Abstract
mtDNA non-synonymous germ line variation (G10398A; p.A114T) has remained equivocal with least mechanistic understanding in showing an association with cancer. This has necessitated showing in-vitro how an over-expression within mitochondria of either of the variants produces higher intracellular ROS, resulting in differential anchorage dependent and independent growth. Both these features were observed to be relatively higher in ND3:114T variant. An elevated amount of intracellular carbonylated proteins and a reduced activity of a key glycolytic enzyme, Pyruvate kinase M2, along with high glucose uptake and lactate production were other pro-cancerous features observed. The retrograde signaling through surplus ROS was generated by post-ND3 over-expression regulated nuclear gene expression epigenetically, involving selectively the apoptotic-DDR-pathways. The feature of ND3 over-expression, inducing ROS mediated pro-cancerous features in the cells in in vitro, was replicated in a pilot study in a limited number of sporadic breast tumors, suggesting the importance of mitochondrial germ-line variant(s) in enabling the cells to acquire pro-cancerous features.
Mitochondrial functioning is essential for several cellular physiological processes including normal cell growth and apoptosis1,2 and requires a well-orchestrated interplay between both nuclear and mitochondrial (mt) genomes3. Proteomic analysis of mitochondria has revealed the presence of thousands of proteins within this organelle. However, only 13 proteins are contributed by the maternally inherited mitochondrial DNA, a 16569 bp long multicopy, double stranded circular molecule, coding for a total of 37 genes (13 polypeptides, 2 rRNA and 22 tRNA)4. Mitochondrial (mt) DNA sequence-data-base confirms its highly polymorphic nature with the occurrence of several pathogenic mutations in general population as frequently observed phenomenon5. Both germline and somatic differences in mt genome have been proposed to provide a differential susceptibility towards various patho-physiological conditions, generally late onset and complex disorders - ranging from mild myopathy to Alzheimer's, Parkinson's, aging and cancer2,6,7,8. Comparative study on whole mitochondrial genome from tumor and adjacent normal tissues of prostate9, head and neck10, bladder11, pancreas12, breast and esophagus13 identified a number of pathogenic mutations, providing a probable advantage in cancers to tumor growth. Case-control comparative studies in diverse cancer types from different populations have also shown an association of mitochondrial DNA single nucleotide polymorphisms (SNPs) with breast13,14, esophagus13,15 prostate & renal16, endometrial17 cancers. However, the functional role of these SNPs in any given cancer has not been studied adequately, except a few using cybrid technology18,19 and their role remains elusive. Scanty studies with codon optimized cloned mitochondrial genes10,11, a method of choice to understand the function of mitochondrial gene variations in vitro, require validation before drawing any conclusion on the functional implication of any of the germline or somatic mitochondrial genome variant. A non-synonymous polymorphism G10398A(p.A114T) within ND3, a key subunit involved in active/de-active transition of the 47 subunit-multimeric-enzyme-complex-I20, is one of the most extensively studied mitochondrial polymorphism at genetic level where both allelic forms of the variation have been found in association with either susceptibility or protection towards Parkinson's disease21, Alzheimer22, bipolar disorder23, metabolic syndrome24, longevity25 and sporadic breast cancer14,26. This has resulted in equivocal conclusions; for example, initial studies on African-American and Indian populations showed a significant association of 10398A (p.114T) with sporadic breast cancer14,26, but later studies on other populations generated both supportive and equivocal opinion on its presence and associated function27,28,29. Despite non-synonymous (ND3:A114T) behavior and proven role of G10398A allele in controlling mitochondrial matrix pH and calcium signaling7, the functional role of this polymorphism in normal cellular physiology and sporadic breast cancer patho-physiology continues to be equivocal. In order to resolve the contrary findings, we proposed to evaluate the functional role of the G10398A (ND3:A114T) germline polymorphism in in-vitro cell culture model, after cloning and stably expressing both G and A nucleotide variants of ND3 gene.
The pathogenic germline or somatic variations in mitochondrial DNA have been proposed to contribute to an imbalanced redox-homeostasis and tumor growth promotion through proteomic alteration, such as oxidative modification of either regulatory proteins like protein tyrosine phosphatase30, thioredoxin and peroxiredoxine family proteins31 or key glycolytic enzyme, pyruvate kinase (PK) M2. The latter is suggestive of providing the rapidly growing tumor cells an advantage for pooling intermediate molecules, required for the synthesis of proteins and nucleic acids; and also to divert the glucose flux into pentose phosphate pathway to generate sufficient reducing potential to survive in elevated oxidative situations32. Here, using a codon optimized system for over expression of both the variants of ND3 gene, we attempted to explore the role of G10398A (ND3:p.A114T) polymorphism on cellular redox homeostasis, tumor cell metabolism involving glucose uptake, lactate production and the activity of terminal glycolytic enzyme, pyruvate kinase M2 (PKM2). We also assessed the effect of the stably expressing germline polymorphic variants on the real time expression and epigenetic regulation of a select set of genes involved in DDR-apoptotic and anti-apoptotic pathways.
Results
mtND3:114A and mtND3:114T stably expressing cells show enhanced complex I activity, differential levels of ROS and oxidative stress
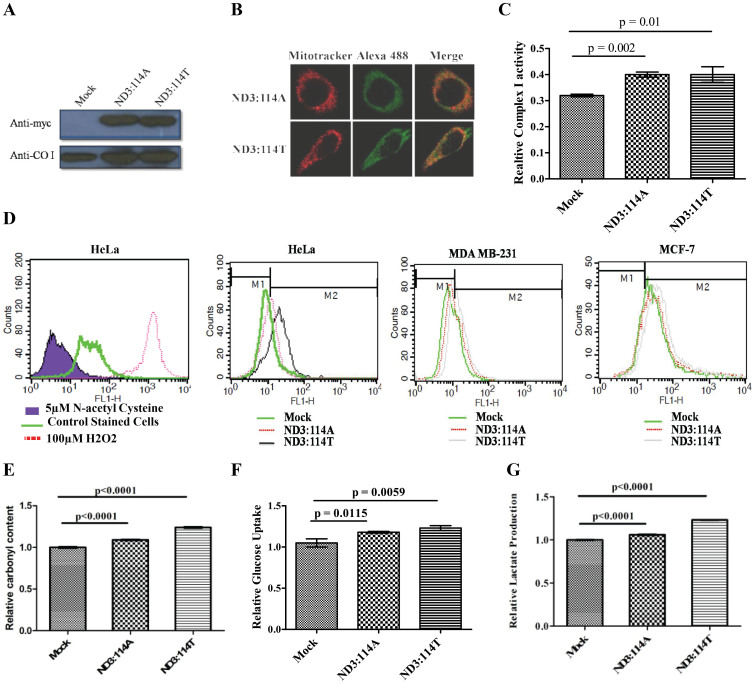
Ten different cell lines were analyzed for their mitochondrial genetic background at 10398 position (Supplementary Figure 1) within the ND3 gene. HeLa cells with 10398G (mtND3:114A), representing M haplogroup and MCF-7 & MDA-MB 231 cell lines with 10398A (mtND3:114T), representing N haplogroup, were stably transfected for exogenous over-expression of mtND3:114A or 114T; along with vector control. After confirming the expression and mitochondrial localization of exogenously expressed protein (Figs. 1A, B), a differentially increased activity of complex I was observed in the mitochondrial enriched fraction of stably transfected cells in comparison to the vector control (Fig. 1 C). The authenticity of the elevated activity of complex I within mitochondria was further confirmed through complex I assay using cytosolic fraction, which did not show any significant change (Supplementary Figure 2). The resultant effect of the increased activity of complex I was apparent in the enhanced ROS producing capacity (26% for ND3:114A, 55% for ND3:114T) in HeLa cells; (14% for ND3:114A and 52% for ND3:114T) and in MDA-MB 231 cells when compared to vector control. This was further confirmed under similar experimental conditions in another cell line MCF-7, where exogenous over expression of ND3:114A generated 18% and ND3:114T generated 55% higher ROS as compared to vector control (Fig. 1 D). Observing a similar phenotype of over expression for both the variants, irrespective of genotype background at 10398 position, we conducted all the experiments using MDA-MB231 cell line, unless stated otherwise. The resultant effect of high production of ROS was checked for oxidative stress through carbonylated protein content in stably transfected cells with: (i) empty vector, (ii) exogenously expressing mtND3:114A and (iii) mtND3:114T; where 2, 4-dinitrophenylhydrazone derivative was approximately 12% higher in mtND3:114A and 28% higher in mtND3:114T expressing cells, as compared to vector control (Fig. 1E). In addition, a slight decrease was observed of mitochondrial ROS target, aconitase, which was non-significant (Suplementry Figure 3). The stably transfected MDA-MB 231 cells expressing either mtND3:114A ormtND3:114T, when assessed for other pro-cancerous phenotype, showed a significantly high glucose uptake and lactate production (Fig. 1F, G).
