Abstract
Soybean seeds are an important source of vegetable oil and biomaterials. The content of individual triacylglycerol species (TAG) in soybean seeds is difficult to quantify in an accurate and rapid way. The present study establishes an approach to quantify TAG species in soybean seeds utilizing an electrospray ionization tandem mass spectrometry with multiple neutral loss scans. Ten neutral loss scans were performed to detect the fatty acyl chains of TAG, including palmitic (P, 16:0), linolenic (Ln, 18:3), linoleic (L, 18:2), oleic (O, 18:1), stearic (S, 18:0), eicosadienoic (20:2), gadoleic (20:1), arachidic (20:0), erucic (22:1), and behenic (22:0). The abundance of ten fatty acyl chains at 46 TAG masses (mass-to-charge ratio, m/z) were determined after isotopic deconvolution and correction by adjustment factors at each TAG mass. The direct sample infusion and multiple internal standards correction allowed a rapid and accurate quantification of TAG species. Ninety-three TAG species were resolved and their levels were determined. The most abundant TAG species were LLL, OLL, LLLn, PLL, OLLn, OOL, POL, and SLL. Many new species were detected and quantified. This shotgun lipidomics approach should facilitate the study of TAG metabolism and genetic breeding of soybean seeds for desirable TAG content and composition.
Soybean (Glycine max (L) Merrill) is an important source of vegetable oil. The major components of vegetable oil are triacylglycerols (TAGs). It is of great interest to detect and quantify TAG molecular species in soybean seeds. A desirable fatty acid composition in seed oil has been targeted for soybean breeding1,2. One TAG molecule consists of one glycerol backbone and three fatty acyl chains. Due to the wide variation of fatty acyl chains attached to the glycerol backbone, in terms of carbon length and degree of unsaturation, the diversity of TAG species is very high. It is therefore a challenge to detect, differentiate, and quantify hundreds of TAG species in soybean seeds.
The complex TAG species in soybean oil can be separated by liquid chromatography (LC) and then be detected by a mass analyzer, a flame ionization detector3 or an ultraviolet-visible detector4. The general ionization sources used for TAG molecules are atmospheric pressure chemical ionization (APCI), electrospray ionization (ESI), and matrix-assisted laser desorption/ionization (MALDI). Various efficient LC-based separation approaches have been coupled with APCI-MS to analyze TAGs in soybean oil (LC-APCI-MS)5,6,7,8,9,10,11,12. In addition, soybean TAGs have been analyzed by mass spectrometry using MALDI as ion source13. The supercritical fluid chromatography/mass spectrometry has also been applied to profile soybean TAGs14. These various methods reported the relative percentage levels of TAG species in soybean seeds based on mass spectra signals, without adjustment of response factors at different TAG masses. Different TAG species have different response to mass analyzer due to the different total carbon number and double bonds; these differences need to be considered in quantification15. It has not been reported for quantification of TAG species in soybean seeds in a precise way, e.g. nmol mg−1 dry weight, although it is very informative in studying TAG metabolism and genetic breeding.
TAGs can be introduced to a mass analyzer without chromatographic separation. TAG adducts, such as lithium adducts and ammonium adducts, can be introduced into tandem mass analyzers by ESI and detected at neutral loss scan modes (NL-ESI-MS/MS), in which the first and third mass analyzers scan ions simultaneously with an offset mass and the second mass analyzer functions as a collision cell to fragment the intact TAG ions for a neutral loss of one fatty acyl chain15,16,17. The ionization efficiency by ESI and the fragmentation efficiency at a collision cell can impact the overall detection sensitivity of a particular TAG species. The sensitivity of fragmentation of TAG ions is different for different fatty acyl chains due to their carbon chain length, degree of unsaturation, and position on glycerol backbones16,17,18. The sensitivity of detection by the mass analyzer is different for different TAG masses (m/z)15. Those differential sensitivities can affect the quantification of TAG species by NL-ESI-MS/MS.
Lithiated adducts of TAG species from mammalian tissues have been detected and quantified by NL-ESI-MS/MS15,19. The sensitivity, or response factors, at each TAG mass (m/z) were extrapolated from a regressive curve that was fitted by analyzing the detection sensitivities of different synthetic TAG species15. The fatty acyl intensities at each TAG mass were corrected by its corresponding response factor at this mass; and the corrected fatty acyl intensities were then utilized to assemble TAG species and calculate their levels15,19. The ammoniated TAGs from soybean seeds were scanned by NL-ESI-MS/MS and the fatty acyl abundance was analyzed20,21; however, the response factors at different TAG masses were not determined and the individual TAG species were not resolved. Recently, the ammoniated TAGs from Arabidopsis seed extracts were scanned by NL-ESI-MS/MS; the fatty acyl abundance was corrected by the response factor at each TAG mass (m/z); and the levels of TAG species were determined22.
Here, we describe a NL-ESI-MS/MS approach to quantify soybean TAG species by detecting ammoniated TAGs. The fatty acyl abundance acquired from neutral loss scans were corrected by response factors at each TAG mass. We further corrected the detection sensitivity for five major fatty acids in soybean seeds based on the results of gas-chromatography. The corrected fatty acyl abundance was utilized to assemble TAG species and quantify their content. The accuracy of the quantification was evaluated by comparison of fatty acid compositions retrieved from the determined TAG levels and the ones determined by gas chromatography.
Results
Detection of ammonium TAG adducts by neutral loss scans
Five types of fatty acids in soybean seeds were detected by gas chromatography with a flame ionization detector (GC-FID, Supplementary Table S1), including palmitic acid (16:0; 9.5%), linolenic acid (18:3, 8.3%), linoleic acid (18:2; 62.6%), oleic acid (18:1; 17.0%), and stearic acid (18:0; 2.6%). The isomeric fatty acid species, such as 18:1Δ9 and 18:1Δ11, were combined in calculation. Only numbers of carbons and double bonds are considered hereafter in the description of fatty acids. The TAG molecular species were described by total carbon number and the number of double bonds in their three acyl chains, by TAG mass (m/z), and by their fatty acyl combinations. For example, the ammonium adduct of TAG 18:2–18:2–18:3, [M + NH4]+, has a TAG mass charge ratio (m/z) of 894.8, a molecule formula of C57H100O6N, and can be expressed as C54:7. The sn-position of fatty acyl chains on glycerol backbone was not specified in this work.
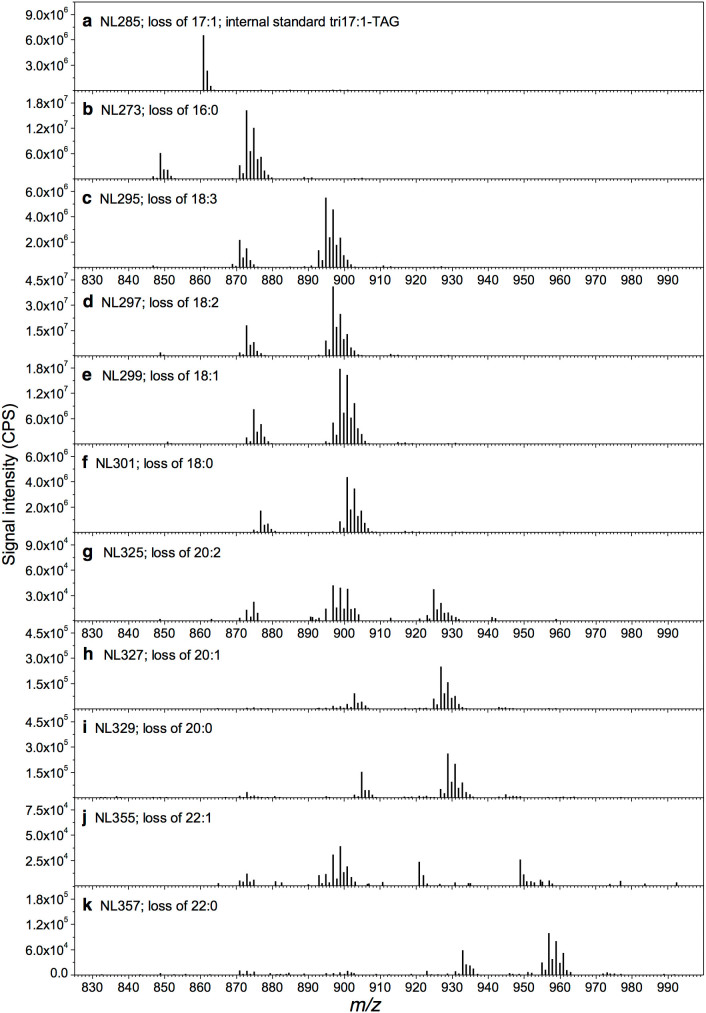
The crude TAG extract from soybean seeds was infused directly into mass analyzers by an electrospray ionization source in a positive mode and the ammoniated TAGs can be detected at multiple neutral loss modes (Fig. 1 and Supplementary Fig. S1). For example, TAG 18:2-18:2-18:3 (m/z 894.8; C57H100O6N) can be scanned at Q1 mass analyzer and fragmentized by collision induced dissociation (CID) at Q2 collision cell. A loss of a neutral fragment, e.g. 295 (18:3 + NH3; C18H33O2N), from a precursor ion, e.g. m/z 894.8 (18:2-18:2-18:3; C57H100O6N), during CID warrants the presence of the precursor ion peak in the neutral loss scan spectrum. The diacylglycerol (DAG) product ions generated in Q2 can be scanned by Q3 mass analyzer (DAG 18:2-18:2; m/z 599.5; C39H67O4). Therefore, Q1 and Q3 mass analyzer scan simultaneously with a mass offset of 295 and the 18:3-containing TAGs can be detected (Fig. 1c). The scans were performed for the fatty acids that were detected by GC-FID, including NL273 (16:0), NL295 (18:3), NL297 (18:2), NL299 (18:1), and NL301 (18:0) (Fig. 1). Five types of trace amount of fatty acids were undetectable by GC-FID but detected by neutral loss scans of NL325 (20:2), NL327 (20:1), NL329 (20:0), NL355 (22:1), and NL357 (22:0; Fig. 1). These five types of fatty acids were detectable in Arabidopsis seeds by neutral loss scans22, suggesting their biological relevance. NL285 scan mode was performed to scan ammoniated tri17:1-TAG, an internal standard that is not naturally occurring and detectable in soybean seeds (Fig. 1a).
