Abstract
Inherited mutations in the Krebs cycle enzyme fumarate hydratase (FH) predispose to hereditary leiomyomatosis and renal cell cancer (HLRCC). Loss of FH activity in HLRCC tumours causes accumulation of the Krebs cycle intermediate fumarate to high levels, which may act as an oncometabolite through various, but not necessarily mutually exclusive, mechanisms. One such mechanism, succination, is an irreversible non-enzymatic modification of cysteine residues by fumarate, to form S-(2-succino)cysteine (2SC). Previous studies have demonstrated that succination of proteins including glyceraldehyde 3-phosphate dehydrogenase (GAPDH), kelch-like ECH-associated protein 1 (KEAP1) and mitochondrial aconitase (ACO2) can have profound effects on cellular metabolism. Furthermore, immunostaining for 2SC is a sensitive and specific biomarker for HLRCC tumours. Here, we performed a proteomic screen on an FH-mutant tumour and two HLRCC-derived cancer cell lines and identified 60 proteins where one or more cysteine residues were succinated; 10 of which were succinated at cysteine residues either predicted, or experimentally proven, to be functionally significant. Bioinformatic enrichment analyses identified most succinated targets to be involved in redox signaling. To our knowledge, this is the first proteomic-based succination screen performed in human tumours and cancer-derived cells and has identified novel 2SC targets that may be relevant to the pathogenesis of HLRCC.
Keywords: fumarate hydratase, succination, cysteine, renal cancer, hereditary leiomyomatosis and renal cell cancer (HLRCC), oncometabolite, biomarker, reactive oxygen species (ROS)
1. Introduction
The accumulation of metabolites resulting from cancer-associated mutations in genes encoding key metabolic enzymes has been proposed to drive oncogenic transformation [1]. Mutations in genes encoding isocitrate dehydrogenase 1 and 2 (IDH1/2), succinate dehydrogenase (SDH) and fumarate hydratase (FH) can lead to high intracellular levels of d-2-hydroxyglutarate (D2HG), succinate and fumarate, respectively [2]. Due to their structural similarity to 2-oxoglutarate (2OG), these oncometabolites have been demonstrated to modulate the activities of 2OG-dependent dioxygenases; a family of enzymes with diverse functions including epigenetic regulation, oxygen sensing, collagen maturation and regulation of translation [3,4,5,6,7,8]. Fumarate is an electrophile that reacts with cysteine residues in susceptible proteins to form S-(2-succino)cysteine (2SC), a biomarker of mitochondrial stress in obesity and diabetes, and of FH-deficiency in hereditary leiomyomatosis and renal cell cancer (HLRCC) patients [9,10,11,12]. Succination is a non-enzymatic and irreversible reaction, which seems to preferentially target cysteine residues with low pKa values [13], although other factors affecting the susceptibility to succination may exist. To date, several studies have detected succinated proteins in a variety of animal and cellular models and have identified functional consequences associated with a range of cellular responses including activation of the Nuclear factor (erythroid-derived 2)-like 2 (NRF2)-mediated antioxidant pathway, inhibition of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and mitochondrial aconitase (ACO2) activity, and amplification of reactive oxygen species (ROS) signaling [13,14,15,16]. Here, we performed a proteomic-based screen using tandem mass spectrometry (MS/MS) to analyse an FH-deficient tumour and two FH-mutant cell lines derived from metastatic renal cancers. We identified 60 succinated proteins, 10 of which were modified at cysteine residues either predicted or experimentally proven to be functionally important. This study has expanded our knowledge of the succinated proteome in FH-associated renal cancer and identified novel 2SC targets, which may contribute to HLRCC tumourigenesis.