Figure 1.
(A) Western blots (Cropped) with anti-myc antibody showing the expression of exogenously expressed ND3:114A and ND3:114T; Anti-COI western shows loading control (Full length blots are provided in Supplementary figure 4); (B) Fluorescence confirming mitochondrial localization of exogenously expressed protein, mitochondria were stained with 25 nM mitotracker, exogenously expressed protein were probed using alexa 488 linked secondary antibody for myc tag; (C) Enzyme activity of mitochondria enriched fraction for complex I from cells transfected with empty vector, ND3:114A and ND3: 114T; (D) Reactive oxygen species in mock, ND3:114A and ND3: 114T transfected cells. HeLa cells treated with 50 μM H2O2 and 5 μM N-acetyl cysteine treated cells were used as positive and negative control, respectively; (E) Relative carbonyl content; (F) Relative glucose uptake; and (G) Relative Lactate production.
ND3 over expression mediated ROS alters expression of apoptosis related and DNA damage genes
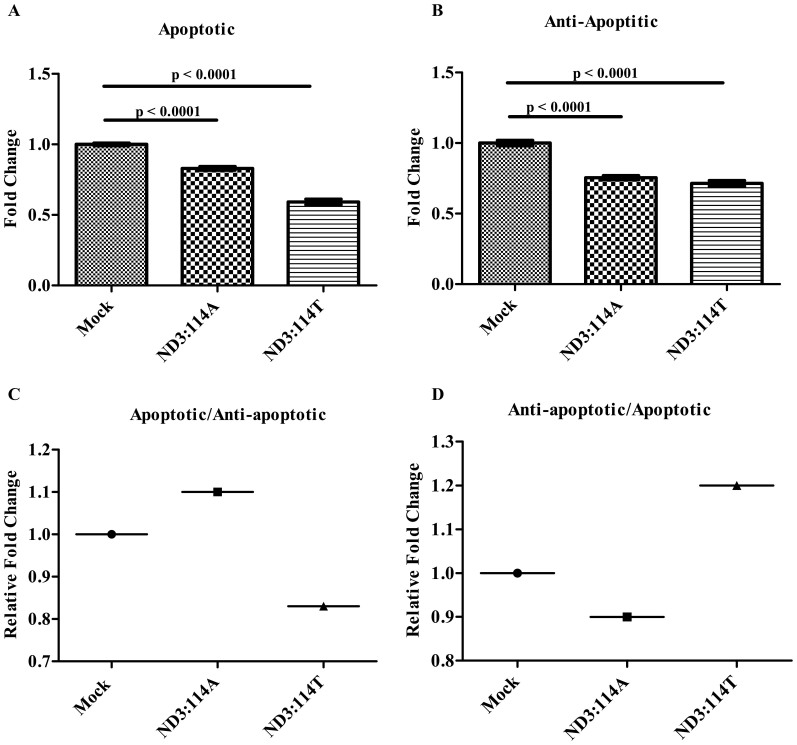
Real time expression of 19 genes involved in pro-apoptotic, anti-apoptotic (both extrinsic and intrinsic) and DNA damage pathway (Fig. 2) were studied to find out a possible role of ND3 over expression mediated high ROS providing pro-cancerous property to cells. The down regulated expression of both extrinsic apoptotic pathway, such as TRAIL (0.84 fold for ND3:114A & 0.54 fold for ND3:114T), DR4 (0.81 fold for ND3:114A & 0.50 fold for ND3:114T), DR5 (0.91 fold for ND3:114A and 0.68 fold for ND3:114T); and intrinsic apoptotic pathway genes involving CASP8 (0.56 fold for ND3:114A and 0.47 fold for ND3:114T), CASP 3 (0.96 fold for ND3:114A and 0.50 fold for ND3:114T) and CYC (0.93 fold for ND3:114A and 0.85 fold for ND3:114T), was suggestive of attaining a pro-cancerous features by the mtND3 over-expressing cells, incidentally supported by glucose uptake-lactate production. Although the down-regulation of apoptotic genes was obvious in both 114A and 114T stably expressing cells in comparison to vector control, however, the trend was more severe in ND3:114T expressing cells (Fig. 2A), supporting our previous correlation of this germline mitochondrial variant prevalence in cancer patients. Interestingly, the expression pattern of genes involved in negative regulation of extrinsic apoptotic pathway (anti-apoptotic genes) involving DCR1(0.65 fold for ND3:114A and 0.70 fold for ND3:114T), DCR2 (0.72 fold for ND3:114A and 0.85 fold for ND3:114T), FLIPL (0.85 fold for ND3:114A and 0.80 fold for ND3:114T) and FLIPS (0.82 fold for ND3:114A and 0.96 fold for ND3:114T) with a relatively increased expression in ND3:114T stably expressing cells (Fig. 2B- when compared between 114A and 114T), supported further the chances of attaining a pro-cancerous state by these cells, although the negative regulator of the intrinsic pathway of the apoptosis like CASP3S (0.76 fold for ND3:114A and 0.64 fold for ND3:114T), CASP8L (0.71 fold for ND3:114A and 0.39 fold for ND3:114T) and BCL-2 (0.75 fold for ND3:114A and 0.66 fold for ND3:114A) did not show the same pattern (Fig. 2B), which remains unexplained. Concomitantly, the expression of DNA damage pathway genes, such as MDM2 (1.15 fold for ND3:114A and 0.55 fold for ND3:114T), P53 (0.62 fold for ND3:114A and 0.79 fold for ND3:114T), P21 (1.03 fold for ND3:114A and 0.57 fold for ND3:114T), H2AX (1.18 fold for ND3:114A and 1.27 fold for ND3:114T), ATM (1.05 fold for ND3:114A and 0.93 fold for ND3:114T), CHK2 (0.64 fold for ND3:114A and 1.20 fold for ND3:114T), (Supplementary Figure. 5) when compared to vector control under similar experimental conditions of over expressed ND3 gene and generation of high ROS, apparently reflected a role in cell survival, as was obvious from the observations made for the down-regulated apoptotic and up-regulated expression of anti-apoptotic genes, especially in ND3:114T over-expressing cells. Analyzing the real time expression of these genes either individually or all together suggested an impact of ND3 over expression mediated ROS on alteration of ratio of apoptotic and anti-apoptotic genes (Fig. 2C & D). For example, in ND3:114T transfected cells, the cumulative fold change for expression of studied apoptotic genes was found 1.71 fold lower than vector control; while there was no apparent change observed in ND3:114A expressing cells (Fig. 2C & D). Interestingly, analysis of expression pattern of genes from anti-apoptotic family showed a similar pattern as that of apoptotic family (0.92 fold for ND3:114A and 0.541 fold for ND3:114T). The expression profile of a representative gene, Caspase 3, with high difference in expression at mRNA level was confirmed at protein level through Western Blot (Supplementary Figure 5*).