Figure 1. Multiple neutral loss scans of ammoniated TAGs in extracts of soybean seeds.
(a) Neutral loss scan of ammoniated tri17:1-TAG (internal standard TAG 17:1-17:1-17:1) in extracts of a seed sample. (b–k) Scanning for precursors of the neutral fragment corresponding to each of the ten fatty acyl chains. The m/z of signals in the spectra was used to determine the identities of each ammonium TAG adduct and the intensity was used to determine TAG amount.
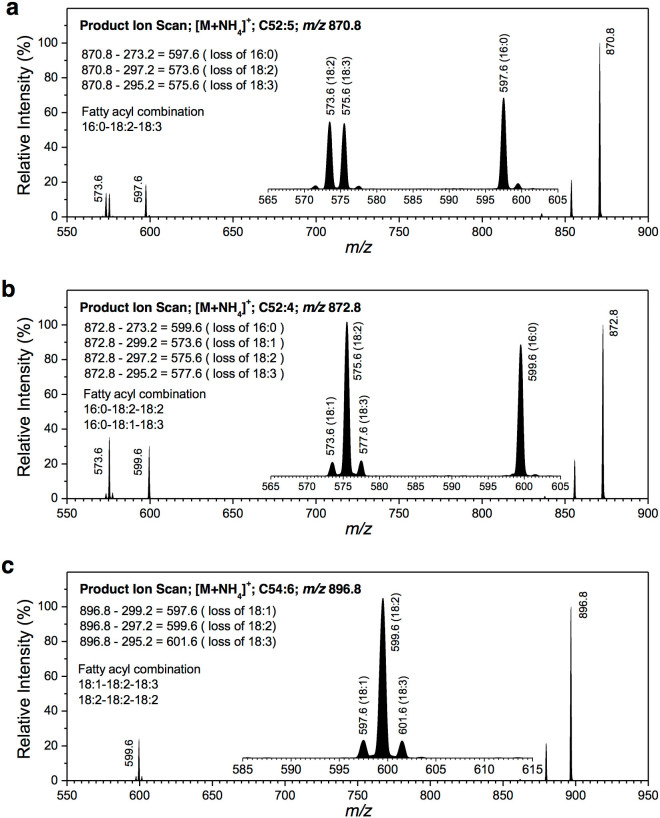
In soybean seed extracts, the majority of fatty acyls of TAG ammonium adducts could undergo neutral loss generally close (Fig. 2). For example, the MS/MS spectrum of C52:5 (m/z 870.8, Fig. 2a) was typical of the characteristic decomposition processes of the [M + NH4]+ ions of TAGs which showed the neutral loss of 273.2 (16:0 + NH3), 297.2 (18:2 + NH3), and 295.2 (18:3 + NH3) to form ions at m/z 597.6, 573.6, and 575.6, respectively. The fatty acyl combinations for m/z 870.8 (C52:5) can be deduced to be 16:0-18:2-18:3. The MS/MS spectrum of m/z 870.8 indicates that the neutral losses of 18:2 and 18:3 were generally close. However, the level of neutral loss of 16:0 tended to be 26% higher than those of 18:2 or 18:3. In addition, the MS/MS spectrum of C52:4 (m/z 872.8, Fig. 2b) showed the neutral loss of 273.2 (16:0 + NH3), 299.2 (18:1 + NH3), 297.2 (18:2 + NH3), and 295.2 (18:3 + NH3) to form ions at m/z 599.6, 573.6, 575.6, and 577.6, respectively. The fatty acyl combinations for m/z 872.8 (C52:4) can be deduced to be 16:0-18:2-18:2 and 16:0-18:1-18:3. The MS/MS spectrum of m/z 872.8 indicates the neutral losses of 18:1 and 18:3 were generally close. However, the efficiency of neutral loss of 16:0 preferred to be 36% higher than expected. Furthermore, the MS/MS spectrum of C54:6 (m/z 896.8, Fig. 2c) showed the neutral loss of 299.2 (18:1 + NH3), 297.2 (18:2 + NH3), and 295.2 (18:3 + NH3) to form ions at m/z 597.6, 599.6, and 601.6, respectively. The fatty acyl combinations for m/z 896.8 (C54:6) can be deduced to be 18:1-18:2-18:3 and 18:2-18:2-18:2. The MS/MS spectrum of m/z 896.8 indicates the neutral losses of 18:1 and 18:3 were generally similar. The amount of fatty acyls of 18:1, 18:2, and 18:3 accounted for 88% of total soybean seed fatty acids while 16:0 for 9.5% (Supplementary Table S1). These results suggest that majority of fatty acyls in TAG ammonium adducts can undergo neutral loss generally close in soybean seed extracts.
Figure 2. Product ion scans of TAG m/zs at 870.8, 872.8, and 896.8 from extracts of soybean seeds.
(a) The theoretical m/z of the singly charged ammoniated ion of C52:5 is 870.8. The generation of product ions of m/z 597.6, 573.6, and 575.6 indicate the neutral losses of 16:0, 18:2, and 18:3, respectively. The potential fatty acyl combination at m/z 870.8 is 16:0-18:2-18:3 with 52 carbons and 5 double bonds on fatty acyl chains. (b) The theoretical m/z of the singly charged ammoniated ion of C52:4 is 872.8. The generation of product ions of m/z 599.6, 573.6, 575.6, and 577.6 indicate the neutral losses of 16:0, 18:1, 18:2, and 18:3, respectively. The potential fatty acyl combinations at m/z 872.8 are 16:0-18:2-18:2 and 16:0-18:1-18:3, which have 52 carbons and 4 double bonds on fatty acyl chains. (c) The theoretical m/z of the singly charged ammoniated ion of C54:6 is 896.8. The generation of product ions of m/z 597.6, 599.6, and 601.6 indicate the neutral losses of 18:1, 18:2, and 18:3, respectively. The potential fatty acyl combinations at m/z 896.8 are 18:1-18:2-18:3 and 18:2-18:2-18:2, which have 54 carbons and 6 double bonds on fatty acyl chains.
Acquirement of fatty acyl signals and isotopic deconvolution
For the spectrum at each NL mode, the background was subtracted; the peaks were smoothed; and the peak areas were integrated using a custom script and Applied Biosystems Analyst software. Based on the overall spectra of the ten NL scans, 46 TAG masses (m/z) were identified that represent C48, C50, C52, C54, C56, C58, and C60 with various degree of unsaturation (Fig. 1 and Supplementary Table S2). Due to the isotopic overlap, the peak signal at each TAG mass needs to be deconvoluted. For example, C54:8 has a monoisotopic peak (M peak) at m/z 892.7 while its second isotopic peak (M + 2 peak) with two 13C isotopic carbons is at m/z 894.8. The M + 2 peak of C54:8 overlaps with the monoisotopic peak of C54:7, which has a m/z 894.8 (Supplementary Fig. S1 and Fig. S2). C54:8 has one more double bond than C54:7, which results in a loss of two hydrogen atoms or a smaller mass of 2 Da.
