Abstract
Glutathione S-transferase Omega (GSTO) plays an important role in the development of cancer. Recently, a number of studies have investigated the association between single nucleotide polymorphisms on GSTO and susceptibility to cancer; however, the results remain inconclusive. We performed a meta-analysis of 20 studies, involving 4770 cases and 5701 controls to identify the strength of association by pooled odds ratios (ORs) with corresponding 95% confidence intervals (CIs). Overall, the pooled results revealed a significantly increased risk of susceptibility for GSTO2 polymorphism (GG vs. AA: OR = 1.20, 95%CI: 1.02–1.41, Pheterogeneity = 0.116), but no significant association was found for GSTO1 polymorphism. Subgroup analysis showed that GSTO2 polymorphism significantly increased cancer risk in Caucasian population (GG vs. AA: OR = 1.32, 95%CI 1.06–1.64, Pheterogeneity = 0.616) and GSTO2 polymorphism was significantly associated with elevated risk of breast cancer (GG vs. AA OR = 1.37, 95%CI: 1.06–1.77; Pheterogeneity = 0.281). This meta-analysis demonstrates that GSTO2 polymorphism may significantly increase cancer risk in Caucasian population and is associated with elevated risk of breast cancer; while GSTO1 polymorphism is not associated with cancer risk.
Glutathione S-transferases (GSTs) are a family of Phase II detoxification enzymes that catalyze the conjugation of glutathione (GSH) to various endogenous and exogenous electrophilic compounds1. Up to now, human cytosolic GST super family contains at least 16 genes subdivided into eight distinct classes designated as: Alpha, Kappa, Mu, Omega, Pi, Sigma, Theta, and Zeta2,3. GSTs possess both enzymatic and non-enzymatic functions and are involved in many important cellular processes, such as, phase II metabolism, stress response, cell proliferation, apoptosis, oncogenesis, tumor progression and drug resistance4. Many studies have explored the association between single nucleotide polymorphisms (SNPs) of GSTs and susceptibility to various cancers. Clinical association studies have shown that genetic alterations within the human GST isozymes may play a key role in cancer susceptibility and treatment5. For example, GSTM1 and GSTP1 genetic polymorphisms are associated with increased risk of breast cancer6 and hepatocellular carcinoma7.
As a member of GSTs, glutathione S-transferase Omega (GSTO) has two members, named GSTO1 and GSTO2. Three polymorphisms in hGSTO genes: hGSTO1*A140D, hGSTO1*E155del and hGSTO2*N142D have been identified8. Numerous case-control studies have been performed to investigate the association between hGSTO1*A140D and hGSTO2*N142D and cancer risk in the last decades. But the results were inconsistent. Several investigators have reported an increased risk of breast cancer, hepatocellular carcinoma, bile duct carcinoma, urothelial cancer, acute lymphoblastic leukemia and non-small cell lung cancer for the GSTO1 A140D9,10,11,12. However, Granja et al.13 and Marahatta et al.14 did not find any significant association in thyroid and colorectal cancers. In addition, there was no evidence for association of GSTO1 or GSTO2 polymorphism and breast cancer risk in the study performed by Irena E. Andonova et al.15. After literature research, we did not find any previous genome-wide association studies (GWAS) relevant to the polymorphism of GSTO.
Therefore, we performed this meta-analysis to explore the association strength of GSTO1 and GSTO2 polymorphism with cancer risk.
Results
Characteristics of eligible studies
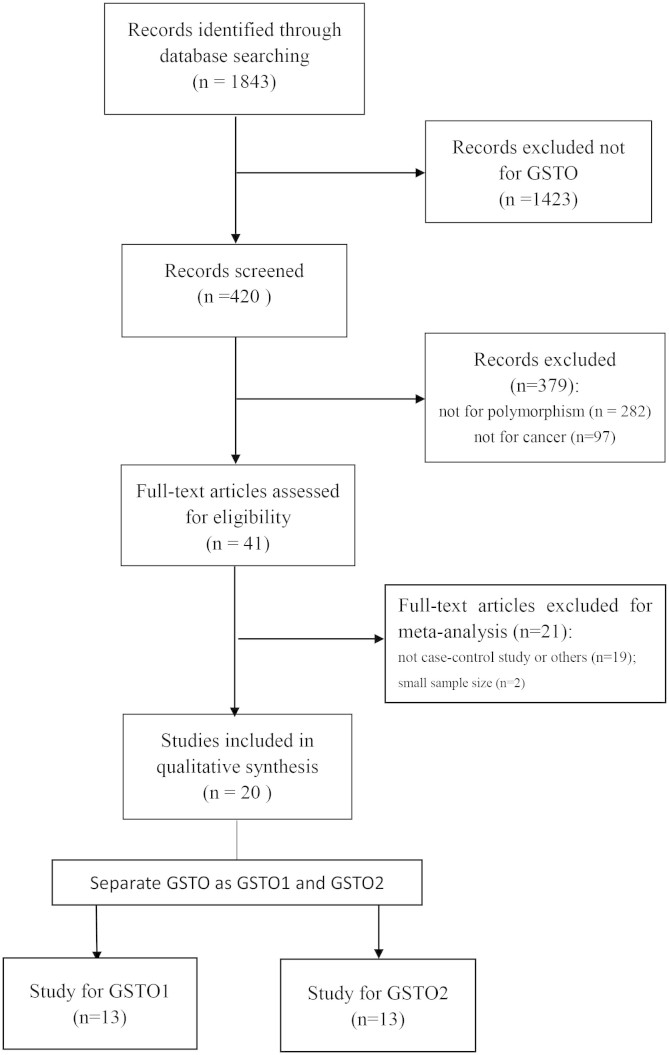
In total, 20 articles9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29 were identified according to the inclusion and exclusion criteria. The flow chart of literature search and study selection was illuminated in Figure 1. After deleting the duplicate articles, 1843 articles were obtained in total. We read the title and abstract, and then screened out 420 studies for GSTO. Among these, 379 articles not for polymorphism (282) or not for cancer (97) were excluded. Then, the remaining 41 articles underwent further identification and 19 articles were not included owing to not case-control-designed study or not enough information on the association between GSTO polymorphism and cancer risk. Two articles14,30 were excluded for the reason of not for cancer susceptibility and small sample size (less than 50). Among the rest of 20 articles, some researched both GSTO1 and GSTO2 polymorphism, so we regarded one study as two separate ones. Specifically, 13 studies for GSTO1 and 13 studies for GSTO2 were analyzed in our meta-analysis. Table 1 shows the detailed characteristics of the eligible studies included in this meta-analysis.
