf1 or Δcnf2.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. And RNA-sequencing data are also available at NCBI's Gene Expression Omnibus (GEO accession number GSE57146).