The peak signal at m/z 894.8 (C54:7) was corrected by subtracting the M + 2 signal of m/z 892.7 (Supplementary Fig. S2e and Supplementary Table S3). The isotopic correction factor at TAG mass M, ZM, was calculated using the following formula: ZM = [(NC * 1.12) * (NC * 1.12)/200 + NO * 0.204)]/100 [1], where NC represents the number of carbon and NO represents the number of oxygen in the DAG product ion from CID of [M + NH4]+. For example, at NL295 (C18H33O2N), the ammonium TAG adduct, C54:8 (m/z 892.7; C57H98O6N), can generate a DAG product ion m/z 597.5 (C39H65O4), in which the NC and NO were 39 and 4, respectively. The calculation of isotopic correction factor was detailed in Supplementary Table S3.
After obtaining ZM, the peak signal at m/z 894.8 (C54:7) can be corrected using the following formula: CIM+2 = IM+2 − CIM * ZM [2], where CIM+2 represents the corrected peak signal (e.g. at m/z 894.8); IM+2 represents the acquired raw mass spectral signal (e.g. at m/z 894.8); CIM represents the corrected signal (e.g. at m/z 892.7); and ZM represent the isotopic correction factor (e.g. at m/z 892.7). Therefore, the contribution of M + 2 peak of m/z 892.7 was removed from the peak signal at m/z 894.8 (C54:7). The detailed process of isotopic convolution was shown using examples of the four most abundant TAG masses scanned at NL295 mode (Supplementary Fig. S2a,e; Supplementary Table S3). The acquired and isotopic deconvoluted signals were shown for the four most abundant TAG masses at scan modes of NL295, NL297, NL299, and NL301 (Supplementary Fig. S2e–h).
Adjustment of fatty acyl signals at each TAG mass
After isotopic deconvolution for peak signals acquired at the NL295 mode, the abundance of 18:3 at four TAG masses was obtained (Supplementary Fig. S2e), and the abundance of 18:3 can also be obtained at the other 42 TAG masses (m/z) using the same isotopic deconvolution process. The detection sensitivities for each of the 46 TAG masses were different22. Such differences were corrected by the adjustment factor at each of the 46 TAG masses. The adjustment factors at the 46 TAG masses, AFTAG, were extrapolated from multiple regressive curves22 (Supplementary Table S2). For example, the adjustment factor for TAG mass, m/z 894.8 (C54:7), was 4.35, which meant that 4.35 moles of C54:7 and 1.0 mole of C51:3 (internal standard; tri17:1-TAG) would generate equal amount of spectra signal when scanned by ESI-MS. Therefore, the abundance of 18:3 at m/z 894.8 (C54:7) was corrected by multiplying 4.35, the adjustment factor at this TAG mass. The detailed adjustment process by AFTAG was demonstrated in Supplementary Table S4.
The 18:3 abundance at each of the 46 TAG mass (m/z) was adjusted by its corresponding AFTAG and was added up to be 72.21 nmol mg−1 dry weight (Supplementary Table S1). The abundance of 18:3 was 60.72 nmol mg−1 dry weight as determined by GC-FID, which was lower than the amount determined by NL-ESI-MS/MS (Supplementary Table S1). This difference of detection sensitivity was further corrected by an adjustment factor for 18:3. The possible reason for a slightly lower detection sensitivity for 18:3 could be due to its slightly preference at sn-2 position, where the fragmentation efficiency could be impeded, or due to the length of carbon chain and degree of unsaturation. Since the neutral loss efficiency is influenced by the fatty acyl length, unsaturation degree, and sn-position on glycerol backbone16,17,18, we adjusted the responses of the major fatty acids in spectrometric scans by gas chromatography analysis. The adjustment factors for a particular fatty acid, AFFA, was defined as a ratio of the fatty acid level determined by GC-FID over the one by NL-ESI-MS/MS. The adjustment factors we measured were comparable with the ones reported previously3 (Supplementary Table S1). The adjustment factors of five major fatty acids in soybean seeds were further utilized to adjust the fatty acyl abundance. The detailed adjustment process was displayed in Supplementary Table S4.
After adjustment by the 46 AFTAG factors vertically and 5 AFFA factors horizontally, a two-dimensional dataset was obtained, in which the horizontal dimension was ten NL modes or fatty acyl abundance and the vertical dimension was 46 TAG masses (Supplementary Table S2). In a vertical view for example, scan mode of NL295 (18:3) showed the most abundant 18:3-containing TAG masses were m/z 892.7 (C54:8), m/z 894.8 (C54:7), and m/z 896.8 (C54:6) (Supplementary Table S2). In a horizontal view, scans of m/z 894.8 (C54:7) generated much stronger signals at NL297 (18:2) and NL295 (18:3) than at the other eight NL scan modes (Supplementary Table S2). The impact of AFTAG was much more profound than AFFA as shown for the adjustment results of the fatty acyl abundance at the five most abundant TAG masses (Supplementary Fig. S3).
Assembly of fatty acyl combinations and determination of TAG levels
The fatty acyl combinations at each of the 46 TAG masses (m/zs) were assembled from a two-dimensional dataset (Supplementary Table S2). For example, TAG mass m/z 872.8 (C52:4) contains various abundance of different fatty acyl chains (Supplementary Table S2). The most abundant fatty acyl chains at C52:4 were 16:0 and 18:2 followed by 18:1 and 18:3 (Fig. 3a). Based on the total carbon number and double bonds on the three acyl chains, two types of fatty acyl combinations were deduced for C52:4, including 16:0–18:2–18:2 and 16:0–18:1–18:3 (Table 1). The level of each fatty acyl combination at a particular TAG mass was calculated by the formulas reported previously22. The level of 16:0–18:2–18:2 was represented by the abundance of 18:2, A18:2. Since 16:0–18:2–18:2 contains two 18:2 acyl chains, the formula to calculate its level was A18:2/2. The level of 16:0–18:1–18:3 was represented by the abundance of 18:1, A18:1, and the abundance of 18:3, A18:3. The formula to calculate the level of 16:0–18:1–18:3 was (A18:1 + A18:3)/2.
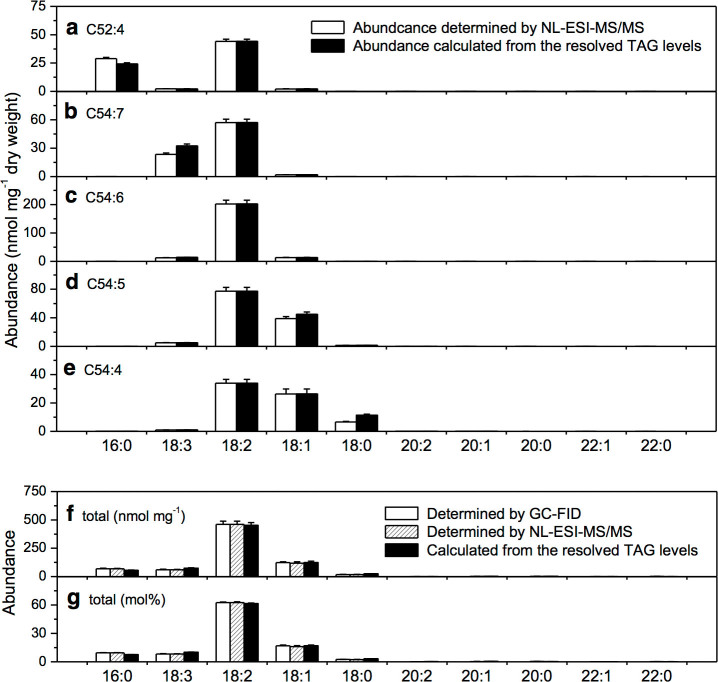
Figure 3. Evaluation of the quantification accuracy for the resolved TAG species.
(a) Fatty acyl abundance at C52:4 determined by NL-ESI-MS/MS and from the levels of resolved TAG species. (b–e) Fatty acyl abundance at C54:7, C54:6, C54:5, and C54:4 determined by NL-ESI-MS/MS and from the levels of resolved TAG species. (f–g) Absolute and relative levels of fatty acids in soybean seeds determined by GC-FID, by NL-ESI-MS/MS, and from the levels of the resolved TAG species. Values were means ± SE (n = 5).