Figure 1. Flow chart of studies in the analysis.
Table 1. Characteristics of eligible studies.
| Author | Year | Country | Ethnicity | Cancer | Sources of control | Genotyping method | Cases | Controls | Case | Control | Phwe | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSTO1 polymorphism | CC | CA | AA | CC | CA | AA | |||||||||
| Granja13 | 2005 | Brazil | mixed | TC | PB | PCR-SSCP | 93 | 173 | 74 | 17 | 2 | 146 | 19 | 8 | <0.001 |
| Bufalo21 | 2006 | Brazil | mixed | TC | PB | PCR-RFLP | 73 | 184 | 59 | 14 | 159 | 25 | |||
| Olsen9 | 2008 | Denmark | Caucasian | BC | PB | PCR-RFLP | 396 | 396 | 121 | 199 | 76 | 123 | 210 | 63 | 0.089 |
| Lima-Jr16 | 2008 | Brazil | mixed | PC | HB | PCR-SSCP | 125 | 100 | 116 | 6 | 3 | 92 | 4 | 4 | <0.001 |
| Pongstaporn10 | 2009 | Thailand | Asian | ALL | HB | PCR-RFLP | 99 | 100 | 59 | 40 | 0 | 76 | 23 | 1 | 0.607 |
| Wang11 | 2009 | Taiwan | Asian | UC | HB | PCR-RFLP | 520 | 520 | 368 | 135 | 17 | 356 | 153 | 11 | 0.243 |
| Andonova15 | 2010 | Germany | Caucasian | BC | PB | MALDI-TOF MS | 1000 | 992 | 396 | 509 | 95 | 429 | 456 | 107 | 0.384 |
| Chariyalertsak17 | 2009 | Thailand | Asian | BC | HB | PCR-RFLP | 101 | 151 | 80 | 20 | 1 | 117 | 33 | 1 | 0.414 |
| Chung18 | 2011 | Taiwan | Asian | UC | HB | PCR-RFLP | 149 | 251 | 107 | 41 | 1 | 166 | 78 | 7 | 0.546 |
| Beebe-Dimmer15 | 2012 | USA | Caucasian | UC | PB | Taqman | 212 | 266 | 97 | 98 | 17 | 123 | 113 | 30 | 0.601 |
| Sanguansin20 | 2012 | Thailand | Asian | HNSCC | HB | PCR-RFLP | 300 | 299 | 231 | 67 | 2 | 234 | 62 | 3 | 0.619 |
| Guzide ADA12 | 2013 | Turkey | Caucasian | LC | PB | PCR-RFLP | 172 | 214 | 82 | 77 | 13 | 104 | 87 | 23 | 0.456 |
| Min-Che Tung29 | 2014 | Taiwan | Asian | UC | HB | PCR-RFLP | 300 | 233 | 230 | 70 | 150 | 79 | |||
| GSTO2 polymorphism | AA | AG | GG | AA | AG | GG | |||||||||
| Morari22 | 2006 | Brazil | mixed | OC | PB | PCR-RFLP | 69 | 222 | 24 | 37 | 8 | 87 | 104 | 31 | 0.993 |
| Leite23 | 2007 | Brazil | mixed | SC | PB | PCR-RFLP | 88 | 124 | 37 | 38 | 13 | 40 | 62 | 22 | 0.811 |
| Masoudi24 | 2009 | Iran | Asian | GC | PB | PCR-RFLP | 67 | 134 | 33 | 29 | 5 | 54 | 61 | 19 | 0.791 |
| Pongstaporn10 | 2009 | Thailand | Asian | ALL | HB | PCR-RFLP | 99 | 100 | 57 | 34 | 8 | 63 | 30 | 7 | 0.208 |
| Wang11 | 2009 | Taiwan | Asian | UC | HB | PCR-RFLP | 520 | 520 | 298 | 175 | 47 | 294 | 195 | 31 | 0.859 |
| Masoudi25 | 2010 | Iran | Asian | BC | PB | PCR-RFLP | 181 | 181 | 74 | 75 | 32 | 80 | 78 | 23 | 0.561 |
| Andonov15 | 2010 | Germany | Caucasian | BC | PB | MALDI-TOF MS | 1004 | 997 | 425 | 456 | 123 | 442 | 453 | 102 | 0.371 |
| Chariyalertsak17 | 2009 | Thailand | Asian | BC | HB | PCR-RFLP | 101 | 151 | 59 | 38 | 4 | 86 | 60 | 5 | 0.155 |
| Chung15 | 2011 | Taiwan | Asian | UC | HB | PCR-RFLP | 149 | 251 | 88 | 59 | 2 | 134 | 104 | 13 | 0.207 |
| Masoud26i | 2011 | Iran | Asian | CRC | PB | PCR-RFLP | 63 | 126 | 30 | 25 | 8 | 52 | 55 | 19 | 0.482 |
| Lesseur27 | 2012 | USA | Caucasian | UC | PB | CIGG | 658 | 928 | 255 | 321 | 82 | 398 | 439 | 91 | 0.057 |
| Sanguansin20 | 2012 | Thailand | Asian | HNSCC | HB | PCR-RFLP | 300 | 299 | 170 | 112 | 18 | 171 | 109 | 19 | 0.771 |
| Sohail28 | 2013 | Pakistan | Asian | BC | PB | PCR-RFLP | 100 | 102 | 0 | 10 | 90 | 7 | 28 | 67 | 0.105 |
PB: population-based; HB: hospital-based; Phwe: Hardy-Winberg equilibrium; TC: thyroid carcinoma; BC: breast cancer; PC: prostate carcinoma; ALL: acute lymphoblastic leukemia; UC: urothelial carcinoma; HNSCC: head and neck squamous cell carcinoma; LC: lung cancer; OC: ovarian carcinoma; SC: skin cancer; GC: gastric cancer CRC: colorectal cancer; CIGG: custom Illumina Golden Gate 96 SNP panel array.