2. Results and Discussion
2.1. HLRCC Tumours and Derived Cell Lines Exhibit High Levels of 2SC
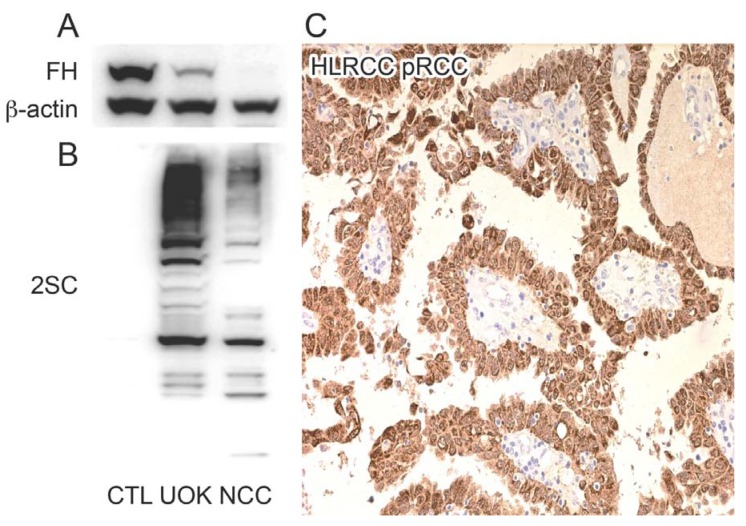
In order to verify the presence of succinated proteins in the FH-mutant tumour-derived cell lines UOK262 [17] and NCC-FH-1 (unpublished cell line kindly provided by Prof. Bin Teh), we performed immunoblotting using an anti-2SC antibody [18]. Strong immunoreactivity for 2SC was observed in both of these HLRCC cell lines but was absent in a normal renal proximal tubular epithelial cell line (RPTC) [19,20] (Figure 1B). Similarly immunohistochemistry detected high levels of 2SC in an FH-mutant (HLRCC) type 2 PRCC though was absent in the stromal tissue (Figure 1C) and sporadic type 2 PRCC (data not shown and as previously described [11]). We therefore performed a proteomic screen to identify succinated proteins in the cell lines and HLRCC tumour described above.
Figure 1.
Hereditary leiomyomatosis and renal cell cancer (HLRCC) tumours and tumour-derived cell lines express high levels of 2SC. (A) Immunoblotting confirms the presence of fumarate hydratase (FH) in the control human renal proximal tubular epithelial (RPTC) cell line (CTL), the presence of the mutant FH allele in the UOK262 (UOK) HLRCC-derived cell line as previously described and absence of FH protein in the NCC-FH-1 (NCC) HLRCC-derived cell line. β-actin levels were used as a loading control for each cell line. (B) Immunoblotting for 2SC clearly shows an absence of 2SC expression in the control cell line but high expression of 2SC in the FH-mutant cell lines (UOK and NCC). (C) Positive 2SC immunoreactivity (indicated by brown staining) is evident in the tumour cells of an FH mutant type 2 PRCC (HLRCC PRCC) but absent in the stromal tissue (blue staining).
2.2. Multiple Proteins Are Succinated in FH-Mutant Cells and Tumours
As previously reported, 2SC modification is very specific to cells with elevated fumarate [11,21,22] and we did not detect this modification in FH wildtype RPTC cells or tumour stromal tissue cells by immunoblotting and immunohistochemistry respectively (Figure 1). However, in the two HLRCC-derived cell lines and tumour, succinated peptides (as determined by a 116 Da mass shift [11]) were detected in 60 different proteins (Table 1). Furthermore, 10 of these proteins were succinated on cysteine residues with potentially regulatory functions (Table 1; red font).
Table 1.
Succinated proteins in HLRCC cancers and derived cell lines.