Table 1. Levels of TAG species in soybean seeds determined by NL-ESI-MS/MS. The fatty acyl abundance at each TAG mass obtained in Supplementary Table S2 was applied in a formula to calculate the levels of TAG species. The derivation of the formula listed below was described previously22. The TAG levels are expressed in absolute level (nmol mg−1 dry mass) and relative level (mol% of total). Values are means ± SE (n = 5).
| m/z | Acyl combination | Abbreviation | nmol mg−1 | mol% | |
|---|---|---|---|---|---|
| 824.8 | C48:0 | 16:0–16:0–16:0 | PPP | 0.014 ± 0.001 | 0.006 ± 0.000 |
| 846.8 | C50:3 | 16:0–16:0–18:3 | PPLn | 0.286 ± 0.013 | 0.117 ± 0.006 |
| 848.8 | C50:2 | 16:0–16:0–18:2 | PPL | 2.462 ± 0.094 | 1.005 ± 0.033 |
| 850.8 | C50:1 | 16:0–16:0–18:1 | PPO | 0.662 ± 0.061 | 0.252 ± 0.019 |
| 852.8 | C50:0 | 16:0–16:0–18:0 | PPS | 0.019 ± 0.002 | 0.008 ± 0.001 |
| 868.7 | C52:6 | 16:0–18:3–18:3 | PLnLn | 0.547 ± 0.028 | 0.224 ± 0.013 |
| 870.8 | C52:5 | 16:0–18:2–18:3 | PLLn | 6.573 ± 0.323 | 2.679 ± 0.105 |
| 872.8 | C52:4 | 16:0–18:2–18:2 | PLL | 22.037 ± 0.957 | 8.970 ± 0.207 |
| 16:0–18:1–18:3 | POLn | 2.202 ± 0.078 | 0.898 ± 0.023 | ||
| 874.8 | C52:3 | 16:0–18:0–18:3 | PSLn | 0.150 ± 0.006 | 0.061 ± 0.003 |
| 16:0–18:1–18:2 | POL | 10.713 ± 0.651 | 4.351 ± 0.149 | ||
| 876.8 | C52:2 | 16:0–16:0–20:2 | PPEi | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 16:0–18:0–18:2 | PSL | 1.881 ± 0.082 | 0.767 ± 0.031 | ||
| 16:0–18:1–18:1 | POO | 2.216 ± 0.336 | 0.897 ± 0.118 | ||
| 878.8 | C52:1 | 16:0–16:0–20:1 | PPG | 0.001 ± 0.000 | 0.001 ± 0.000 |
| 16:0–18:0–18:1 | PSO | 0.446 ± 0.044 | 0.181 ± 0.013 | ||
| 880.8 | C52:0 | 16:0–16:0–20:0 | PPA | 0.004 ± 0.001 | 0.002 ± 0.001 |
| 16:0–18:0–18:0 | PSS | 0.040 ± 0.005 | 0.017 ± 0.002 | ||
| 890.7 | C54:9 | 18:3–18:3–18:3 | LnLnLn | 0.343 ± 0.023 | 0.140 ± 0.009 |
| 892.7 | C54:8 | 18:2–18:3–18:3 | LLnLn | 4.768 ± 0.323 | 1.942 ± 0.107 |
| 894.8 | C54:7 | 18:1–18:3–18:3 | OLnLn | 1.920 ± 0.077 | 0.784 ± 0.031 |
| 18:2–18:2–18:3 | LLLn | 28.556 ± 1.844 | 11.611 ± 0.523 | ||
| 896.8 | C54:6 | 18:0–18:3–18:3 | SLnLn | 0.137 ± 0.010 | 0.056 ± 0.003 |
| 18:1–18:2–18:3 | OLLn | 13.450 ± 0.794 | 5.454 ± 0.079 | ||
| 18:2–18:2–18:2 | LLL | 62.779 ± 4.121 | 25.440 ± 0.656 | ||
| 898.8 | C54:5 | 16:0–18:3–20:2 | PLnEi | 0.108 ± 0.008 | 0.043 ± 0.001 |
| 18:0–18:2–18:3 | SLLn | 1.416 ± 0.116 | 0.575 ± 0.035 | ||
| 18:1–18:1–18:3 | OOLn | 3.472 ± 0.345 | 1.404 ± 0.098 | ||
| 18:1–18:2–18:2 | OLL | 37.893 ± 2.640 | 15.333 ± 0.341 | ||
| 900.8 | C54:4 | 16:0–18:3–20:1 | PLnG | 0.072 ± 0.007 | 0.029 ± 0.003 |
| 16:0–18:2–20:2 | PLEi | 0.093 ± 0.010 | 0.037 ± 0.003 | ||
| 18:0–18:1–18:3 | SOLn | 0.845 ± 0.055 | 0.344 ± 0.018 | ||
| 18:0–18:2–18:2 | SLL | 10.548 ± 0.659 | 4.272 ± 0.033 | ||
| 18:1–18:1–18:2 | OOL | 12.753 ± 1.738 | 5.140 ± 0.553 | ||
| 902.8 | C54:3 | 16:0–18:3–20:0 | PLnA | 0.038 ± 0.003 | 0.015 ± 0.001 |
| 16:0–18:2–20:1 | PLG | 0.213 ± 0.012 | 0.086 ± 0.003 | ||
| 16:0–18:1–20:2 | POEi | 0.018 ± 0.001 | 0.007 ± 0.000 | ||
| 18:0–18:0–18:3 | SSLn | 0.000 ± 0.000 | 0.000 ± 0.001 | ||
| 18:0–18:1–18:2 | SOL | 5.510 ± 0.340 | 2.234 ± 0.038 | ||
| 18:1–18:1–18:1 | OOO | 3.086 ± 0.859 | 1.234 ± 0.320 | ||
| 904.8 | C54:2 | 16:0–18:2–20:0 | PLA | 0.362 ± 0.010 | 0.147 ± 0.005 |
| 16:0–18:1–20:1 | POG | 0.071 ± 0.011 | 0.028 ± 0.004 | ||
| 16:0–18:0–20:2 | PSEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:0–18:2 | SSL | 0.237 ± 0.037 | 0.098 ± 0.014 | ||
| 18:0–18:1–18:1 | SOO | 1.257 ± 0.181 | 0.507 ± 0.059 | ||
| 906.9 | C54:1 | 16:0–16:0–22:1 | PPEr | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 16:0–18:1–20:0 | POA | 0.093 ± 0.016 | 0.037 ± 0.005 | ||
| 16:0–18:0–20:1 | PSG | 0.001 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:0–18:1 | SSO | 0.113 ± 0.011 | 0.065 ± 0.005 | ||
| 908.9 | C54:0 | Not detectable | |||
| 920.8 | C56:8 | 18:3–18:3–20:2 | LnLnEi | 0.003 ± 0.001 | 0.001 ± 0.000 |
| 922.8 | C56:7 | 18:3–18:3–20:1 | LnLnG | 0.016 ± 0.004 | 0.007 ± 0.002 |
| 18:2–18:3–20:2 | LLnEi | 0.061 ± 0.010 | 0.025 ± 0.004 | ||
| 924.8 | C56:6 | 18:3–18:3–20:0 | LnLnA | 0.012 ± 0.004 | 0.005 ± 0.001 |
| 18:2–18:3–20:1 | LLnG | 0.229 ± 0.027 | 0.093 ± 0.010 | ||
| 18:2–18:2–20:2 | LLEi | 0.199 ± 0.013 | 0.081 ± 0.0023 | ||
| 18:1–18:3–20:2 | OLnEi | 0.012 ± 0.001 | 0.005 ± 0.000 | ||
| 926.8 | C56:5 | 18:2–18:3–20:0 | LLnA | 0.226 ± 0.016 | 0.091 ± 0.005 |
| 18:2–18:2–20:1 | LLG | 0.798 ± 0.035 | 0.322 ± 0.011 | ||
| 18:1–18:3–20:1 | OLnG | 0.072 ± 0.017 | 0.029 ± 0.006 | ||
| 18:1–18:2–20:2 | OLEi | 0.062 ± 0.009 | 0.025 ± 0.004 | ||
| 18:0–18:3–20:2 | SLnEi | 0.001 ± 0.000 | 0.000 ± 0.000 | ||
| 928.8 | C56:4 | 16:0–18:3–22:1 | PLnEr | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 16:0–20:2–20:2 | PEiEi | 0.001 ± 0.001 | 0.000 ± 0.000 | ||
| 18:1–18:3–20:0 and 18:2–18:2–20:0 | OLnA and LLA | 0.821 ± 0.047 | 0.330 ± 0.009 | ||
| 18:0–18:3–20:1 and 18:1–18:2–20:1 | SLnG and OLG | 0.425 ± 0.037 | 0.171 ± 0.011 | ||
| 18:0–18:2–20:2 and 18:1–18:1–20:2 | SLEi and OOEi | 0.018 ± 0.003 | 0.007 ± 0.001 | ||
| 930.9 | C56:3 | 16:0–18:3–22:0 | PLnB | 0.017 ± 0.005 | 0.007 ± 0.002 |
| 16:0–18:2–22:1 | PLEr | 0.002 ± 0.001 | 0.001 ± 0.000 | ||
| 16:0–20:1–20:2 | PGEi | 0.019 ± 0.005 | 0.008 ± 0.002 | ||