Among the 20 articles, 5 of them were studies of Caucasian9,12,15,19,27, 10 studies were of Asian10,11,17,18,20,24,25,26,28,29 and the rest were study of different races (white and non-white groups)13,16,21,22,23. Cancer cases were all diagnosed histologically or pathologically in these studies. Polymerase chain reaction restriction fragment length polymorphism (PCR–RFLP) assay, TaqMan genotyping assay, matrix assisted laser desorption/ionizationtime-of-flight mass spectrometry (MALDI-TOF MS) assay, a custom Illumina Golden Gate 96SNP panel array, and polymerase chain reaction–single strand conformation polymorphism (PCR-SSCP)-sequencing approach were used as genotyping methods in 15, 1, 1, 1 and 2 articles respectively. Blood sample was used for genotyping in all studies. In our meta-analysis, 2 studies about GSTO1*A140D polymorphism9,13 were deviated from HWE.
The GSTO1 polymorphism
13 eligible studies, involving 3540 cancer cases and 3879 controls, were pooled for the analysis of GSTO1 polymorphism. No significant association of GSTO1 polymorphism with cancer risk was observed in any of the five comparison models (Table 2). Similarly, In the subgroup analysis by ethnicity, sources of control or cancer types, we did not find any significant association between the GSTO1 polymorphism and cancer risk, except that an increased cancer risk was found in the heterozygote comparison model (CA vs. CC: OR = 1.30, 95%CI 1.03–1.65, Pheterogeneity = 0.985) for other cancers and a decreased cancer risk was found in the dominant model (AA/AC vs. CC: OR = 0.82, 95%CI 0.70–0.98, Pheterogeneity = 0.985) for urothelial carcinoma.
Table 2. Meta-analysis results of GSTO1 polymorphism.
| N | Case/Control | AA vs CC | Ph | CA vs CC | Ph | AA vs CA/CC | Ph | AA/CA vs CC | Ph | A vs C | Ph | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 12 | 3240/3646 | 0.95(0.78,1.16) | 0.692 | 1.09(0.97,1.21) | 0.176 | 0.92(0.76,1.11) | 0.474 | 1.01(0.87,1.18) | 0.049 | 1.02(0.94,1.11) | 0.601 |
| Cancer | ||||||||||||
| BC | 3 | 1497/1539 | 1.05(0.82,1.34) | 0.639 | 1.12(0.96,1.31) | 0.357 | 1.00(0.80,1.26) | 0.293 | 1.11(0.96,1.29) | 0.618 | 1.06(0.95,1.18) | 0.888 |
| UC | 3 | 881/1037 | 0.87(0.54,1.39) | 0.147 | 0.91(0.74,1.11) | 0.293 | 0.86(0.55,1.36) | 0.117 | 0.82(0.70,0.98) | 0.162* | 0.91(0.78,1.07) | 0.563 |
| Others | 6 | 862/1070 | 0.65(0.37,1.14) | 0.992 | 1.30(1.03,1.65)* | 0.985 | 0.61(0.35,1.07) | 0.985 | 1.22(0.98,1.52) | 0.391 | 1.07(0.88,1.30) | 0.346 |
| Sources of control | ||||||||||||
| PB | 6 | 1946/2225 | 1.01(0.81,1.25) | 0.328 | 0.98(0.82,1.18) | 0.568 | 0.92(0.75,1,12) | 0.264 | 1.12(0.99,1.27) | 0.852 | 1.04(0.94,1.14 | 0.887 |
| HB | 6 | 1294/1421 | 0.99(0.54,1.82) | 0.473 | 1.08(0.59,2.00) | 0.107 | 0.95(0.54,1.65) | 0.500 | 0.92(0.71,1.21) | 0.026 | 0.98(0.84,1.14) | 0.240 |
| Ethnicity | ||||||||||||
| Asian | 5 | 1169/1231 | 0.99(0.54,1.82) | 0.473 | 1.03(0.77,1.39) | 0.062 | 1.02(0.56,1.86) | 0.432 | 0.93(0.69,1.24) | 0.014 | 0.99(0.84,1.16 | 0.166 |
| Caucasian | 4 | 1780/1868 | 0.97(0.78,1.21) | 0.445 | 1.13(0.99,1.30) | 0.682 | 0.93(0.76,1.14) | 0.218 | 1.10(0.96,1.26) | 0.842 | 1.03(0.94,1.14) | 0.788 |
| Mixed | 3 | 291/457 | 0.54(0.18,1.61) | 0.867 | 1.60(0.86,3.00) | 0.600 | 0.51(0.17,1.52) | 0.812 | 1.31(0.85,2.03) | 0.685 | 1.01(0.63,1.61) | 0.483 |
| Sample size | ||||||||||||
| Large | 4 | 2216/1439 | 1.08(0.85,1.36)* | 0.606 | 1.00(0.79,1.25) | 0.118 | 1.03(0.83,1.28) | 0.284 | 0.95(0.76,1.18) | 0.022 | 1.04(0.95,1.15) | 0.885 |
| Small | 8 | 1024/2207 | 0.64(0.42,0.98)* | 0.946 | 0.57(0.37,0.88)* | 0.879 | 0.62(0.41,0.93)* | 0.948 | 1.08(0.90,1.29) | 0.239 | 0.96(0.83,1.13) | 0.327 |
N: number of studies included; OR: odds ratio; Ph: p value for heterogeneity; BC: breast cancer; UC: urothelial carcinoma; PB: population-based; HB: hospital-based;
*OR with statistical significance.