| Uniprot ID | Protein Symbol | Protein Name | Succination Site | Source |
|---|---|---|---|---|
| P53396 | ACLY | ATP-citrate synthase | C20 | T, N |
| Q09666 | AHNAK | Neuroblast differentiation-associated protein AHNAK | C1833 | T, N |
| P15121 | AKR1B1 | Aldose reductase | C299 | T |
| P54886-1 | ALDH18A1 | Isoform Long of Delta-1-pyrroline-5-carboxylate synthase | C612 | T, U, N |
| Q5TYW2 | ANKRD20A1 | Ankyrin repeat domain-containing protein 20A1 | C789 | N |
| P04083 | ANXA1 | Annexin A1 | C324 | T, U |
| P16615-1 | ATP2A2 | Isoform 1 of Sarcoplasmicendoplasmic reticulum calcium ATPase 2 | C997 | N |
| Q86VP6-1 | CAND1 | Isoform 1 of Cullin-associated NEDD8-dissociated protein 1 | C942 | U |
| P20810-2 | CAST | Isoform 2 of Calpastatin | C408, C413 | N |
| P23528 | CFL1 | Cofilin-1 | C139 | T, U |
| Q9NX63 | CHCHD3 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial | C112 | T |
| Q8N5K1 | CISD2 | CDGSH iron-sulfur domain-containing protein 2 | C92 | T |
| Q00610-1 | CLTC | Isoform 1 of Clathrin heavy chain 1 | C870 | U, N |
| P55060-1 | CSE1L | Isoform 1 of Exportin-2 | C272 | U, N |
| Q6NSH3 | CT45A5 | Cancertestis antigen family 45 member A5 | C22 | U |
| Q9UBR2 | CTSZ | Cathepsin Z | C92 | U |
| Q9H773 | DCTPP1 | dCTP pyrophosphatase 1 | C162 | T |
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | C200 | U |
| P33316-2 | DUT | Isoform 2 of Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial | C3 | N |
| P07814 | EPRS | Bifunctional aminoacyl-tRNA synthetase | C105 | T, U |
| O95571 | ETHE1 | Protein ETHE1, mitochondrial | C170 | T, U, N |
| P21333-2 | FLNA | Isoform 2 of Filamin-A | C717, C2543 | T, U, N |
| O75369-1 | FLNB | Isoform 1 of Filamin-B | C2501 | N |
| Q9HA64 | FN3KRP | Ketosamine-3-kinase | C24 | T |
| P02794 | FTH1 | Ferritin heavy chain | C91 | T |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | C152 | T, U, N |
| P07203 | GPX1 | Glutathione peroxidase 1 | C202 | T |
| P53701 | HCCS | Cytochrome c-type heme lyase | C39 | U, N |
| P00492 | HPRT1 | Hypoxanthine-guanine phosphoribosyltransferase | C106 | T |
| Q14197 | ICT1 | Peptidyl-tRNA hydrolase ICT1, mitochondrial | C82 | N |
| Q9NWZ3 | IRAK4 | Isoform 1 of Interleukin-1 receptor-associated kinase 4 | C13 | U, N |
| O14880 | MGST3 | Microsomal glutathione S-transferase 3 | C150, C151 | T, N |
| P46013-1 | MKI67 | Isoform Long of Antigen KI-67 | C1285 | U |
| P35579-1 | MYH9 | Isoform 1 of Myosin-9 | C988 | T, U, N |
| Q9NX24 | NHP2 | HACA ribonucleoprotein complex subunit 2 | C18 | T |
| P53384-1 | NUBP1 | Isoform 1 of Cytosolic Fe-S cluster assembly factor NUBP1 | C22, C25 | T |
| Q9H6K4-1 | OPA3 | Isoform 1 of Optic atrophy 3 protein | C164 | N |
| Q99497 | PARK7 | Protein DJ-1 | C106 | T, N |
| Q9Y570-1 | PPME1 | Isoform 1 of Protein phosphatase methylesterase 1 | C381 | N |
| Q06830 | PRDX1 | Peroxiredoxin-1 | C173 | T |
| P30048 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial | C108 | T |
| P30041 | PRDX6 | Peroxiredoxin-6 | C91 | T, U, N |
| Q15185 | PTGES3 | Prostaglandin E synthase 3 | C58 | T, U, N |
| P49023-2 | PXN | Isoform Alpha of Paxillin | C535, C538 | U |
| P63000-1 | RAC1 | Isoform A of Ras-related C3 botulinum toxin substrate 1 | C178 | T |
| P54727 | RAD23B | UV excision repair protein RAD23 homolog B | C390 | U |
| P50914 | RPL14 | Ribosomal protein L14 variant | C42 | T |
| P05386 | RPLP1 | 60S acidic ribosomal protein P1 | C61 | T |
| P31947-1 | SFN | Isoform 1 of 14-3-3 protein sigma | C38 | T, U, N |
| Q15005 | SPCS2 | Signal peptidase complex subunit 2 | C17, C26 | T, U, N |
| P42224-1 | STAT1 | Isoform Alpha of Signal transducer and activator of transcription 1-alphabeta | C492 | N |
| Q00059 | TFAM | Transcription factor A, mitochondrial | C246 | T, N |
| Q12931 | TRAP1 | Heat shock protein 75 kDa, mitochondrial | C573 | N |
| Q9H4B7 | TUBB1 | Tubulin beta-1 chain | C12 | T, U, N |
| P10599 | TXN | Thioredoxin | C73 | T |