| 18:0–18:3–20:0 | SLnA | 0.0046 ± 0.003 | 0.002 ± 0.001 | ||
| 18:1–18:2–20:0 | OLA | 0.559 ± 0.061 | 0.224 ± 0.018 | ||
| 18:0–18:2–20:1 | SLG | 0.268 ± 0.038 | 0.112 ± 0.015 | ||
| 18:1–18:1–20:1 | OOG | 0.026 ± 0.026 | 0.010 ± 0.010 | ||
| 18:0–18:1–20:2 | SOEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 932.9 | C56:2 | 16:0–18:2–22:0 | PLB | 0.182 ± 0.014 | 0.074 ± 0.007 |
| 16:0–18:1–22:1 | POEr | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 16:0–20:1–20:1 | PGG | 0.213 ± 0.025 | 0.086 ± 0.007 | ||
| 16:0–20:0–20:2 | PAEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:2–20:0 | SLA | 0.295 ± 0.015 | 0.121 ± 0.006 | ||
| 18:1–18:1–20:0 | OOA | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:1–20:1 | SOG | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:0–20:2 | SSEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 934.9 | C56:1 | 16:0–18:1–22:0 | POB | 0.054 ± 0.006 | 0.022 ± 0.002 |
| 16:0–18:0–22:1 | PSEr | 0.001 ± 0.000 | 0.000 ± 0.000 | ||
| 16:0–20:0–20:1 | PAG | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–18:1–20:0 | SOA | 0.057 ± 0.004 | 0.023 ± 0.002 | ||
| 18:0–18:0–20:1 | SSG | 0.001 ± 0.001 | 0.001 ± 0.001 | ||
| 936.9 | C56:0 | Not detectable | |||
| 950.8 | C58:7 | 18:3–18:3–22:1 | LnLnEr | 0.002 ± 0.002 | 0.001 ± 0.001 |
| 18:3–20:2–20:2 | LnEiEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 952.8 | C58:6 | 18:3–18:3–22:0 | LnLnB | 0.004 ± 0.004 | 0.001 ± 0.001 |
| 18:2–18:3–22:1 | LLnEr | 0.002 ± 0.001 | 0.001 ± 0.001 | ||
| 18:3–20:1–20:2 | LnGEi | 0.001 ± 0.001 | 0.000 ± 0.000 | ||
| 954.8 | C58:5 | 18:2–18:3–22:0 | LLnB | 0.145 ± 0.008 | 0.058 ± 0.002 |
| 18:1–18:3–22:1 and 18:2–18:2–22:1 | 0.014 ± 0.009 | 0.005 ± 0.003 | |||
| 18:1–20:2–20:2 | 0.001 ± 0.001 | 0.000 ± 0.000 | |||
| 18:2–20:2–20:1 and 18:3–20:1–20:1 | |||||
| 956.9 | C58:4 | 18:0–18:3–22:1 | SLnEr | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 18:1–18:2–22:1 | OLEr | 0.001 ± 0.001 | 0.000 ± 0.000 | ||
| 18:1–18:3–22:0 and 18:2–18:2–22:0 | OLnB and LLB | 0.482 ± 0.025 | 0.195 ± 0.011 | ||
| 18:1–20:1–20:2 and 18:2–20:1–20:1 | OGEi and LGG | 0.002 ± 0.001 | 0.001 ± 0.000 | ||
| 18:2–20:0–20:2 and 18:3–20:0–20:1 | LAEi and LnAG | ||||
| 958.9 | C58:3 | 18:0–18:3–22:0 and 18:1–18:2–22:0 | SLnB and OLB | 0.270 ± 0.027 | 0.108 ± 0.007 |
| 18:0–18:2–22:1 and 18:1–18:1–22:1 | OLEr and OOEr | 0.001 ± 0.000 | 0.000 ± 0.000 | ||
| 18:1–20:0–20:2 and 18:2–20:0–20:1 | OAEi and LAG | ||||
| 18:3–20:0–20:0 and 18:0–20:1–20:2 | LnAA and SGEi | 0.005 ± 0.001 | 0.002 ± 0.001 | ||
| 18:1–20:1–20:1 and 18:2–20:0–20:1 | OGG and LAG | ||||
| 960.9 | C58:2 | 18:0–18:2–22:0 | SLB | 0.047 ± 0.014 | 0.019 ± 0.006 |
| 18:1–18:1–22:0 | OOB | 0.144 ± 0.025 | 0.058 ± 0.008 | ||
| 18:0–18:1–22:1 | SOEr | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:0–20:0–20:2 | SAEi | 0.087 ± 0.015 | 0.036 ± 0.005 | ||
| 18:1–20:0–20:1 | OAG | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:2–20:0–20:0 | LAA | 0.010 ± 0.002 | 0.004 ± 0.001 | ||
| 962.9 | C58:1 | Not detectable | |||
| 964.9 | C58:0 | Not detectable | |||
| 980.9 | C60:6 | 18:3–20:2–22:1 | LnEiEr | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 20:2–20:2–20:2 | EiEiEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 982.9 | C60:5 | 18:3–20:2–22:0 | LnEiB | 0.020 ± 0.003 | 0.008 ± 0.001 |
| 20:1–20:2–20:2 | GEiEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 984.9 | C60:4 | 18:2–20:2–22:0 and 18:3–20:1–22:0 | LEiB and LnGB | 0.000 ± 0.000 | 0.000 ± 0.000 |
| 18:1–20:2–22:1 and | OEiEr | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 18:2–20:1–22:1 and 18:3–20:0–22:1 | LGEr and LnAEr | ||||
| 20:0–20:2–20:2 and 20:1–20:1–20:2 | AEiEi and GGEi | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 986.9 | C60:3 | 18:1–20:2–22:0 | OEiB | 0.109 ± 0.015 | 0.044 ± 0.005 |
| 18:2–20:1–22:0 | LGB | 0.185 ± 0.038 | 0.074 ± 0.012 | ||
| 18:3–20:0–22:0 | LnAB | 0.001 ± 0.001 | 0.000 ± 0.000 | ||
| 20:1–20:1–20:1 | GGG | 0.000 ± 0.000 | 0.000 ± 0.000 | ||
| 988.9 | C60:2 | Not detectable | |||
| 990.9 | C60:1 | Not detectable | |||
| 993.0 | C60:0 | Not detectable |
The nomenclature of the fatty acid was abbreviated to be one letter. A, arachidic acid (20:0); B, behenic acid (22:0); Er, erucic acid (22:1); Ei, eicosadienoic acid (20:2); G, gadoleic acid (20:1); L, linoleic acid (18:2); Ln, linolenic acid (18:3); O, oleic acid (18:1); P, palmitic acid (16:0); S, stearic acid (18:0).
Ninety-three individual TAG species, identified by their fatty acyl combinations, were resolved and their levels were calculated (Table 1). For those that cannot be resolved into individual species but with the same TAG mass, they were combined together as a group in quantification (Table 1). The TAG levels were expressed in absolute amount (nmol mg−1) and molar percentage (mol%). Of the TAG species detected, 99.2% belonged to the resolved 93 individual species and 0.8% belonged to the 9 TAG groups (Table 1). The most abundant eight TAG species accounted for 80% TAGs that were detected in soybean seeds, including 18:2–18:2–18:2 (LLL, 25.4%), 18:1–18:2–18:2 (OLL, 15.3%), 18:2–18:2–18:3 (LLLn, 11.6%), 16:0–18:2–18:2 (PLL, 9.0%), 18:1–18:2–18:3 (OLLn, 5.5%), 18:1–18:1–18:2 (OOL, 5.1%), 16:0–18:1–18:2 (POL, 4.4%), and 18:0–18:2–18:2 (SLL, 4.3%).