Figure 2.
Cumulative Fold change ratio of Apoptotic/Anti-Apoptotic genes represented in Bars: Relative Cumulative Fold change ratio of -(A) Apoptotic genes (TRAIL + DR4 + DR5 + CASP8 + CASP3 + CYCC); and (B) Anti-Apoptotic genes (DCR1 + DCR2 + FLIPL + FLIPS + Bcl2 + CASP3S + CASP8L); Ratio of - (C) Apoptotic/Anti-Apoptotic genes; and (D) Anti-Apoptotic/Apoptotic genes.
Differential expression of genes in response to ND3 over-expression is mediated by epigenetic regulation
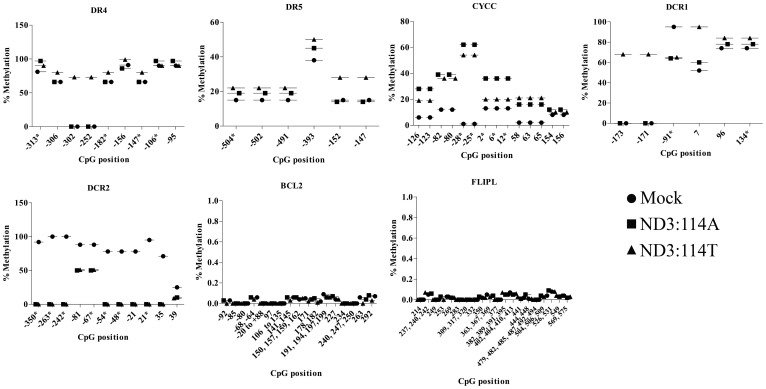
The impact of ND3 over-expression mediated ROS was further assessed on the methylation status of 256CpG sites distributed within the promoter regions of randomly selected 8 genes: DR4, DR5, BCL2, DCR1, DCR2, FLIP CASP8 & CYCS (Ensemble ID provided in supplementary Table 2) to find out if methylation played a role in differential expression of these genes. An expected co-relation between real time expression and promoter methylation was observed at selective CpG islands in the studied regions. For example, hypermethyaltion at −313, −306, −302, −252 −182 −156, −147, −106 and −95 positions for DR4; −504, −502, −491 and −393 positions for DR5; and −126, −123, −82, −80, −28, −25, +2, +6, +12, +58, +63, +65, +154, and +156 positions for CYCC, of promoter regions (Fig. 3) was strongly co-related with down regulated expression of these genes in response to ND3 over expression. The bioinformatics analysis for transcription factor binding at these positions further supported the conclusions drawn (supplementary Table 3). For example, deleting the CpG island at −106 or −95 of DR4 resulted in the loss of binding sites for transcription factor E47, ADR1 and HSF. Similarly, hypermethylated −173, −171 and +7 positions for DCR1 correlated in a similar manner. However, a hypomethylated situation at positions −350, −263, −242, −81, −67, −54, −48, −21, +21, +35 and +39 for DCR2 and at a single CpG position 55 for BCL2 did not show a clear correlation with their enhanced expression. The methylation status for CASP8 and FLIP was observed similar throughout the studied CpG islands, which probably leaves a scope for an extensive investigation involving other CpG islands within their promoter regions. Similarly, identically methylated or hypo methylated status at −84 and −60 positions for DR4; −152 and −147 position for DR5 and −91, +96 and +134 positions for DCR1, was not in accordance with their real time expression, probably suggesting the role of alternative CpG positions for the regulation of gene expression. To check if the methylation pattern could be reversed in the presence of an anti-oxidant, an analysis of methylation pattern at CpG islands within promoter of selected set of genes after treatment of cells with N-acetyl cystein was assessed. The trend of methylation within studied CpG islands showed prevention of hypermethylation at the sites which otherwise were found in hypermethylated state. For example, all the observed hypermethylated CpG sites for CYCC, DR5 and DR4 were found to be hpomethylated after NAC treatment. Similarly, for DCR2, the CpG sites at −350, −54, −48, 35 and 39 showed methylation restoration after NAC treatment, which was hypomethylated in ND3 over-expression situation. Although other CpG sites at −263, −243, −81 and −67 position did not show the corresponding change. For DCR1, CpG at −249, −237, −235, 7 and 134 showed methylation restoration after NAC treatment, which was hypomethylated in ND3 over-expression situation. Whereas, CpG at −173, −171, −91 and 96 did not show the corresponding change, though. The reversal under anti-oxidant conditions at most of the CpG sites did support the observation we made. A complete list of methylation status is provided in dataset 1.
Figure 3. Differential methylation pattern in promoter regions of representative apoptotic and anti-apoptotic genes in response to ND3 Over expression mediated ROS.
The CpG islands with a minimum of 5% difference are shown. (*) denotes bio-informatically predicted important sites for transcription factor binding. A complete set of the bio-informatically predicted important CpG sites is detailed in dataset 1.