The GSTO2 polymorphism
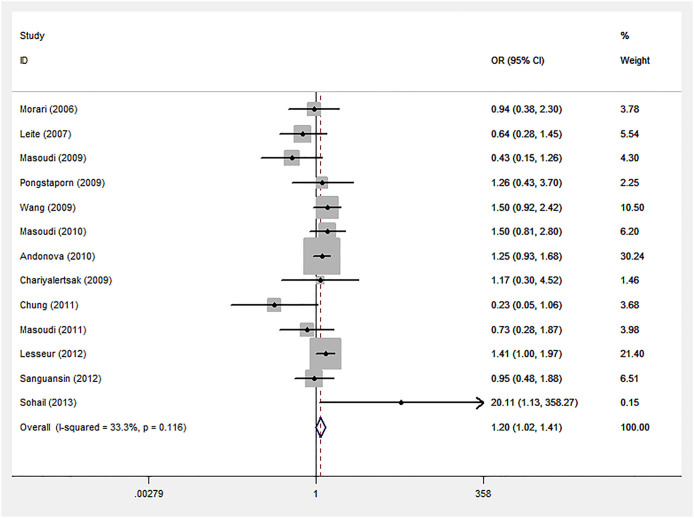
By pooling 13 eligible studies with 3399 cancer cases and 4135 controls, we observed a significantly increased risk of cancer susceptibility in homozygote comparison model (GG vs. AA: OR = 1.20, 95%CI: 1.02, 1.41, Pheterogeneity = 0.116; Figure 2) for GSTO2 polymorphism, but no significant association was found in other comparison models (Table 3).
Figure 2. Forest plot of homozygote comparison for overall comparison (GG vs. AA).
Table 3. Meta-analysis results of GSTO2 polymorphism.
| N | Case/Control | GG vs AA | Ph | AG vs AA | Ph | GG vs AG/AA | Ph | GG/AG vs AA | Ph | G vs A | Ph | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 13 | 3399/4135 | 1.20(1.02,1.41)* | 0.116 | 1.01(0.92,1.12) | 0.735 | 1.18(0.91,1.52) | 0.019 | 1.04(0.95,1.15) | 0.331 | 1.04(0.91,1.19) | 0.002 |
| Cancer type | ||||||||||||
| BC | 4 | 1386/1431 | 1.37(1.06,1.77)* | 0.281 | 1.04(0.89,1.23) | 0.697 | 1.76(0.98,3.18) | 0.015 | 1.10(0.95,1.28) | 0.294 | 1.38(0.93,2.04) | 0.001 |
| GC | 2 | 130/260 | 0.57(0.28,1.16) | 0.469 | 0.78(0.50,1.23) | 0.978 | 0.65(0.33,1.27) | 0.456 | 0.73(0.48,1.12) | 0.807 | 0.76(0.55,1.04) | 0.616 |
| UC | 3 | 1327/1699 | 1.18(0.68,2.07) | 0.066 | 1.01(0.86,1.17) | 0.249 | 1.19(0.70,2.02) | 0.071 | 1.05(0.91,1.21) | 0.167 | 1.03(0.85,1.25) | 0.092 |
| Others | 4 | 556/745 | 0.89(0.59,1.35) | 0.780 | 1.03(0.81,1.31) | 0.388 | 0.90(0.60,1.33) | 0.939 | 1.01(0.80,1.27) | 0.365 | 0.98(0.82,1.17) | 0.505 |
| Sources of control | ||||||||||||
| PB | 8 | 2230/2814 | 1.11(0.82,1.51) | 0.092 | 1.05(0.93,1.18) | 0.52 | 1.22(0.87,1.69) | 0.011 | 1.08(0.97,1.22) | 0.154 | 1.08(0.89,1.32) | <0.001 |
| HB | 5 | 1169/1321 | 1.12(0.80,1.57) | 0.217 | 0.94(0.80,1.12) | 0.832 | 1.15(0.82,1.59) | 0.199 | 0.97(0.83,1.14) | 0.77 | 1.00(0.88,1.14) | 0.515 |
| Ethnicity | ||||||||||||
| Asian | 9 | 1580/1864 | 1.03(0.68,1.56) | 0.074 | 0.94(0.81,1.09) | 0.879 | 1.18(0.75,1.85) | 0.006 | 0.90(0.91,1.29) | 0.452 | 0.86(0.69,1.08) | 0.019 |
| Caucasian | 2 | 1662/1925 | 1.32(1.06,1.64)* | 0.616 | 1.09(0.94,1.25) | 0.550 | 1.26(1.02,1.55)* | 0.757 | 1.13(0.99,1.29) | 0.515 | 1.12(1.02,1.24)* | 0.556 |
| Mixed | 2 | 157/346 | 0.76(0.41,1.39) | 0.539 | 0.93(0.62,1.42) | 0.121 | 0.81(0.46,1.40) | 0.992 | 0.89(0.60,1.33) | 0.135 | 0.89(0.68,1.18) | 0.276 |
| Sample size | ||||||||||||
| Large | 4 | 2482/2744 | 1.31(1.08,1.59)* | 0.708 | 1.04(0.92,1.16) | 0.531 | 1.28(1.06,1.53)* | 0.649 | 1.08(0.97,1.21) | 0.640 | 1.10(1.01,1.19)* | 0.780 |
| Small | 9 | 917/1391 | 0.95(0.69,1.29) | 0.101 | 0.95(0.78,1.14) | 0.686 | 1.04(0.63,1.72) | 0.004 | 0.95(0.79,1.13) | 0.270 | 1.04(0.80,1.35) | <0.001 |
N: number of studies included; OR: odds ratio; Ph: p value for heterogeneity; BC: breast cancer; UC: urothelial carcinoma; GC: gastrointestinal cancer; PB: population-based; HB: hospital-based;
*OR with statistical significance.