Evaluation of the determined TAG levels
The calculation accuracy for the levels of TAG species was evaluated at the five most abundant TAG masses (Fig. 3). The fatty acyl abundance at TAG m/z 872.8 (C52:4) was obtained by NL-ESI-MS/MS (Supplementary Table S2 and Fig. 3a), from where the levels of two TAG species, 16:0–18:2–18:2 and 16:0–18:1–18:3, were calculated (Table 1). From the known levels of these two TAG species, the fatty acyl abundance of 16:0, 18:1, 18:2, and 18:3, was calculated. It was comparable for the fatty acyl composition that was determined by NL-ESI-MS/MS and that retrieved from the levels of the two resolved TAG species at m/z 872.8 (Fig. 3a). It was also the case for TAG masses at m/z 894.8 (C54:7), m/z 896.8 (C54:6), m/z 898.8 (C54:5), and m/z 900.8 (C54:4; Fig. 3b–e). In addition, it was comparable for the fatty acyl composition determined by GC-FID, by NL-ESI-MS/MS, and from the levels of the resolved TAG species (Fig. 3f–g). The total TAG amount, or oil content, was comparable with those measured by methods of nuclear magnetic resonance and near infrared reflectance spectroscopy (Supplemental Table S5)23,24,25,26,27. NL-ESI-MS/MS method detected many TAG species with less abundance and the levels of major TAG species were compared among various other mass spectrometric methods (Supplemental Table S6)3,4,7,11,12.
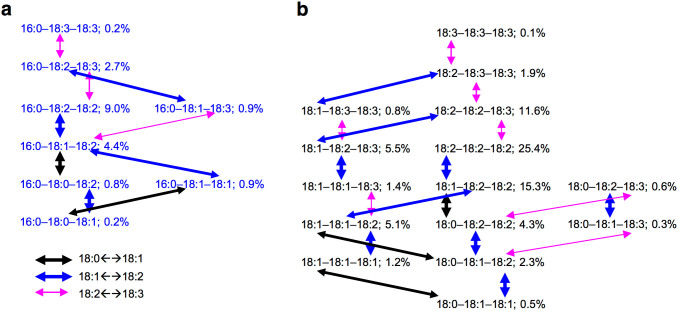
Three general trends were found by comparing the level between two TAGs that differ in one fatty acyl chain (Fig. 4). First, the levels tended to be about three-fold higher for 18:1-containing than 18:0-containing TAGs when the other two acyl chains were same in the two TAG species. For example, the level of 18:1–18:2–18:2 was higher than the level of 18:0–18:2–18:2 (15.3% vs 4.3%; Fig. 4b). Second, the level tended to be approximately three-fold higher for 18:2-containing than 18:1-containing TAGs when the other two acyl chains were same in the two TAG species. For example, the level of 18:1–18:2–18:3 was higher than the level of 18:1–18:1–18:3 (5.5% vs 1.4%; Fig. 4b). Third, the level tended to be approximately six-fold lower for 18:3-containing than 18:2-containing TAGs when the other two acyl chains were the same in the two TAG species. For example, the level of 18:2–18:3–18:3 was higher than the level of 18:2–18:2–18:3 (1.9% vs 11.6%; Fig. 4b). Considering the desaturation of fatty acyl chain esterified on lipid carriers, these trends implied that the fatty acyl desaturases catalyzing 18:0 → 18:1 and 18:1 → 18:2 were more active than the ones catalyzing 18:2 → 18:3 for the fatty acyl chains destined to TAG assembly.
Figure 4. Comparison of the levels of two TAG species that differ in one acyl chain.
(a) Eight 16:0-containing TAG species that accounted for 19% of the total TAGs. (b) Fifteen TAG species with fatty acyl chains of 18:0, 18:1, 18:2, or 18:3 that accounted for 76% of the total TAGs. Dark arrow points to the different fatty acyl chains between two TAG species (18:0 or 18:1); Blue arrow points to the different fatty acyl chains between two TAG species (18:1 or 18:2); and pink arrow points to the different fatty acyl chains between two TAG species (18:2 or 18:3).
The patterns of the fatty acyl combinations were analyzed (Fig. 5). The synthesis of fatty acids, 16:0, 18:0, and 18:1, takes place in plastids (designated as A pool here); the synthesis of fatty acids, 18:2 and 18:3, involves acyl desaturation (B pool); and the synthesis of fatty acids, 20:2, 20:1, 20:0, 22:1, and 22:0, involves acyl elongation (C pool)28. NL scans showed that the pool size was 28%, 71%, and 1% for A, B, and C, respectively (Supplementary Table S2). From these pool sizes, a predicted randomized combination of fatty acyl chains was calculated (Fig. 5). A measured combination of fatty acyl chains was obtained by sorting the levels of the resolved TAG species in Table 1. The analysis showed that the amount of TAGs assembled using 18:2 and 18:3, or type B-B-B, was 36.6%, while it would be 39.6% if 18:2 and 18:3 were randomly selected from the fatty acid pools to assemble one TAG molecule (Fig. 5). The amount of the type of TAGs, A-B-B, assembled using one acyl chain of 16:0, 18:0, or 18:1 and other two acyl chains of 18:2 or 18:3, was determined to be 38.5%, while it would be 42.4% if the fatty acids were randomly selected from the fatty acid pools to form this type of TAG (Fig. 5). These results indicate the selection of three fatty acids is not random on the assembly of one TAG molecule.
Figure 5. Pattern comparison between the TAGs whose fatty acyl combinations were randomly selected and the ones that were measured by NL-ESI-MS/MS.
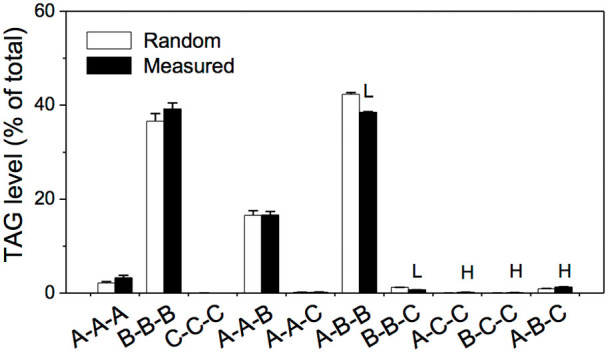
Letter A refers to 16:0, 18:0, or 18:1; B refers to 18:2 or 18:3; C refers to 20:2, 20:1, 20:0, 22:1, or 22:0. The resolved TAGs listed in Table 1 accounts for 99.2 mol% while the combined TAG groups account for 0.8 mol%. The levels of resolved TAGs were applied to count the different TAG types. The sn-position of acyl chains are not specified in the description of TAG molecules in this work. A chi-square test has been performed to compare the difference between observed group pattern and expected group pattern and the p value is 0.996, which is larger than 0.05 and indicates that the difference is not significant. HSignificantly higher and LSignifcantly lower, each at P < 0.05, compared with the randomized acyl combination and measured combination, based on Student's t test. Values were means ± SE (n = 5).
Discussion
Various LC-MS methods had been described to determine the TAG compositions in soybean seeds due to its great economic values3,4,5,6,7,8,9,10,11,12. However, a quantitative documentation of TAG species in absolute amount, e.g. nmol mg−1, is still lacking for soybean seeds. A quantitative TAG profiling method for soybean seeds is described here based on NL-ESI-MS/MS. The general steps include mass spectral acquisition, isotopic deconvolution, adjustment of fatty acyl signals, assignment of fatty acyl chains, and calculation of TAG levels. This method does not require prior derivatization or chromatographic separation. Ninety-three individual TAG species were resolved and quantified, including the minor ones that are not detected by LC-MS methods. The detailed quantitative information on individual TAG species could facilitate the genetic breeding of soybean for desirable oil content and composition.
Our MS/MS spectra suggest that the fatty acyls of 18:1, 18:2, 18:3, which accounts for 88% of soybean fatty acids, can undergo neutral loss generally close from ammonium adducts of TAGs in soybean seed extracts, while 16:0, which accounts for 9.5% of total soybean fatty acids, tended to be 26–36% higher of neutral loss efficiency than expected (Fig. 2; Supplementary Table S1). The reason could be that 16:0 has a preference at sn-1 position, from where dissociation of fatty acyl chain is more efficient than at sn-2 position of intact ammonium adducts of TAGs. Since the neutral loss efficiency is influenced by the fatty acyl length, unsaturation degree, and sn-position on glycerol backbone16,17,18, we adjusted the responses of the fatty acids in spectrometric scans by gas chromatography analysis. The adjustment factors we measured are similar to those reported previously3.