ND3 over-expression mediated ROS regulate metabolic reprogramming and induction of pro-cancerous features
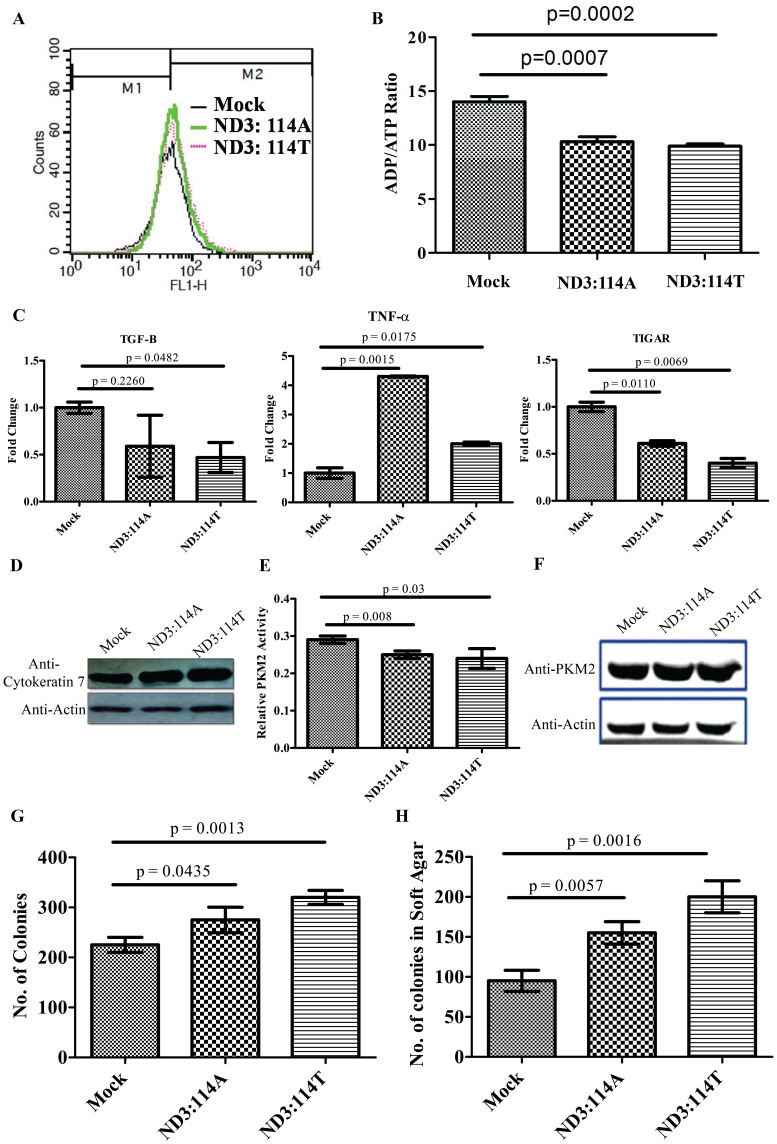
In the background of activation of studied pro-cancerous feature of: glucose uptake and lactate production, six additional independent parameters of cell survival, transformation and metabolic reprogramming of cancer cells were analyzed with respect to the altered ratio of apoptotic/anti-apoptotic genes mediated by enhanced ROS in cells expressing either of the variants of ND3. The cell survival signal and enhanced proliferation, relatively higher in ND3:114T, was observed in cells expressing either of the variant, evident from elevated mitochondrial membrane potential; where cells over-expressing ND3:114A showed 16% higher membrane potential and cells over-expressing ND3:114T showed 22% higher membrane potential than mock transfected cells (Fig. 4A) and lower ADP/ATP ratio (Fig. 4B). In addition, the relative expression of cell growth inhibitory cytokine TGF-β decreased by 0.59 fold for ND3:114A and 0.47 fold for ND3:114T; while the relative expression of pro-inflammatory growth promoting cytokine TNF-α was higher by 4.3 fold for ND3:114A and 2 fold for ND3:114T. The relative expression of ROS mediated apoptosis inducer, TIGAR, also decreased in the same manner (0.61 fold for ND3:114A and 0.40 fold for ND3:114T) (Fig. 4C). Altogether, effect of this alteration on cell transformation for cancer promotion was evident from the level of cell surface marker cytokeratin 7 (Fig. 4D). While monitoring pro-cancerous metabolism, the activity of pyruvate kinase (PK)-M2 when checked showed down regulation by 13% for ND3:114A and 17.24% forND3:114T as compared to vector control (Fig. 4E), without affecting its expression (Fig. 4F). The resultant effect of this altered expression/activity of regulatory and metabolic factors was found to promote both anchorage independent and dependent growth (Fig. 4G and H). Based on an extensive in-vitro observation of ROS promoting nature of either of the variants of ND3, we investigated the endogenous status of mitochondrial coded ND3 at transcript level in four breast tumor tissue samples, in a preliminary pilot study. This was attempted to find out if the observations made in-vitro of the elevated ROS, due to over-expression of ND3, could be true for tumors which usually show an elevated ROS as one of the hallmarks. When compared for the ND3 expression status between the studied tumors and matched normal tissues, we observed a very high level of ND3 at mRNA level ranging from 25 to 648 fold (Supplementary Figure 6), except for one tumor tissue with ND3 expression approximately five fold lower, apparently supporting the phenomenology observed in in-vitro experiments, resulting in generating pro-cancerous features in cells.
Figure 4.
(A) FACS analysis for mitochondrial membrane potential; (B)Bars representing relative ADP/ATP ratio; (C) Real Time fold change for TGF-β, TNF-α and TIGAR; (D) Croped Western blot for cell surface marker Cytokeratin 7 (Full length blots are provided in Supplementary figure 3); (E) Bars showing relative Pyruvate Kinase (PK) M2 activity; (F) Western blot of PKM2 and actin in cells transfected with empty vector, ND3:114A and ND3:114T (G) Bars representing number of colonies in colony forming assay & (H) Bars showing number of colonies in soft agar assay.
Discussion
Mitochondria play a central role in cellular redox and energy homeostasis; in addition to controlling apoptotic and metabolic pathways. The energy generating pathway, which is the major source of reactive oxygen species (ROS) production in mitochondria, involves enzyme complexes located in inner mitochondrial membrane; and is bi-genomic in nature with only some of its subunits coded by mitochondrial DNA. Variations within mtDNA, both germline13,14,29,33 and somatic13,34,35,36, have been equivocally associated with susceptibility towards cancer; and their differential expression has been the focus of several studies to understand mitochondrial genome biology and function in cancer37.
One of the major challenges in exploring the possible role of specific mitochondrial genomic variation(s) and over-expressed mitochondrial gene(s) in a cell is the biased nature of the codons of mitochondrial genes. This has been overcome by codon optimization and artificial gene synthesis, followed by an exogenous expression within the mitochondria of a cell10,11. Although alternative approach of cybrids38 has been one of the routinely used choices but has limitations of ignoring other mitochondrial genetic variations within the DNA of enriched mitochondria, besides the absence of a proper control. We have used codon optimized commercially synthesized ND3 gene cloned in mitochondrial localization vector system to study the functional role of one of its non-synonymous germline polymorphism G10398A (ND3: p.A114T), which was initially found associated with sporadic breast cancer susceptibility in Indian13,14 and African26 population. However, the mechanism for such an association has remained elusive and equivocal. The patho-physiology of cancer in association with mtDNA variations is suggested to be a manifestation of elevated ROS, reported as a mitogenic mediator and as an inducer of apoptosis39,40,41. In addition, elevated ROS regulates retrograde signaling42,43 and epigenetic modifications44; as reported in tumors when compared to matched normal tissues45. Despite all these advancements, the basic reasons for the elevation of mitochondrial ROS in age dependent late onset and complex disorder like cancer is hardly explained in the mitochondrial genomic background. Further, the involvement of mitochondrial ROS in tumorigenic transformation and growth promotion as a result of epigenetic and metabolic reprogramming of cancer cells is gaining importance and needs attention.