| P09936 | UCHL1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | C90 (T), C152 (T, U) | T, U |
| P45880-2 | VDAC2 | Isoform 2 of Voltage-dependent anion-selective channel protein 2 | C76 | T, U, N |
| Q9Y277-1 | VDAC3 | Isoform 1 of Voltage-dependent anion-selective channel protein 3 | C65 | T, U, N |
| P08670 | VIM | Vimentin | C328 | T |
| P54577 | YARS | Tyrosyl-tRNA synthetase, cytoplasmic | C424 | U |
2SC targets identified an HLRCC tumour and tumour-derived cell lines. Succinated proteins are listed alphabetically by gene symbol, with succinated cysteine residues indicated. Targets highlighted in red indicate cysteine residues with either predicted or experimentally proven roles in protein function [23]. T = HLRCC tumour, U = UOK262 cell line, N = NCC-FH-1 cell line.
2.3. Succination Preferentially Targets Proteins Involved in Redox Regulation
Hierarchical clustering and gene ontology analyses suggest that most of the succination targets identified are involved in redox homeostasis or subject to ROS regulation (Table 2). This is unsurprising considering the critical roles of cysteine residues in cellular oxidative stress regulation (reviewed in [24]). Previous studies have demonstrated the functional consequences of protein succination, including inhibition of the metabolic enzymes GAPDH and ACO2 [15,16], loss of KEAP1 function resulting in constitutive NRF2 expression in type 2 papillary RCC (PRCC) and Fh1-deficient mice [25,26] and modification of reduced glutathione (GSH) in FH-deficient cells resulting in the amplification of ROS-dependent signaling [14]. In this study, we identified succination of Cys-106 at the active site of Parkinson disease protein 7 (PARK7) (also known as protein DJ-1) (Figure 2), a redox-sensitive protein that is implicated in cellular protection against oxidative stress in Parkinson’s disease (reviewed in [27]), mediates cellular responses to hypoxia [28] and can regulate metabolic pathways in RCC [29]. Interestingly PARK7 is closely associated with NRF2 through interaction with Superoxide dismutase 1 (SOD1) [30] (Figure 3). Further, NRF2 and PARK7 are prognostic factors in lung cancer and upregulated in inflammatory multiple sclerosis lesions [31,32]. Other succinated cysteine residues that may be critical to protein functions (highlighted in red font in Table 1) include Cys-299 of Aldose reductase (AKR1B1), which catalyzes the NADPH-dependent reduction of a wide variety of carbonyl-containing compounds to their corresponding alcohols with a broad range of catalytic efficiencies. Evidence suggests that Cys-299 may regulate the kinetic and inhibition properties of AKR1B1, but does not participate in catalysis [33]. Peroxiredoxins are antioxidant enzymes that control cytokine-induced peroxide levels and thereby mediate signal transduction in mammalian cells [34]. We observed succination of peroxiredoxin 1 and 3 (PRDX1 and PRDX3) at Cys-173 and Cys-108, respectively, both of which participate in intermolecular disulfide formation and are crucial to their molecular function [34,35]. Succination was also detected in thioredoxin at a crucial residue Cys-73, which acts as a donor for nitrosylation of target proteins and therefore its succination may potentially impair the S-nitrosylating activity of thioredoxin [36]. Isoform 1 of Cytosolic Fe-S cluster assembly factor Nucleotide binding protein 1 (NUBP1) is succinated at residues Cys-22 and Cys-25, both of which are involved in iron-sulfur cluster (4Fe-4S) assembly [37]. Succination of NUBP1 may lead to enzymatic inhibition, similar to that observed in ACO2 [15]. Further, microsomal glutathione S-transferase 3 (MGST3) is succinated at Cys-150 and Cys-151, both of which are also targets of S-palmitoylation [38]. Site-directed mutagenesis of cysteines for some of the above proteins provides experimental evidence for their functional roles (Table 3). Therefore modification by succination may be predicted to cause either a loss or gain of function in these proteins.