The ammonium adducts of TAGs in soybean seed extracts had been analyzed by NL-ESI-MS/MS and the fatty acyl abundance was reported in unit of mass spectra signal mg−1 dry weight20,21. The method described here is built upon it with several aspects of significant improvement. Different TAG species have different responses to mass analyzer due to their varied total fatty acyl carbons and degree of unsaturation15. The adjustment factors are applied in this current method to obtain the fatty acyl abundance in levels of nmol mg−1 dry weight. In addition, this method resolves individual TAG species at each particular TAG mass. Furthermore, neutral loss scans were performed for five types of abundant fatty acids as well as another five types of fatty acids present in trace amount in soybean seeds, which allows detection of more types of TAG species. Collectively, the method described here is able to quantify levels of individual TAG species in an absolute content expressed in unit of nmol mg−1 dry weight.
The quantitative measurement of individual TAG species is potentially useful in study of oil synthesis. Vegetable oils provide approximately 25% of dietary calories to the developed world and are an increasingly important source for renewable bioenergy29. It is largely unknown how TAG species are assembled and deposited28. Identification of individual TAG species and quantification of its content would facilitate the study of mechanism of synthesis and the ways to enhance the oil accumulation. It remains unclear how the synthesis of a particular type of TAG is achieved and its impact on seed oil content. Our analysis reveals that the level of TAGs with polyunsaturated fatty acids, 18:2 and 18:3, tends to be higher than expected if 18:2 and 18:3 were randomly selected from fatty acid pools to assemble one TAG molecule. The amount of the type of TAGs assembled using one acyl chain of 16:0, 18:0, or 18:1, which have plastidic origin, and the other two acyl chains of 18:2 or 18:3, which are produced in endoplasmic reticulum, tends to be lower than expected if those three fatty acyl chains were randomly selected from fatty acid pools to assemble one TAG molecule. In addition, by quantifying individual TAG species, we found a pattern among TAG species with the degree of unsaturation. The flows of 18:0 → 18:1 and 18:1 → 18:2 were from TAGs with lower levels to relatively higher ones, while the flow of 18:2 → 18:3 is from TAGs with higher levels to lower ones (Fig. 4). These findings reveal the specific activity of fatty acid desaturases on TAG assembly. Furthermore, our method may provide a tool to characterize the mechanism of TAG assembly by profiling the alterations of specific TAG species in genetic mutants. TAG in plants can be synthesized via different pathways, such as de novo synthesis via the Kennedy pathway, acyl editing, and PC-derived DAGs28,29. Combining the TAG species analysis with mutants of genes in different pathways will help to delineate their specific contributions on TAG assembly.
This method resolves TAG species into fatty acyl chains, but the information is lacking for a specific fatty acyl position on the glycerol backbone. In addition, the double bond positions on the fatty acyl chains are not specified. The scan modes detect the losses of ten fatty acyl chains, and there could have trace amount of fatty acids with chains shorter than 16 carbons or longer than 22 carbons that are not included in the scans here. The adjustment factor at each TAG mass was extrapolated from regression curves. The adjustment factors for minor fatty acids with chain length over 20 carbons were not measured for correction of the fatty acyl abundance. More accurate quantification can be achieved if more TAG standards are commercially available. However, this shotgun lipidomics approach achieves a fairly high accuracy for profiling soybean seed TAGs as evaluated by GC-FID (Fig. 3). This rapid and high throughput method should facilitate the study of TAG metabolism in soybean seeds and be adaptable for other oilseed crops in genetic breeding.
Methods
Plant growth and seed harvest
Soybean plants were grown in an air-conditioned, climate controlled green house (Argus Control System). The growth conditions were set as following: heating set point of 20°C, cooling set point of 21°C; 50% minimum humidity; 16 h-light/8 h-dark cycle; supplemental light threshold of 566 μmol m−2 s−1 (supplemental lights turn off when outside solar radiation is 566 μmol m−2 s−1 or above); 100% shading with shade cloth when solar radiation is 1415 μmol m−2 s−1 or above, 50% shading at 1132 μmol m−2 s−1 or above. Plants were watered regularly and fertilized with Jack's® 15-16-17 Peat-lite water-soluble fertilizer once a week. The mature seeds were harvested and dried at room temperature for over three weeks before TAG extraction and analysis.
Analysis of fatty acids by gas chromatography
Soybean seeds were pulverized using a mortar and pestle at room temperature. Approximately 10 mg fine powder was placed in a glass tube with Teflon-lined screw caps. To each sample, 400 μmol heptadecanoic acid (17:0) and 2.5 mL methanol (1.5% H2SO4 and 0.2% butylated hydroxytoluene) was added. The samples were incubated for 1 h at 90°C for oil extraction and transmethylation. After reaction, 2.0 mL water and 2.0 mL hexane were added and mix well by vortex. After centrifugation at 600 rpm for 10 min, 0.7 mL of the upper phase were transferred to a vial for analysis of fatty acid methyl esters (FAMEs) by gas-chromatography. The direct transmethylation for GC analysis can be performed for Arabidopsis seeds, which contain oil approximately 35%30. Soybean seeds contain approximately 20% of oil23,24,25,26,27. The majority of fatty acyl chains in soybean seeds are esterified onto TAGs and no significant difference in fatty acid composition was found between direct transmethylation and TAGs purified by thin layer chromatography.
The Shimadzu gas chromatograph machine was installed a Supercowax-10 (0.25 mm × 30 m) column, a split/splitess injector, and a flame ionization detector. One μL sample was injected at a split ratio of 50:1. The carrier gas was helium with a flow rate of 20 mL min−1. The oven temperature was maintained at 170°C for 1 min and then ramped to 210°C at a 3°C min−1. FAMEs from soybean seeds were identified by comparing their retention times with FAMEs in a standard mixture.
TAG extraction and preparation of profiling samples
TAGs in soybean seed were extracted following a modified Bligh and Dyer method31. Briefly, soybean seeds were ground into fine powder with a mortar and pestle. Approximately 10 mg fine soybean seed powder in a glass tube with Teflon-line screw cap were added 4.0 mL chloroform:methanol (2:1, v/v), shaked vigorously for 1 hour at room temperature, and centrifuged for 10 min at 600 rpm. Then 100 uL supernatant was mixed with internal standard in a profiling vial and infused directly into mass spectrometer for multiple neutral loss scans. The profiling samples contained 100 μL total TAG extract, 0.575 nmol tri17:1-TAG (Internal standard; Avanti Polar Lipids), and 840 μL of chloroform: methanol: 300 mM ammonium acetate in water (300:665:35, v/v/v). The final volume was adjusted to be 1.2 mL by chloroform for each profiling sample. The concentrations ranged from 15 pmol uL−1 to 350 pmol uL−1 of total TAGs were tested and linearized responses were observed previously22. In this study, the concentration of total TAGs that was directly infused into mass spectrometer was approximately 100 pmol uL−1, a concentration laid in the linearized range of the detection sensitivity. Samples containing internal standard only were run along with the TAG samples and used to correct chemical and instrumental noises.
Mass spectrometric analysis of TAGs
Lipid samples were introduced by continuous infusion into the ESI source on a triple quadrupole mass spectrometer (API4000, Applied Biosystems, Foster City, CA). One mL sample were taken by an autosampler (LC Mini PAL, CTC Analytics AG, Zwingen, Switzerland) fitted with 1.0 mL sample loop. The infusion rate was 30 μL min−1. TAGs were detected as ammonium ions, [M + NH4]+, by 11 NL scans sequentially. The scans targeted losses of fatty acyl chains as neutral fragments with NN3, including NL285 (17:1), NL273 (16:0), NL295 (18:3), NL297 (18:2), NL299 (18:1), NL301 (18:0), NL325 (20:2), NL327 (20:1), NL329 (20:0), NL355 (22:1), and NL357 (22:0).
The scan speed was 100 m/z units s−1. The collision gas was nitrogen and collision energy was +25 V. The collision gas pressure was set on “low”. The declustering potential was +100 V; the entrance potential was +14 V; and the exit potential was +14 V. The source temperature (heated nebulizer) was 100°C; the interface heater was on; +5.5 kV was applied to the electrospray capillary; the curtain gas was set at 20 (arbitrary units); and the tow ion source gases were set at 45 (arbitrary units). The mass analyzers were adjusted to a resolution of 0.7 u full width at half height. Thirty-nine continuum scans were averaged in the multiple channel analyzer mode.