Here, we analyzed the differential capacity of a mitochondrial genomic variation on redox homeostasis, high ROS mediated methylation, altered pro-apoptotic/anti-apoptotic homeostasis and modulation of cancer cell metabolism in mtND3:114A and mtND3:114T stably expressing cells, which showed an enhanced - complex I activity, differential levels of ROS and oxidative stress. This was true for both the mitochondrial genetic backgrounds at 10398 position within the ND3 gene, using HeLa cells (mtND3:114A), representing M haplogroup and MCF-7 & MDA-MB 231 cell lines with 10398A (mtND3:114T), representing N haplogroup, with a relatively higher level in the latter. The resultant effect of high production of ROS and the oxidative stress through carbonylated protein content was also higher in general for proteins involved in cell proliferation pathways. The ROS mediated carbonylation of a number of regulatory and metabolic proteins, generally involved in cell proliferation pathways, have been shown to modulate their activity at post-translational level in a way to provide gain/loss of function. For example, the signaling mediated through platelet derived or epidermal growth factor have been found to begin with an oxidative burst, where inhibiting ROS results in inhibition of normal tyrosine kinase pathway induced by growth factor addition46. Another such example is inhibition of metabolic enzyme pyruvate kinase M2, where exogenous supply of diamide results in its inhibition through cysteine corbonylation and subsequent growth promotion, both in vitro and in vivo32. Although such modulation through endogenous mitochondrial ROS is not well known, the similar effect of ND3 over-expression mediated ROS on total protein carbonylation and reduced activity of pyruvate kinase M2 from our study provides a direct proof of the involvement of mitochondrial ROS. In addition, similar effect as that of insulin mediated ROS signaling47 or compromised PKM2 activity as a virtue of mis-sense mutations in PKM248 on high glucose uptake and lactate production, along with the potential to form anchorage dependent/independent colonies, confirms the pro-cancerous potential provided by the mitochondrial ROS. To investigate the probable mechanism for the pro-cancerous phenotypes, the pattern of real time expression of a select set of genes involved in pro-apoptotic, anti-apoptotic and DNA damage pathways, whose expression is generally altered in cancer45,49 was studied in cells with high ROS generated as a consequence of mtND3 over expression. The down regulated expression of both extrinsic apoptotic pathway, such as TRAIL, DR4, DR5; and intrinsic apoptotic pathway genes, such as CASP8, CASP3 and CYC supported our previous observation of the preferential prevalence of the germline mitochondrial variant, 10398A (mtND3:114T), in cancer patients13,14. Although the down-regulation of apoptotic genes was obvious in both 114A and an 114T stably expressing cells in comparison to vector control; however, the trend was more severe in ND3:114T expressing cells, suggesting the involvement of both the germline backgrounds in providing susceptibility to cancer, depending on the aberrant genomic background or expression status of critical nuclear genes. Another level of evidence to favour the pro-cancerous role played by mtND3 gene over-expression mediated ROS signaling of specific nuclear genes was observed in the expression pattern of anti-apoptotic pathway like DCR1, DCR2, FLIPL, FLIPS, BCL2, CASP3S, CASP8L with a relatively increased expression in ND3:114T stably expressing cells, supporting further the chances of attaining a pro-cancerous state by these cells. Concomitantly, the expression of DNA damage pathway genes, such as MDM2, P53, P21, H2AX, ATM, CHK2, when compared to vector control under similar experimental conditions of over expressed mtND3 gene and generation of high ROS, apparently reflected a role in cell survival and proliferation with high mitochondrial membrane potential and decreased ADP/ATP ratio. It is already known that treatment of cells with a classical uncoupler like carbonyl cyanide m-chlorophenlhydrazone (CCCP) reduces mitochondrial membrane potential and enhances ROS production50. This illustrates that, treatment with uncoupler reduces mitochondrial membrane potential and there is an inverse relation between uncoupler mediated reduced mitochondrial membrane potential and ROS. This effect is a proposed mechanism of TRAIL mediated enhanced killing of cells through elevated ROS, demonstrating that the enhanced mitochondrial membrane potential and elevated ROS in ND3 over-expressed situation are independent phenomena, where observation of high ROS could primarily be due to an altered complex I activity. An expected co-relation between real time expression and promoter methylation (hyper/hypo) at selective CpG islands in the studied regions of a select set of genes was confirmatory for real time expression and the role of ND3 over-expression mediated ROS. Although a more extensive investigation is required for real time expression and methylation study for a global impact of mitochondria generated ROS on the genes involved in critical pro-cancerous pathways, yet the epigenetic modifications, mainly differential methylation pattern for several genes, has been observed in many tumor tissues45,51, nevertheless suggestive of the role mitochondrial genomic background in the ND3 gene at 10398 position could play in promoting pro-cancerous features in a cell. In the background of activation of studied pro-cancerous features, the status of pro-inflammatory growth promoting cytokine TNF-α, growth inhibitory TGF-β and ROS mediated apoptosis inducer, TIGAR when investigated, interestingly depicted an up-regulated expression of TNF-α and down-regulated expression of TGF-β and TIGAR, providing further support to anti-apoptotic processes and the generation of a variety of pro-cancerous features as evident from elevated level of cell surface marker cytokeratin 7 and the observations made of high glucose uptake and lactate production, and the formation of anchorage independent/dependent colonies. Interestingly, all these pro-cancerous features were acquired by a cell under a given germline condition of mitochondria with a potential to produce high ROS.
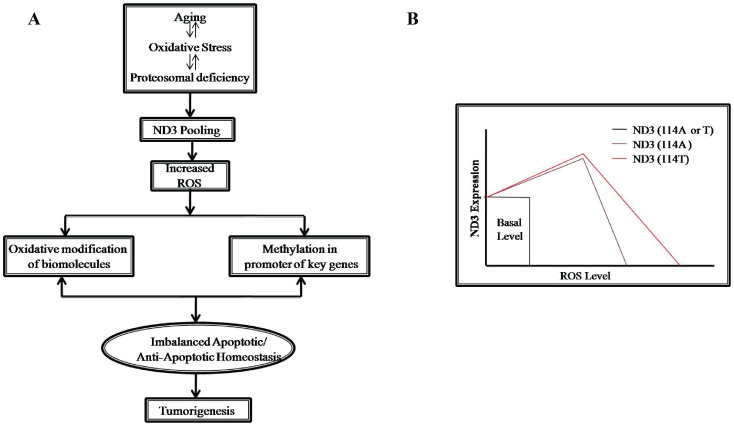
Based on the extensive in-vitro observation of mtND3 over-expression mediated ROS generation and the known concept of elevated ROS as one of the hallmarks of tumor tissues, a preliminary attempt to find out if the mtND3 expression status differed between tumor and matched normal tissues; a very high level of expression was observed of ND3 at mRNA level, ranging from 25 to 648 fold, except for one tumor tissue with ND3 expression approximately five fold lower, apparently supporting the phenomenology observed in in-vitro experiments documented by us which favored generation of pro-cancerous features in cells. In the background of these observations and recent reports showing modulation of mitochondrial diseases as a consequence of compromised activity of Ubiquitin/proteosomal system52, we propose a model where pooling of mitochondrial DNA coded genes, mainly ND3, due to age dependent/Independent proteosomal deficiency lead to elevated ROS; which in turn destabilize apoptotic/anti-apoptotic ratio through epigenetic regulation, facilitating precancerous transformation of cells (Fig. 5A). Further, as proposed in this study, these effects are more pronounced in ND3:114T situation, providing an extra susceptibility towards cancer development (Fig. 5B). Unraveling in future the role of other aberrantly-expressed mitochondrial genes and their interaction with or influence on the diverse nuclear genomic backgrounds, influencing diverse signaling mechanisms within a cell to tilt the balance towards attaining an aberrant status, would be of interest to explore.
Figure 5.
(A) Proposed model for ND3 over expression mediated ROS elevation and its effect in tumorigenesis; and (B) Proposed model for higher susceptibility towards sporadic breast cancer in mitochondrial haplogroup N (i.e, mitochondrial DNA 10398A or ND3:114T) in the germline genomic background of high producer of ND3 with a potential to produce relatively higher ROS than 10398G or ND3:114A (haplogroup M).