Table 2.
Gene ontology analyses of succinated proteins.
| Category | Term | Count | % | p Value |
|---|---|---|---|---|
| GOTERM_BP | GO:0000302~response to reactive oxygen species | 7 | 11.67 | 3.50 × 10−7 |
| GOTERM_BP | GO:0042542~response to hydrogen peroxide | 6 | 10.00 | 2.06 × 10−6 |
| GOTERM_BP | GO:0042743~hydrogen peroxide metabolic process | 5 | 8.33 | 2.14 × 10−6 |
| GOTERM_MF | GO:0051920~peroxiredoxin activity | 4 | 6.67 | 3.35 × 10−6 |
| GOTERM_BP | GO:0034614~cellular response to reactive oxygen species | 5 | 8.33 | 4.57 × 10−6 |
| GOTERM_MF | GO:0016684~oxidoreductase activity, acting on peroxide as acceptor | 5 | 8.33 | 7.57 × 10−6 |
| GOTERM_MF | GO:0004601~peroxidase activity | 5 | 8.33 | 7.57 × 10−6 |
| GOTERM_BP | GO:0034599~cellular response to oxidative stress | 5 | 8.33 | 1.98 × 10−5 |
| GOTERM_BP | GO:0042744~hydrogen peroxide catabolic process | 4 | 6.67 | 3.31 × 10−5 |
| GOTERM_BP | GO:0006979~response to oxidative stress | 7 | 11.67 | 3.32 × 10−5 |
| GOTERM_MF | GO:0016209~antioxidant activity | 5 | 8.33 | 3.59 × 10−5 |
| GOTERM_BP | GO:0070301~cellular response to hydrogen peroxide | 4 | 6.67 | 3.96 × 10−5 |
| GOTERM_BP | GO:0045454~cell redox homeostasis | 5 | 8.33 | 9.06 × 10−5 |
| GOTERM_BP | GO:0010035~response to inorganic substance | 7 | 11.67 | 1.15 × 10−4 |
| GOTERM_BP | GO:0006800~oxygen and reactive oxygen species metabolic process | 5 | 8.33 | 1.15 × 10−4 |
| GOTERM_BP | GO:0019725~cellular homeostasis | 9 | 15.00 | 3.23 × 10−4 |
| GOTERM_BP | GO:0042592~homeostatic process | 11 | 18.33 | 4.20 × 10−4 |
| GOTERM_BP | GO:0055114~oxidation reduction | 10 | 16.67 | 5.67 × 10−4 |
Functional classification of succinated proteins using DAVID Bioinformatics Resources [40,41,42] ranked by statistical significance. Proteins were classified using the gene ontology functional annotation for biological processes (GOTERM_BP) and molecular function (GOTERM_MF). p-values < 0.001 were considered to be highly significant.