Fifteen types commercial TAG species were utilized to build up regression curves for adjustment factors, including C42:0 (tri14:0), C45:0 (tri15:0), C48:3 (tri16:1), C48:0 (tri16:0), C51:0 (tri17:0), C54:6 (tri18:2), C54:3 (tri18:1), C54:0 (tri18:0), C57:6 (tri19:2), C57:3 (tri19:1), C57:0 (tri19:0), C60:9 (tri20:3), C60:6 (tri20:2), C60:3 (tri20:1), and C60:0 (tri20:0; Nu-Chek Prep, Inc.)22. The principle and detailed process were described previously for determining adjustment factors of various TAG masses22. The adjustment factors reported here were measured on a triple quadrupole mass spectrometer (API4000, Applied Biosystems, Foster City, CA), due to the sensitivity, mass-range, and triple quadrupole design. The parameters and settings provided here are intended for use with that instrument, other instruments may also be used with adaptations where necessary.
TAG pattern analysis
The fatty acyl abundance was obtained from the spectra of neutral loss scans after isotopic deconvolution and correction by adjustment factors at each TAG mass (m/z). The fatty acids, 16:0, 18:1, and 18:0, were designated as A, 18:2 and 18:3 as B. The other 6 types fatty acids were designated as C, including 20:2, 20:1, 20:0, 22:1, and 22:0. If three fatty acyl chains were randomly selected from A, B, or C, the expected amounts of each combination would be: A–A–A = a*a*a; B–B–B = b*b*b; C–C–C = c*c*c; A–A–B = 3*a*a*b; A–A–C = 3*a*a*c; A–B–B = 3*b*b*a; B–B–C = 3*b*b*c; A–C–C = 3*c*c*a; B–C–C = 3*c*c*b; A–B–C = 6*a*b*c, where a, b, and c represents the percentage of A, B, and C of total fatty acyl chains, respectively. The types of fatty acyl combinations were also sorted from the determined levels of TAG species listed in Table 1. The sn-position of acyl chains on glycerol backbone are not specified in the description of TAG molecules in this work.
Author Contributions
E.B. performed soybean seed extractions. M.L. and E.B. carried out lipid profiling and data processing. M.L. designed the experiments and supervised the research. M.L. and X.W. wrote the manuscript.
Supplementary Material
Supplementary Information For
Acknowledgments
The authors are grateful for the critical reading by Brian Fanella. Work by X.W. was supported by the U.S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences (BES), Materials Sciences and Engineering Division under Award # DE-SC0001295. Work by M.L. was supported by the National Science Foundation (MCB-0922879). The authors are grateful for Dr. David Osborn at the Center for Nanoscience at the University of Missouri-St. Louis for assistance on maintenance of mass spectrometry instrument and for Kevin Reilly at the Donald Danforth Plant Science Center's Plant Growth Facility for assistance on soybean growth.
References
- Wilson R. F. The role of genomics and biotechnology in achieving global food security for high-oleic vegetable oil. J. Oleo. Sci. 61, 357–367 (2012). [DOI] [PubMed] [Google Scholar]
- Cardinal A. J. et al. Mapping the low palmitate fap1 mutation and validation of its effects in soybean oil and agronomic traits in three soybean populations. Theor. Appl. Genet. 127, 97–111 (2014). [DOI] [PubMed] [Google Scholar]
- Byrdwell W. C., Neff W. E. & List G. R. Triacylglycerol analysis of potential margarine base stocks by high-performance liquid chromatography with atmospheric pressure chemical ionization mass spectrometry and flame ionization detection. J. Agric. Food Chem. 49, 446–457 (2001). [DOI] [PubMed] [Google Scholar]
- Lerma-García M. J. et al. Use of triacylglycerol profiles established by high performance liquid chromatography with ultraviolet-visible detection to predict the botanical origin of vegetable oils. J. Chromatogr. A. 1218, 7521–7527 (2011). [DOI] [PubMed] [Google Scholar]
- Byrdwell W. C., Emken E. A., Neff W. E. & Adlof R. O. Quantitative analysis of triglycerides using atmospheric pressure chemical ionization-mass spectrometry. Lipids 31, 919–935 (1996). [DOI] [PubMed] [Google Scholar]
- Byrdwell W. C. & Neff W. E. Dual parallel electrospray ionization and atmospheric pressure chemical ionization mass spectrometry (MS), MS/MS and MS/MS/MS for the analysis of triacylglycerols and triacylglycerol oxidation products. Rapid Commun. Mass Spectrom. 16, 300–319 (2002). [DOI] [PubMed] [Google Scholar]
- Jakab A., Héberger K. & Forgács E. Comparative analysis of different plant oils by high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A. 976, 255–263 (2002). [DOI] [PubMed] [Google Scholar]
- Jakab A. et al. Differentiation of vegetable oils by mass spectrometry combined with statistical analysis. Rapid Commun. Mass Spectrom. 16, 2291–2297 (2002). [DOI] [PubMed] [Google Scholar]
- List G. R. et al. Triacylglycerol structure and composition of hydrogenated soybean oil margarine and shortening base stocks. J. Agric. Food Chem. 53, 4692–4695 (2005). [DOI] [PubMed] [Google Scholar]
- Dugo P. et al. Comprehensive two-dimensional liquid chromatography combined with mass spectrometric detection in the analyses of triacylglycerols in natural lipidic matrixes. J. Chromatogr. A. 1112, 269–275 (2006). [DOI] [PubMed] [Google Scholar]
- Fasciotti M. & Pereira Netto A. D. Optimization and application of methods of triacylglycerol evaluation for characterization of olive oil adulteration by soybean oil with HPLC-APCI-MS-MS. Talanta 81, 1116–1125 (2010). [DOI] [PubMed] [Google Scholar]
- Wei F. et al. Online profiling of triacylglycerols in plant oils by two-dimensional liquid chromatography using a single column coupled with atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A. 1312, 69–79 (2013). [DOI] [PubMed] [Google Scholar]
- Kaufman M. & Wiesman Z. Pomegranate oil analysis with emphasis on MALDI-TOF/MS triacylglycerol fingerprinting. J. Agric. Food Chem. 55, 10405–10413 (2007). [DOI] [PubMed] [Google Scholar]
- Lee J. W. et al. Application of supercritical fluid chromatography/mass spectrometry to lipid profiling of soybean. J. Biosci. Bioeng. 113, 262–268 (2012). [DOI] [PubMed] [Google Scholar]
- Han X. & Gross R. W. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Chem. 295, 88–100 (2001). [DOI] [PubMed] [Google Scholar]
- Hsu F. F. & Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 10, 587–99 (1999). [DOI] [PubMed] [Google Scholar]
- Murphy R. C. et al. Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal. Biochem. 366, 59–70 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F. F. & Turk J. Electrospray ionization multiple-stage linear ion-trap mass spectrometry for structural elucidation of triacylglycerols: assignment of fatty acyl groups on the glycerol backbone and location of double bonds. J. Am. Soc. Mass Spectrom. 21, 657–669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R. H., Wang M., Fang X. & Han X. Simulation of triacylglycerol ion profiles: bioinformatics for interpretation of triacylglycerol biosynthesis. J. Lipid Res. 54, 1023–1032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. Enhanced seed viability and lipid compositional changes during natural ageing by suppressing phospholipase Dα in soybean seed. Plant Biotechnol. J. 10, 164–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Welti R., Schapaugh W. T. & Trick H. N. Phospholipid and triacylglycerol profiles modified by PLD suppression in soybean seed. Plant Biotechnol. J. 9, 359–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Quantitative profiling and pattern analysis of triacylglycerol species in Arabidopsis seeds by electrospray ionization mass spectrometry. Plant J. 77, 160–172 (2014). [DOI] [PubMed] [Google Scholar]
- Goettel W. et al. Identification and characterization of transcript polymorphisms in soybean lines varying in oil composition and content. BMC Genomics. 15, 299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. & Hildebrand D. Changes in oil content of transgenic soybeans expressing the yeast SLC1 gene. Lipids. 44, 945–951 (2009). [DOI] [PubMed] [Google Scholar]
- Bolon Y. T. et al. Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156, 240–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A. T., Shannon J. G. & Bilyeu K. D. Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor. Appl. Genet. 125, 503–515 (2012). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol. Genet. Genomics. In press (2014). [DOI] [PubMed] [Google Scholar]
- Bates P. D., Stymne S. & Ohlrogge J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364 (2013). [DOI] [PubMed] [Google Scholar]
- Chapman K. D. & Ohlrogge J. B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P. D. Glycerolipid analysis methods. Acyl-Lipid Metabolism. In: The Arabidopsis Book 8, e0133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E. G. & Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information For