Methods
Cell culture and characterization of the germ-line status at 10398 nucleotide position
HeLa, MCF-7, MDA-MB 231, HepG2, H1299, A459, MiaPaCa, PANK1, HEK293T and SiHa cell lines were grown in DMEM medium supplemented with 10% Fetal Bovine Serum, containing penicillin/streptomycin (1%) in a humidified incubator at 37°C and 5% CO2. Genomic DNA from cells grown in a monolayer was isolated, using Genelute mammalian genomic DNA miniprep kit (Sigma). Mitochondrial DNA region, encompassing ND3 gene and involving 10398 position, was amplified by primer sets: forward 5′- CCTTTTACCC CTACCATGAG-3′ and reverse 5′- ATTATTCCTTCTAGGCATAGTA-3′ and subjected to RFLP study after restriction digestion by DdeI restriction endonuclease. The amplicon with “G” nucleotide at 10398 position of mitochondrial DNA created an additional site for DdeI restriction enzyme, generating3 fragments (185 bp, 86 bp and 38 bp) as compared to only two (223 bp and 86 bp) when “A” nucleotide was present at the same position53 (supplementary Figure 1). DNA sequencing of ND3 gene was performed using Big Dye terminator v3.1 cycle sequencing kit on ABI 3100 (Applied Biosystems, Foster City) automated sequencer after the amplification of ND3 gene, using 5′-CTTACCACAAGGCACACCTACA-3′ (forward) and 5′-GGCACAATATTGGCTAAGAGGG-3′ (reverse) primers, followed by sequencing, using 5′- TAATATTTCACTTTACATCCA- 3′ primer to check the presence of any endogenous variations. In HeLa cells, a non-synonymous variation at position 28 within the gene, changing 10th amino acid from Asparagine to Aspartic acid (ND3:N10D), was observed. The cell line, however, was included in the study since it was the only line available out of the pool of 10 cell lines, representing “M” haplogroup.
Cloning and site directed mutagenesis
10398A variant of ND3 gene (ND3:114T) was commercially synthesized using long range gene synthesis (GenScript USA Inc.) in nuclear format. The gene was sub cloned into Sal1 and Not1 site of mitochondria targeting plasmid vector, pCMV-myc-mito (In-vitrogen, USA), with N-terminal mitochondrial targeting sequence (MTS) and C-terminal myc epitope. Site directed mutagenesis was carried out by long range PCR method, using quick change site directed mutagenesis kit (Stratagene, USA) to generate 10398G variant (ND3:114A). Positive clones of ND3:114T and ND3:114A were screened by single colony PCR and insert release; and finally confirmed by DNA sequencing with ABI Big Dye cycle sequencing kit (Applied Biosystem, USA). The primers used for site directed mutagenesis were, 5′- GGCCTGGACTGGGCCGAGTGAGCGG-3′ (forward) and 5′-CCGCTCACTCGGCCCAG TCCAGGCC-3′ (reverse).
Transfection, generation of stable cell lines, immuno-fluorescence and Western blot
Before transfection, 2 × 106 cells were seeded in 6 well plate. Plasmid constructs were linearized, using EcoR1 restriction endonuclease and transfection was carried out at 60–80% confluency of cells by Lipofectamine™ LTX reagent (Invitrogen USA) according to manufacturer's protocol. For generating stable lines expressing ND3:114A, ND3:114T, mock (empty vector transfected), the cells were splitted after 48 hrs of transfection at a ratio of 1:10; and grown in complete medium, containing 400–1000 μg/ml G418, for 15 days with a regular change of medium every alternate day. Mitochondrial localization of exogenously expressed ND3 variants was confirmed using fluorescence microscopy. In brief, cells (1 × 105) were reseeded over cover slips in a 6-well plate. Cells were stained with 25 nM mitotracker red at 37°C for 45 minutes, followed by fixing with 3.7% paraformaldehyde for 20 minutes at room temperature. Permeabilization and blocking were carried out with 5% chicken serum in phosphate buffer saline containing 0.1% triton × 100, for 1 hr and incubated with rabbit anti-myc primary antibody overnight at 4°C. This was followed by 1 hr incubation with alexa 488 linked chicken anti-rabbit secondary antibodies. Excess of secondary antibody was washed off with phosphate buffer saline and cells were stained with 1 μg/ml DAPI for 12 minutes at room temperature. Mounting was done using Prolong Gold Antifade Reagent (Invitrogen) and images captured with Olympus XL 100 fluorescence microscope.
For Western blot, proteins (10–30 μg) from whole cell lysate or mitochondria-rich-fraction was separated on 12% SDS-PAGE, transferred to nitrocellulose membrane (mdi, USA) overnight at 4°C; and probed with anti-myc, anti-PKM2, anti-COIV and anti-β-actin (Cell signaling Technology, USA) primary antibodies. This was followed by incubation with appropriate secondary antibody for 1 hour at room temperature and detected using enhanced chemiluminescence kit (Thermo Scientific, USA).
Preparation of mitochondria enriched fraction and NADH Ubiquinone oxidoreductase (mitochondrial complex I) assay
Mitochondria rich fraction was prepared from both ND3 variants and mock transfected stable cells, using mitochondrial isolation kit (Thermo Scientific, Rockford, USA), according to manufacturer's protocol. The mitochondrial pellet was suspended at a concentration of 1 μg/μl and stored at −80°C till complex I activity was assayed. The assay was performed after freeze-thawing thrice the mitochondrial solution to disrupt the mitochondrial membrane, followed by the protocol of Janseen et.al (2007)54. Here, 2, 6-dichloroindophenol (DCIP) was used as terminal acceptor of electron. In brief, the electron produced as a result of NADH oxidation by complex I was taken up by artificial substrate decyclubiquinone which subsequently delivers the electron to DCIP. The reduction of DCIP was followed spectro-photometrically at 600 nm for 5 minutes. The whole reaction was carried out in 1 ml incubation volume containing 25 mM potassium phosphate, 3.5 mg/ml BSA, 60 μM DCIP, 70 μM decyclubiquinone, 1 μM antimycin-A, and 20 μM NADH. Reaction was started by adding 40 μg of mitochondrial enriched fraction and monitored for 5 minutes at room temperature.
ADP/ATP ratio measurement
The ADP/ATP ratio was measured through ApoSENSOR™ ADP/ATP Ratio Bioluminescence Assay Kit, according to manufacturer's instructions. In brief, 5000 cells seeded in 96 well plate were lysed in buffer supplied with the product and assayed ADP/ATP ratio. The assay utilizes the enzyme luciferase to catalyze the formation of light from ATP and luciferin, where the light was measured using a luminometer. ADP level was measured by its conversion to ATP that was subsequently detected using the same reaction.
Reactive oxygen species assessment
The amount of intracellular oxidant production was measured using the cell permeable fluorescent dye DCFH-DA (Sigma Aldrich). Mechanism of fluorescence included hydrolysis of DCFH-DA by cellular esterases to non-fluorescent DCFH which was further oxidized to fluorescent dichlorofluorescin (DCF) by the action of cellular oxidants. Higher fluorescence in samples indicated a higher generation of reactive oxygen species. A day before the experiment, stably transfected cells were grown in monolayer under similar experimental conditions. For measuring intracellular ROS, cells were washed twice with 1 ml of PBS followed by trypsinization and re-suspending in 1 ml of fresh complete medium, containing a final concentration of 10 μM of DCFH-DA. The cells were incubated at 37°C for 45 minutes in humidified CO2 incubator. Followed by incubation, cells were washed with 1 ml of PBS and finally re-suspended in 200 μl of PBS. The fluorescence was measured using fluorescent activated cell sorting (BD Biosciences, USA) system. Cells pretreated with 200 μM H2O2 for 30 minutes were used as a positive control; and for the negative control, cells were treated with 200 μM ascorbic acid for 3 hours.