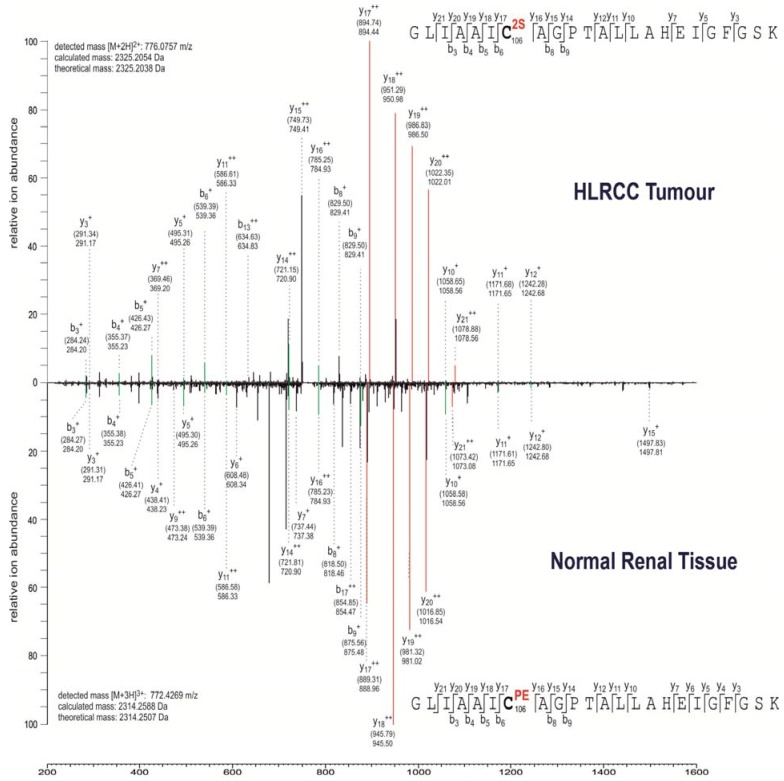
Figure 2.
MS/MS spectra showing succination of the active site (C106) of PARK7/DJ-1.
MS/MS spectra showing either succination (2SC) or pyridylethylation (PEC) at cysteine 106 in the 100GLIAAICAGPTALLAHEIGFGSK122 peptide of human DJ-1 derived from human HLRCC tumour tissue and normal renal tissue, respectively. Fragment ions are indicated for the peptide sequence and are labelled as follows: b: N-terminal fragment ion; y: C-terminal fragment ion; +: singly charged fragment ion and ++: doubly charged fragment ion. Both theoretical mass and detected mass (in brackets) are given for each assigned fragment ion. Matching fragment ion peaks between the two peptide species that do not contain the modified residue are highlighted in green, whereas peptide fragments of different mass that contain the modified residue are highlighted in red.
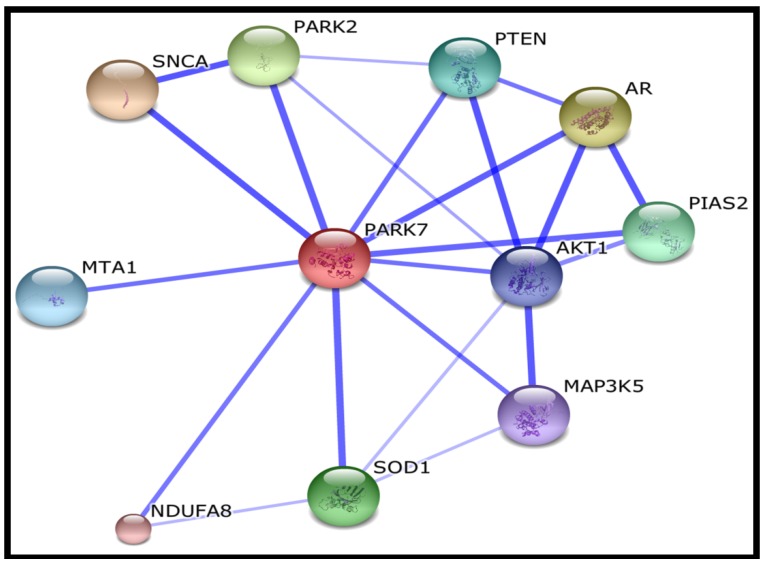
Figure 3.
Protein interactions of the redox-sensitive succination target PARK7/DJ-1.
PARK7 acts as a positive regulator of androgen receptor- dependent transcription, prevents aggregation of SNCA, protects neurons against oxidative stress and cell death and potentially functions as a redox-sensitive chaperone and as a sensor for oxidative stress. Figure generated by STRING v9.1 [47,48].
Table 3.