Then we performed subgroup analyses to investigate the effect of ethnicity, cancer types and sources of control. As for cancer types, there was a statistically increased cancer risk for breast cancer (GG vs. AA: OR = 1.37, 95%CI: 1.06–1.77; Pheterogeneity = 0.281; Figure S1). As for ethnicity, increased cancer risk was found in Caucasian in the homozygote comparison model (GG vs. AA: OR = 1.32, 95%CI 1.06–1.64, Pheterogeneity = 0.616; Figure S2), recessive comparison model (GG vs. AG/AA: OR = 1.26, 95%CI 1.02–1.55, Pheterogeneity = 0.757; Figure S3) and allelic comparison model (G vs. A: OR = 1.12, 95%CI 1.02–1.24, Pheterogeneity = 0.556; Figure S4).
Heterogeneity
Heterogeneity between studies in each comparison model was shown in Table 2 and 3. No significant heterogeneity was found for GSTO1 polymorphism, but for GSTO2, obvious heterogeneity was detected in two comparison models (GG vs. GA/AA, P = 0.019; G vs. A, P = 0.002). Meta-regression revealed that ethnicity, cancer types, sample size and sources of control did not contributed to the source of heterogeneity (τ2 > 0.05).
Sensitivity analysis
To examine the stability and reliability of our meta-analysis results, we performed sensitivity analyses by repeatedly deleting the single studies each time from pooled analysis. Our analysis showed that the omission of individual studies did not materially alter the results because the recalculated ORs and 95%CIs were not quantitatively changed, suggesting that the results were robust and convincing. (Figures not shown).
Publication bias
Begg's funnel plot and Egger's test were performed to assess the publication bias of literatures, and we did not find any publication bias for GSTO1 or GSTO2 polymorphism in all compassion models.
Discussion
GSTs are a family of phase II detoxifying enzymes that catalyze the conjugation of glutathione to a wide variety of electrophilic compounds. Besides detoxifying electrophilic xenobiotics such as chemical carcinogens, environmental pollutants, and antitumor agents, these transferases inactivate endogenous alpha, beta-unsaturated aldehydes, quinones, epoxides, and hydroperoxides formed as secondary metabolites during oxidative stress31. GSTs play important roles in the protection of cells against foreign compounds and cellular stress, and may consequently play a role in the development of cancer32. And due to high expression of GSTs in tumors when compared to normal tissues and their high level in plasma from cancer patients, these enzymes were considered to be cancer markers33. Recently, many studies have demonstrated the association between SNPs of GSTs and cancer risk. Study conducted by Safarinejad, M.R et al.34 suggested that the GSTP1 polymorphism and its combination with GSTM1, and GSTT1 may be associated with bladder cancer susceptibility. Yang et al.35 also found that GSTT1 null genotype contributes to lung cancer risk in Asian populations.
Unlike other GSTs, GSTO has an active site cysteine that is able to form a disulfide bond with GSH and exhibits glutathione dependent dehydroascorbate reductase and thiol transferase activities, reminiscent of glutaredoxin and thioredoxin enzymes36. Expression of GSTO is abundant in a wide range of normal tissues, including the liver, colon, heart, ovary, pancreas, prostate and spleen. The widespread distribution of GSTO suggests that it has important biological functions8. GSTO participates in cellular signalling and overexpression of GSTO has been reported to be linked with the induction of apoptosis involving the development of cancer37. Additionally, GSTO was shown to promote activation of the pro-inflammatory cytokine, interleukin-1β (IL-1β) by post-translational processing38. Thus, association between GSTO and cancer risk has been explored in some studies. Marahatta et al.14 demonstrated that GSTO1*A140D polymorphism could play an important role for the development of cholangiocarcinoma, breast cancer and hepatocellular carcinoma, and Mohammad Masoudi26 and his colleagues indicated that GSTO2 NN genotype increase the risk of colorectal cancer. On the contrary, GSTO1 and GSTO2 variants were not associated with breast cancer risk in some study17.