Protein carbonyl estimation
The amounts of oxidized proteins were assessed using spectrophotometer by measuring the amount of 1, 2-Dinitrophenylhydrazone formed. 1, 2-Dinitrophenylhydrazine binds very specifically to the carbonylated amino acids within the proteins generated through their oxidation to form the product 1, 2-Dinitrophenylhydrazone, which shows absorbance at 360 nm. For measuring protein carbonyl content in cells stably transfected with ND3:114A or ND3: 114T, an equal amount of whole cell lysate was precipitated using 10% TCA. The protein precipitate was treated with 2N HCl, containing 10 mM Dinitrophenyl hydrazine, at 15°C for one hour. The reaction mixture was centrifuged and washed three times with ethanol-ethyl acetate (1:1). The final precipitate was dissolved in 6 M guanidine chloride or 8 M urea and the absorbance measured at 360 nm. The carbonyl content was measured as nmoles/mg of protein using a molar extinction co-efficient of 22000−1.
Aconitase assay
Aconitase activity was measured through Isocitrate-NADP coupled reaction55. In brief, a 3 ml reaction mixture containing Tris (36 mM), citric acid (0.07 mM), β-Nicotinamide Adenine Dinucleotide Phosphate (0.18 mM), Manganese Sulfate (1.3 mM), Ferrous Ammonium Sulfate (0.0008 mM), L-Cysteine (0.08 mM) and Isocitric Dehydrogenase (0.7 unit) was pre-equilibrated at 30°C. The activity of Aconitase was measured through a continuous spectrophotometric rate determination method at a wavelength of 340 nm using 50 μg of mitochondrial enriched fraction.
Glucose consumption and lactate production assays
Stably transfected cells were grown in 24 well plate at a density of 5 × 105. 96 hour after seeding, media was collected from the wells and glucose uptake was analyzed using glucose (Hexokinase) assay kit (Sigma aldrich) as per the manufacturer's instructions. Lactate production was analyzed using lactate assay kit (BioVision, USA) by following manufacturer's specifications. All the measurements were normalized to cell numbers as described earlier47.
Pyruvate kinase M2 (PKM2) activity assay
Stably transfected cells were lysed using Qproteome mammalian cell lysis buffer (Qiagen) containing appropriate protease inhibitor cocktail (Qiagen). Protein concentration was estimated using pierce BCA kit. 5 μg of whole cell lysate was used for PKM2 activity assay in a NADH-LDH coupled reaction as described earlier56.
RNA Isolation, cDNA preparation and Real time for nuclear gene expression
RNA from stably transfected cells, and a representative set of tumor and matched normal tissues was isolated by standard phenol-chloroform extraction. 4 μg of RNA was used to synthesize cDNA, using reverse transcription kit (Applied Biosystems, Foster City, CA, USA). Commercially available Taqman Gene expression Assay system (Applied Biosystems, Foster City, CA, USA) was used to quantitate transcript levels of genes related to DNA damage response, apoptotic and metabolic pathways: TRAIL, DR4, DR5, DCR1, DCR2, CASP3, CASP3S, CASP8, CASP8L, FLIPL, FLIPS, BCL2, CYCS, ATM, TP53, CHEK2 and H2AX, MDM2, P21, TGF-B, TIGAR and ND3, in cells stably transfected with ND3:114A or ND3: 114T or mock (vector alone). GAPDH, B-Actin, PUM1, and MRPL19 (Applied Biosystems) were used as endogenous controls. Quantitative real-time PCR was carried out using an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Threshold cycle (Ct) numbers were established by using SDS 1.1 RQ software (Applied Biosystems). All the reactions were carried out in duplicate. The normalization factor obtained from GeNorm software was used to compute normalized expression for the target genes as described earlier45.
Bisulfite conversion, In vitro transcription, T cleavage assay, Mass Spectrometry and Methylation study
A set of genes whose expression differed from mock transfected stable cells, was selected for further methylation studies. Primers were designed using epi-designer software (Sequenom, San Diego, CA, USA) to cover the regions with enriched CpG sites. The selected amplicons were mostly located in the promoter region of the genes or covered the promoter and the first exon. AT7-promoter tag was added to the reverse primer and a 10 mer-tag sequence was added to the forward primer to balance the PCR primer length (supplementary Table 1). Genomic DNA from stably transfected cells was isolated using QIA amp DNA mini kit. The bisulfite conversion was carried out according to manufacturer's protocol, using EZ-96 DNA Methylation Kit (Zymo Research, Orange, CA, USA). The C/T conversion reaction was performed using the PCR program as follows: 95°C for 30 sec and 50°C for 15 min, which was repeated for 46 cycles. The bisulfite treated genomic DNA was amplified using Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany) (4 min at 95°C followed by 45 cycles of 20 sec at 95°C, 30 sec at 62°C, and 1 min at 72°C with a 3 minute final extension). Methylated and unmethylated positive control human DNA was procured from Sequenom. Fully methylated DNA was mixed with pure unmethylated DNA in a ratio of 100:0, 60:40, 40:60, and 0:100. Unincorporated dNTPs were dephosphorylated by adding 1.7 μl H2O and 0.3U shrimp alkaline phosphatase (Sequenom, San Diego, CA, USA), followed by incubation at 37°C for 40 min and heat inactivation of shrimp alkaline phosphatase at 85°C for 5 min. In general, 2 μl of the PCR product was directly used as a template in a 5 μl transcription reaction. T7 RNA and DNA Polymerase (20U) (Sequenom, San Diego, CA, USA) was used to incorporate dTTP in the transcripts. Ribonucleotides were used at 1 mmol/l and the dNTP substrate at 2.5 mmol/l. RNaseA enzyme (Sequenom, San Diego, CA, USA) was added in the same step to cleave the in vitro transcripts (T-cleavage assay). The T cleavage reaction was carried out at 37°C for 3 hours and further diluted with H2O to a final volume of 27 μl. Conditioning of the phosphate backbone was achieved by adding 6 mg of Clean Resin (Sequenom, San Diego, CA, USA) before performing MALDI-TOF MS on Sequenom Mass ARRAY. 22 nl of cleavage reaction was robotically dispensed onto a silicon matrix pre-loaded chips (SpectroCHIP; Sequenom, San Diego, CA, USA), and the mass spectra obtained using the MassARRAY. Methylation ratios were generated by the MALDI-TOF and EpiTYPER software v1.0 (Sequenom, San Diego, CA, USA). The assay was able to discriminate between the methylated and unmethylated components of the positive control according to the ratios.
Author Contributions
R.K.S., A.S., P.K., S.M. and R.C. performed the experiments. R.N.K.B. conceptualized; R.K.S. and R.N.K.B. designed the experiments and wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
mtDNA germ line variation mediated ROS generates retrograde signaling and induces pro-cancerous metabolic features
Dataset 1
Acknowledgments
R.K.S. was supported by fellowship from Council of Scientific and Industrial Research, India.