Mutational studies on cysteine residues observed to be succinated in FH-mutant tumours and cancer cells.
| Uniprot ID | Protein Symbol | Succination Site (S) | Mutational Data | References |
|---|---|---|---|---|
| O14880 | GAPDH | C150, C151 | C → S: Abolishes S-acylation; when associated with S-151. C → S: Abolishes S-acylation; when associated with S-150 |
[38] |
| Q99497 | PARK7 | C106 | C → A: Abolishes oxidation, association with mitochondria and protease activity. No effect on chaperone activity. Reduced binding to OTUD7B. C → A: Reduced localization in lipid rafts; when associated with A-46. C → D: Abolishes oxidation and association with mitochondria. No effect on chaperone activity. C → S: No effect on mitochondrial translocation. Reduced protease activity. |
[49,50,51,52,53,54,55] |
| P10599 | TXN | C73 | C → D: Strongly reduced S-nitrosylation of CASP3. C → S: Loss of nitrosylation, and loss of S-nitrosylating activity towards CASP3. Retains interaction with APEX1 and transcription activation; when associated with S-62 and S-69. C → S: Retains its reducing activity. |
[36,56,57] |
| P09936 | UCHL1 | C90 | C → S: Abolishes enzymatic activity. | [58,59,60] |
Site-directed mutagenesis of cysteine residues that are targeted for succination, has provided evidence of their functional roles, which may be affected by succination.
3. Experimental Section
3.1. Cell Lines and Human Tissue Samples
Cell lines were cultured as previously described [39]. The UOK262 cell line was a kind gift from Dr Marston Linehan [17], and the NCC-FH-1 cell line from Professor Bin Teh (characterized by Choon Kiat Ong, Min Han Tan, Bernice Wong and Victoria Perrier-Trudova). The RPTC cell line was a kind gift from Professors Albert Ong and Lorraine Racusen. Anonymized human tumour and normal samples were collected with full ethical approval (MREC 05/Q1605/66) as approved by the Oxford Centre for Histopathology Research.
3.2. Proteomics and Mass Spectrometry
Tumour and normal kidney tissue and cells were homogenized and sonicated in Urea-SDS buffer and protein extracts separated by SDS-PAGE and processed for trypsin digestion and MS/MS analyses as previously described [15]. Database searches were performed against UniProt/SwissProt [23] or International Protein Index [43] database using Mascot [44] or CPFP 1.3.0 [45]. For label-free quantitation of succinated peptides, samples were analyzed in three technical replicates.
3.3. Immunohistochemistry (IHC) and Immunoblotting (IB)
Analyses of tumours and cell lines by IHC and IB were performed as previously described [11,46]. The 2SC antibody was a kind gift from Dr Norma Frizzell, the fumarase antibody from Nordic Labs (Copenhagen, Denmark), and the β-actin antibody purchased from Abcam (Cambridge, UK).
 |
SNCA | synuclein, alpha (non A4 component of amyloid precursor) |
 |
AR | androgen receptor |
 |
PARK2 | Parkinson disease (autosomal recessive, juvenile) 2, parkin |
 |
SOD1 | superoxide dismutase 1, soluble |
 |
PIAS2 | protein inhibitor of activated STAT, 2 |
 |
PTEN | phosphatase and tensin homolog; Tumour suppressor |
 |
MTA1 | metastasis associated 1 |
 |
AKT1 | v-akt murine thymoma viral oncogene homolog 1 |
 |
MAP3K5 | mitogen-activated protein kinase kinase kinase 5 |
 |
NDUFA8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
3.4. Gene Ontology Analysis
Functional classification of succinated proteins was performed using DAVID Bioinformatics Resources v6.7 (National Institute of Allergy and Infectious Diseases (NIAID), NIH, Frederick, MD, USA) [41,42]. Protein network analysis for PARK7 was generated using STRING v9.1 [47,48].