Given the inconsistent results from individual studies, we decided to explore the association between GSTO polymorphisms and cancer risk. In the present meta-analysis, 20 eligible studies including 4770 cases and 5701 controls, were identified and analyzed. Our results showed that there was no significant association between the GSTO1 polymorphism and susceptibility to cancer. Similarly, subgroup analyses by cancer type, source of control or ethnicity did not suggest a significantly different result. As for GOTS2 polymorphism, we can observe an increased risk of overall cancer and breast cancer. In addition, it is worth noting that the association between GSTO2 polymorphism and cancer risk was significant in Caucasian populations. We demonstrated an increased cancer risk in Caucasian for GSTO2 polymorphism, specifically in the homozygote comparison model, recessive comparison model and allelic comparison model. However, in our meta-analysis for GSTO2, only two study were conducted in Caucasian race totally. Corina Lesseur et al.27 found bladder cancer risk overall was associated with GSTO2 Asn142Asp. Whereas Irena E. Andonova et al.15 did not find any evidence for GSTO2 in breast cancer risk. So, a conclusion the GOST2 polymorphism increasing the cancer risk in Caucasian may not be convincing that much.
As we mentioned before, two studies9,13 were deviated from HWE and two study could not be calculated for HWE due to its incomplete data21,29. Traditionally speaking, any study that deviated from HWE should have been removed. However, Minelli et al.39 pointed out that unless there are other grounds for doubting the quality of the study, studies that appear to deviate from HWE should be investigated further rather than just excluded. Until now, it is still inconclusive whether studies deviated from HWE should be included or excluded in conducting meta-analysis40. When deleting the four studies in the sensitivity analysis, the pooled results did not change significantly. Meta-regression results revealed that ethnicity, cancer types, sources of control and sample size did not contributed to the source of heterogeneity.
Several limitations should be acknowledged in this meta-analysis. Firstly, the studies were full text in English and some inevitable publication bias might exist in the publications. Secondly, our results were based on single-factor estimates without adjustment for other risk factors such as age, family history and environment factors, should be conducted if possible. These factors may explain the heterogeneity. Beyond that, the number of studies for subgroup analysis was small.
In conclusion, we demonstrate that GSTO2 polymorphism may significantly increase cancer risk in Caucasian population and is associated with elevated risk of breast cancer; while GSTO1 polymorphism is not associated with cancer risk. To further confirm the results, large scale case-control studies with different ethnic groups and multiple cancer types are needed.
Methods
Identification of eligible studies
We extracted Eligible case-control studies by searching databases and manual search of references of relative reviews and articles. To identify all the studies that examined the association of GSTO polymorphism and cancer risk, we conducted a computerized literature search of Embase, Web of Science, PubMed and China National Knowledge Infrastructure (CNKI). The combination of the following key words were used as search terms: GST (e.g.: “Glutathione S-transferase”); cancer (e.g.: “carcinoma”, “tumor” or “neoplasms”) and polymorphism (e.g.: “single nucleotide polymorphism”, “SNP” or “variation”). There was no limitation of research and the last research was carried out on Aug 13, 2014. To explore potentially additional studies, we also examined the references of articles and reviews.
Inclusion and exclusion criteria
The following criteria were used for the literature selection: (a) information on the association of cancer risk with GSTO1 or GSTO2 polymorphism; (b) participants more than fifty; (c) sufficient genotype data to calculate the odds ratios (ORs) with 95% confidence intervals (CIs). The major exclusion criteria were: (a) overlapping study populations; (b) non case-control design; (c) without detailed data on genotype distribution. Titles and abstracts of searching records were screened and full text papers were further evaluated to confirm the eligibility. According to the inclusion criteria, two reviewers (Xu and Wang) extracted eligible studies independently, and disagreement between the two reviewers was settled by discussing with the third reviewer (Qiu).
Data extraction
According to the selection criteria mentioned above, the following date was extracted from each study independently by two authors (Xu and Wang): name of first author, year of publication, country where the study was conducted, ethnicity of participants, methods for genotyping, sources of control, cancer types, genotype frequency in cases and controls. Different ethnicities were defined as Asian, Caucasian and mixed races. All eligible studies were categorized as population-based (PB) and hospital-based (HB) according to the sources of control. Cancer types were classified as breast cancer, urothelial carcinoma (including bladder cancer), gastrointestinal cancer (gastric cancer and colorectal cancer) only for GSTO2 and other cancers (thyroid carcinoma, gastric cancer, colorectal cancer, ovarian cancer, basal cell skin carcinoma, prostate carcinoma, head and neck cancer, acute lymphoblastic leukemia and non-small cell lung cancer). Chi-square test was used to examine the Hardy–Weinberg equilibrium (HWE) based on the two polymorphisms genotyping distribution in controls (p < 0.05 indicated significant deviation from HWE). Two reviewers reached consensus on each item.
Statistical analysis
We utilized ORs with 95% CIs to assess the strength of the association between GSTO1 or GSTO2 polymorphism and cancer risk. The estimated pooled ORs were achieved by calculating a weighted average of OR from each study. Pooled ORs were calculated for homozygote comparison (AA vs. CC for GSTO1; GG vs. AA for GSTO2), heterozygote comparison (AC vs. CC for GSTO1; GG vs. GA for GSTO2), allelic comparison (A vs. C for GSTO1; G vs. A for GSTO2), recessive model (AA vs. AC/CC for GSTO1; GG vs. GA/AA for GSTO2) and dominant model (AA/AC vs. CC for GSTO1; GG/GA vs. AA for GSTO2), respectively. A 95% CI without 1 for OR indicating a significant increased or reduced cancer risk.