References
- Galluzzi L., Kepp O., Trojel-Hansen C. & Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ Res 111, 1198–1207 (2012). [DOI] [PubMed] [Google Scholar]
- Nunnari J. & Suomalainen A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J. D. & Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9, 383–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981). [DOI] [PubMed] [Google Scholar]
- Elliott H. R., Samuels D. C., Eden J. A., Relton C. L. & Chinnery P. F. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet 83, 254–260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon V. S. & Fenech M. Mutations that affect mitochondrial functions and their association with neurodegenerative diseases. Mutat Res 759, 1–13 (2014). [DOI] [PubMed] [Google Scholar]
- Kazuno A. A. et al. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet 2, e128 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. A mitochondrial bioenergetic etiology of disease. J Clin Invest 123, 1405–1412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros J. A. et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A 102, 719–724 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. et al. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci U S A 104, 7540–7545 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Hoque M. O., Upadhyay S. & Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res 68, 700–706 (2008). [DOI] [PubMed] [Google Scholar]
- Lam E. T. et al. Mitochondrial DNA sequence variation and risk of pancreatic cancer. Cancer Res 72, 686–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochhait S., Bhatt A., Sharma S., Singh Y. P., Gupta P. & Bamezai R. N. Concomitant presence of mutations in mitochondrial genome and p53 in cancer development - a study in north Indian sporadic breast and esophageal cancer patients. Int J Cancer 123, 2580–2586 (2008). [DOI] [PubMed] [Google Scholar]
- Darvishi K., Sharma S., Bhat A. K., Rai E. & Bamezai R. N. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett 249, 249–255 (2007). [DOI] [PubMed] [Google Scholar]
- Li X. Y., Guo Y. B., Su M., Cheng L., Lu Z. H. & Tian D. P. Association of mitochondrial haplogroup D and risk of esophageal cancer in Taihang Mountain and Chaoshan areas in China. Mitochondrion 11, 27–32 (2011). [DOI] [PubMed] [Google Scholar]
- Booker L. M. et al. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol 175, 468–472; discussion 472–463 (2006). [DOI] [PubMed] [Google Scholar]
- Liu V. W. et al. Mitochondrial DNA variant 16189T > C is associated with susceptibility to endometrial cancer. Hum Mutat 22, 173–174 (2003). [DOI] [PubMed] [Google Scholar]
- Arnold R. S. et al. An inherited heteroplasmic mutation in mitochondrial gene COI in a patient with prostate cancer alters reactive oxygen, reactive nitrogen and proliferation. Biomed Res Int 2013, 239257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vioque E., Bornstein B., Gallardo M. E., Fernandez-Moreno M. A. & Garesse R. The pathogenicity scoring system for mitochondrial tRNA mutations revisited. Mol Genet Genomic Med 2, 107–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin A. et al. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem 283, 20907–20913 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt J. M. et al. Mitochondrial Polymorphisms Significantly Reduce the Risk of Parkinson Disease. The American Journal of Human Genetics 72, 804–811 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt J. M. et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett 365, 28–32 (2004). [DOI] [PubMed] [Google Scholar]
- Kato T., Kunugi H., Nanko S. & Kato N. Mitochondrial DNA polymorphisms in bipolar disorder. J Affect Disord 62, 151–164 (2001). [DOI] [PubMed] [Google Scholar]
- Juo S. H. et al. A common mitochondrial polymorphism 10398A > G is associated metabolic syndrome in a Chinese population. Mitochondrion 10, 294–299 (2010). [DOI] [PubMed] [Google Scholar]
- Niemi A. K. et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet 13, 166–170 (2005). [DOI] [PubMed] [Google Scholar]
- Canter J. A., Kallianpur A. R., Parl F. F. & Millikan R. C. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res 65, 8028–8033 (2005). [DOI] [PubMed] [Google Scholar]
- Covarrubias D., Bai R. K., Wong L. J. & Leal S. M. Mitochondrial DNA variant interactions modify breast cancer risk. J Hum Genet 53, 924–928 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzotti A., Kraft P., Hankinson S. E., Hunter D. J., Buring J. & Cox D. G. The mitochondrial A10398G polymorphism, interaction with alcohol consumption, and breast cancer risk. PLoS One 4, e5356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka A. M. et al. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res Treat 121, 511–518 (2010). [DOI] [PubMed] [Google Scholar]
- Leslie N. R., Bennett D., Lindsay Y. E., Stewart H., Gray A. & Downes C. P. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J 22, 5501–5510 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H. & Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem 151, 255–261 (2012). [DOI] [PubMed] [Google Scholar]
- Anastasiou D. et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R. K., Leal S. M., Covarrubias D., Liu A. & Wong L. J. Mitochondrial genetic background modifies breast cancer risk. Cancer Res 67, 4687–4694 (2007). [DOI] [PubMed] [Google Scholar]
- McMahon S. & Laframboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis 35, 1046–1054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larman T. C. et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A 109, 14087–14091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Mambo E. & Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene 25, 4663–4674 (2006). [DOI] [PubMed] [Google Scholar]
- Nikolaev A. I., Martynenko A. V., Kalmyrzeav B. B. & Tarantul V. Z. Altered expression of mitochondrial GENES. Mol Med Rep 5, 1526–1530 (2001). [Google Scholar]
- Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene 25, 4768–4776 (2006). [DOI] [PubMed] [Google Scholar]
- Ogrunc M. et al. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ 21, 998–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Induction of apoptosis in human multiple myeloma cell lines by ebselen via enhancing the endogenous reactive oxygen species production. Biomed Res Int 2014, 696107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. et al. ROS signaling: the new wave? Trends Plant Sci 16, 300–309 (2011). [DOI] [PubMed] [Google Scholar]
- Butow R. A. & Avadhani N. G. Mitochondrial signaling: the retrograde response. Mol Cell 14, 1–15 (2004). [DOI] [PubMed] [Google Scholar]
- Liu Z. & Butow R. A. Mitochondrial retrograde signaling. Annu Rev Genet 40, 159–185 (2006). [DOI] [PubMed] [Google Scholar]
- He J. et al. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep 13, 1116–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R. et al. Investigation of DNA damage response and apoptotic gene methylation pattern in sporadic breast tumors using high throughput quantitative DNA methylation analysis technology. Mol Cancer 9, 303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol 194, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M. A. et al. Insulin enhances metabolic capacities of cancer cells by dual regulation of glycolytic enzyme pyruvate kinase M2. Mol Cancer 12, 72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M. A. et al. Missense mutations in pyruvate kinase M2 promote cancer metabolism, oxidative endurance, anchorage independence, and tumor growth in a dominant negative manner. J Biol Chem 289, 8098–8105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail R. S. et al. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res 60, 6744–6749 (2000). [PubMed] [Google Scholar]
- Chaudhari A. A., Seol J. W., Kang S. J. & Park S. Y. Mitochondrial transmembrane potential and reactive oxygen species generation regulate the enhanced effect of CCCP on TRAIL-induced SNU-638 cell apoptosis. J Vet Med Sci 70, 537–542 (2008). [DOI] [PubMed] [Google Scholar]
- Kim J. H. et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res 21, 1028–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A. et al. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab 19, 642–652 (2014). [DOI] [PubMed] [Google Scholar]
- Torroni A. et al. Classification of European mtDNAs from an analysis of three European populations. Genetics 144, 1835–1850 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A. J. et al. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem 53, 729–734 (2007). [DOI] [PubMed] [Google Scholar]
- Morrison J. F. The activation of aconitase by ferrous ions and reducing agents. Biochem J 58, 685–692 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar K., Gupta V., Koul A., Alam N., Bhat R. & Bamezai R. N. Differential behavior of missense mutations in the intersubunit contact domain of the human pyruvate kinase M2 isozyme. J Biol Chem 284, 11971–11981 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mtDNA germ line variation mediated ROS generates retrograde signaling and induces pro-cancerous metabolic features
Dataset 1