The statistical significance of pooled ORs was determined by Z-test (P < 0.05 indicated statistically significant). We used a chi-square based Q-test to check the heterogeneity among the studies. Q-test results of P < 0.10 suggested significant heterogeneity among studies, so the pooled OR of all studies was calculated using the random-effects model based on DerSimonian-Laird method41; Otherwise, the fixed-effects model based on Mantel-Haenszel method was conducted42. Afterwards, subgroup analyses were conducted to test the effects of ethnicity, cancer type and sources of control. Meta-regression was performed to detect the source of heterogeneity by cancer type, ethnicity, sources of control and sample size (studies with less 500 participants were categorized as “small”, and studies with more than 500 participants were categorized as “large”). Between study, variance Tau-squared (τ2) value was used to evaluate the degree of heterogeneity and the τ2 was used to describe the extent of heterogeneity explained43. Sensitivity analysis conducted by sequential deleting a single study each time was carried out to identify the effect of data from each study on pooled ORs.
Both the Begg's funnel plot and the Egger's linear regression test were used to evaluate publication bias across the literatures and a p < 0.05 was considered significant44. All p values were two sided. All of the statistical analyses were performed using STATA software version 12.0 (STATA Corporation, College Station, TX, USA).
Author Contributions
Conceived and designed the experiments: Y.T.X., J.W., R.Y. and L.X. Performed the experiments: Y.T.X., J.W., M.T.Q., L.X., J.W., R.Y. and L.X. Analyzed the data: Y.T.X., J.W. and M.T.Q. Contributed reagents/materials/analysis tools: Y.T.X., J.W., M.T.Q., R.Y. and L.X. Wrote the paper: Y.T.X., J.W., M.T.Q., J.W. and R.Y. Access to full-text articles: L.X. and J.W.
Supplementary Material
Supplementary Information
Acknowledgments
This study is founded by the Natural Science Foundation of China (81372321, 81201830), University Grant of Jiangsu Province (13KJB320010), and Jiangsu Provincial Special Program of Medical Science (BL2012030).
References
- Townsend D. M. & Tew K. D. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 22, 7369–7375 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D. & McLellan L. I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res 31, 273–300 (1999). [DOI] [PubMed] [Google Scholar]
- Board P. G. et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J Biol Chem 275, 24798–24806 (2000). [DOI] [PubMed] [Google Scholar]
- Lo H. W. & Ali-Osman F. Genetic polymorphism and function of glutathione S-transferases in tumor drug resistance. Curr Opin Pharmacol 7, 367–374 (2007). [DOI] [PubMed] [Google Scholar]
- Townsend D. & Tew K. Cancer drugs, genetic variation and the glutathione-S-transferase gene family. Am J Pharmacogenomics 3, 157–172 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi M. et al. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomark Med 6, 797–803 (2012). [DOI] [PubMed] [Google Scholar]
- Song K., Yi J., Shen X. & Cai Y. Genetic polymorphisms of glutathione S-transferase genes GSTM1, GSTT1 and risk of hepatocellular carcinoma. PLoS One 7, e48924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbread A. K., Tetlow N., Eyre H. J., Sutherland G. R. & Board P. G. Characterization of the human Omega class glutathione transferase genes and associated polymorphisms. Pharmacogenetics 13, 131–144 (2003). [DOI] [PubMed] [Google Scholar]
- Olsen A., Autrup H., Sorensen M., Overvad K. & Tjonneland A. Polymorphisms of glutathione S-transferase A1 and O1 and breast cancer among postmenopausal Danish women. Eur J Cancer Prev 17, 225–229 (2008). [DOI] [PubMed] [Google Scholar]
- Pongstaporn W., Pakakasama S., Sanguansin S., Hongeng S. & Petmitr S. Polymorphism of glutathione S-transferase Omega gene: association with risk of childhood acute lymphoblastic leukemia. J Cancer Res Clin Oncol 135, 673–678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H. et al. A significantly joint effect between arsenic and occupational exposures and risk genotypes/diplotypes of CYP2E1, GSTO1 and GSTO2 on risk of urothelial carcinoma. Toxicol Appl Pharmacol 241, 111–118 (2009). [DOI] [PubMed] [Google Scholar]
- Ada T. G. et al. Association between glutathione S-transferase omega 1 A140D polymorphism in the Turkish population and susceptibility to non-small cell lung cancer. Arh Hig Rada Toksikol 64, 61–67 (2013). [DOI] [PubMed] [Google Scholar]
- Granja F., Morari E. C., Assumpcao L. V. & Ward L. S. GSTO polymorphism analysis in thyroid nodules suggest that GSTO1 variants do not influence the risk for malignancy. Eur J Cancer Prev 14, 277–280 (2005). [DOI] [PubMed] [Google Scholar]
- Marahatta S. B., Punyarit P., Bhudisawasdi V., Paupairoj A., Wongkham S. & Petmitr S. Polymorphism of glutathione S-transferase omega gene and risk of cancer. Cancer Lett 236, 276–281 (2006). [DOI] [PubMed] [Google Scholar]
- Andonova I. E. et al. No evidence for glutathione S-transferases GSTA2, GSTM2, GSTO1, GSTO2, and GSTZ1 in breast cancer risk. Breast Cancer Res Treat 121, 497–502 (2010). [DOI] [PubMed] [Google Scholar]
- Lima M. M. Jr et al. Lack of association of GSTT1, GSTM1, GSTO1, GSTP1 and CYP1A1 polymorphisms for susceptibility and outcome in Brazilian prostate cancer patients. Folia Biol (Praha) 54, 102–108 (2008). [DOI] [PubMed] [Google Scholar]
- Chariyalertsak S., Purisa W. & Sangrajrang S. Role of glutathione S-transferase omega gene polymorphisms in breast-cancer risk. Tumori 95, 739–743 (2009). [DOI] [PubMed] [Google Scholar]
- Chung C. J., Pu Y. S., Su C. T., Huang C. Y. & Hsueh Y. M. Gene polymorphisms of glutathione S-transferase omega 1 and 2, urinary arsenic methylation profile and urothelial carcinoma. Sci Total Environ 409, 465–470 (2011). [DOI] [PubMed] [Google Scholar]
- Beebe-Dimmer J. L. et al. Genetic variation in glutathione S-transferase omega-1, arsenic methyltransferase and methylene-tetrahydrofolate reductase, arsenic exposure and bladder cancer: a case-control study. Environ Health 11, 43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguansin S., Petmitr S., P O. C. & Pongstaporn W. Association of glutathione S-transferase omega gene polymorphisms with progression of head and neck cancer. Mol Biol Rep 39, 10915–10920 (2012). [DOI] [PubMed] [Google Scholar]
- Bufalo N. E. et al. Smoking and susceptibility to thyroid cancer: an inverse association with CYP1A1 allelic variants. Endocr Relat Cancer 13, 1185–1193 (2006). [DOI] [PubMed] [Google Scholar]
- Morari E. C. et al. Role of glutathione-S-transferase and codon 72 of P53 genotypes in epithelial ovarian cancer patients. J Cancer Res Clin Oncol 132, 521–528 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite J. L. et al. Influence of the glutathione s-transferase gene polymorphisms on the susceptibility to basal cell skin carcinoma. Rev Med Chil 135, 301–306 (2007). [DOI] [PubMed] [Google Scholar]
- Masoudi M., Saadat I., Omidvari S. & Saadat M. Genetic polymorphisms of GSTO2, GSTM1, and GSTT1 and risk of gastric cancer. Mol Biol Rep 36, 781–784 (2009). [DOI] [PubMed] [Google Scholar]
- Masoudi M., Saadat I., Omidvari S. & Saadat M. Additive effects of genetic variations of xenobiotic detoxification enzymes and DNA repair gene XRCC1 on the susceptibility to breast cancer. Breast Cancer Res Treat 120, 263–265 (2010). [DOI] [PubMed] [Google Scholar]
- Masoudi M., Saadat I., Omidvari S. & Saadat M. Association between N142D genetic polymorphism of GSTO2 and susceptibility to colorectal cancer. Mol Biol Rep 38, 4309–4313 (2011). [DOI] [PubMed] [Google Scholar]
- Lesseur C. et al. A case-control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicol Lett 210, 100–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A. et al. Effects of glutathione-S-transferase polymorphisms on the risk of breast cancer: a population-based case-control study in Pakistan. Environ Toxicol Pharmacol 35, 143–153 (2013). [DOI] [PubMed] [Google Scholar]
- Tung M. C. et al. Combined effects of GSTO1 and SULT1A1 polymorphisms and cigarette smoking on urothelial carcinoma risk in a Taiwanese population. J Formos Med Assoc 113, 640–647 (2014). [DOI] [PubMed] [Google Scholar]
- Pongstaporn W. et al. Genetic alterations in chromosome 10q24.3 and glutathione S-transferase omega 2 gene polymorphism in ovarian cancer. J Exp Clin Cancer Res 25, 107–114 (2006). [PubMed] [Google Scholar]
- Cheng T. C. et al. Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: a multigenic study on cancer susceptibility. Int J Cancer 113, 345–353 (2005). [DOI] [PubMed] [Google Scholar]
- Autrup H. Gene-environment interaction in environment carcinogens. Cancer as an Environmental Disease 20, 71–101 (2004). [Google Scholar]
- Gate L. & Tew K. D. Glutathione S-transferases as emerging therapeutic targets. Expert Opin Ther Targets 5, 477–489 (2001). [DOI] [PubMed] [Google Scholar]
- Safarinejad M. R., Safarinejad S., Shafiei N. & Safarinejad S. Association of genetic polymorphism of glutathione S-transferase (GSTM1, GSTT1, GSTP1) with bladder cancer susceptibility. Urol Oncol 31, 1193–1203 (2013). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. GSTT1 null genotype contributes to lung cancer risk in asian populations: a meta-analysis of 23 studies. PLoS One 8, e62181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini J., Amirante A., Zemzoumi K. & Serra E. Characterization of an omega-class glutathione S-transferase from Schistosoma mansoni with glutaredoxin-like dehydroascorbate reductase and thiol transferase activities. Eur J Biochem 269, 5512–5521 (2002). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Cloning, expression and characterization of human glutathione S-transferase Omega 2. Int J Mol Med 16, 19–27 (2005). [PubMed] [Google Scholar]
- Laliberte R. E. et al. Glutathione s-transferase omega 1-1 is a target of cytokine release inhibitory drugs and may be responsible for their effect on interleukin-1beta posttranslational processing. J Biol Chem 278, 16567–16578 (2003). [DOI] [PubMed] [Google Scholar]
- Minelli C., Thompson J. R., Abrams K. R., Thakkinstian A. & Attia J. How should we use information about HWE in the meta-analyses of genetic association studies? Int J Epidemiol 37, 136–146 (2008). [DOI] [PubMed] [Google Scholar]
- Xue H. et al. A meta-analysis of interleukin-10 -592 promoter polymorphism associated with gastric cancer risk. PLoS One 7, e39868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A. & Whitehead J. A general parametric approach to the meta-analysis of randomized clinical trials. Stat Med 10, 1665–1677 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information