4. Conclusions
Recent technological advances in mass spectrometry have facilitated high throughput screening of post-translational modifications in proteins. Here, we analysed cell lines and a tumour derived from HLRCC patients to try to identify the scope of the succinated proteome under FH-deficient settings. Succination of cysteine residues is a candidate mechanism for FH-associated oncogenesis, which is corroborated by genetic and biochemical evidence generated from the analyses of Fh1 knockout mice and gene expression profiling and exome sequencing of type 2 RCC [25,26,61]. Interestingly, some peptides were detected in only one or two of the three FH-mutant samples. However, this is most probably due to the heterogeneity of RCC [62,63]; both the UOK262 and NCC-FH-1 cell lines were generated from RCC metastases from different patients ([17] and personal communication with Dr Teh). Furthermore, due to lack of antibody-based enrichment technologies, the “succinome” thus identified is biased towards abundant proteins in a cell line specific manner. Therefore we cannot rule out the possibility that the same succinated proteins were present in all samples, albeit at different levels, some below the threshold of detection. Given that 2SC was originally identified in diabetic rats and induced as a marker of mitochondrial stress in adipocytes resulting from glucotoxicity [12,16,18,64], it is likely that succination resulting from elevated levels of intracellular fumarate targets proteins involved in a diverse range of cellular and biological processes, including microtubule dynamics [65], the NRF2-mediated antioxidant response [24,25], altered metabolism [15,16,39] and as identified here, redox homeostasis. Further studies are required to test the functionality and biological consequences of these modifications.
Acknowledgments
We are grateful to Marston Linehan for the UOK262 cell line [17], Professors Albert Ong and Lorraine Racusen for providing the RPTC cell line [19,20] and Norma Frizzell for providing the 2SC antibody [18]. We are grateful to Choon Kiat Ong, Min Han Tan, Bernice Wong and Victoria Perrier-Trudova for the generation and characterization of the NCC FH-1 cell line. Patrick J. Pollard is in receipt of a Cancer Research UK project grant (A13349) and The European Research Council have provided financial support under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 310837 to Patrick J. Pollard.
Author Contributions
Experimental design was conceived by Patrick J. Pollard, Ming Yang and Nicola Ternette. Bin Tean Teh provided the NCC-FH-1 cell line. Patrick J. Pollard, Ming Yang, Nicola Ternette, Huizhong Su, Raliat Dabiri and Julie Adam performed experiments and data analyses. Benedikt M. Kessler is the head of the Core Proteomics facility and provided advice on technical and experimental procedures. All authors contributed to the writing and critical evaluation of the manuscript.
Conflicts of Interest
Patrick J. Pollard has previously filed for a patent co-application with Norma Frizzell [USC-268-P(849)] covering immunohistochemical screening with a 2SC antibody for determination of FH mutations.
References
- 1.Yang M., Soga T., Pollard P.J. Oncometabolites: Linking altered metabolism with cancer. J. Clin. Investig. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullen A.R., DeBerardinis R.J. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol. Metab. 2012;23:552–559. doi: 10.1016/j.tem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng T., Yamamoto A., Wilkins S.E., Sokolova E., Yates L.A., Munzel M., Singh P., Hopkinson R.J., Fischer R., Cockman M.E., et al. Optimal translational termination requires c4 lysyl hydroxylation of erf1. Mol. Cell. 2014;53:645–654. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz M.J., Acevedo J.M., Loenarz C., Galagovsky D., Liu-Yi P., Perez-Pepe M., Thalhammer A., Sekirnik R., Ge W., Melani M., et al. Sudestada1, a drosophila ribosomal prolyl-hydroxylase required for mrna translation, cell homeostasis, and organ growth. Proc. Natl. Acad. Sci. USA. 2014;111:4025–4030. doi: 10.1073/pnas.1314485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loenarz C., Sekirnik R., Thalhammer A., Ge W., Spivakovsky E., Mackeen M.M., McDonough M.A., Cockman M.E., Kessler B.M., Ratcliffe P.J., et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. USA. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton R.S., Liu-Yi P., Formenti F., Ge W., Sekirnik R., Fischer R., Adam J., Pollard P.J., Wolf A., Thalhammer A., et al. Ogfod1 catalyzes prolyl hydroxylation of rps23 and is involved in translation control and stress granule formation. Proc. Natl. Acad. Sci. USA. 2014;111:4031–4036. doi: 10.1073/pnas.1314482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loenarz C., Schofield C.J. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury R., Sekirnik R., Brissett N.C., Krojer T., Ho C.H., Ng S.S., Clifton I.J., Ge W., Kershaw N.J., Fox G.C., et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature. 2014;510:422–426. doi: 10.1038/nature13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas S.A., Storey K.B., Baynes J.W., Frizzell N. Tissue distribution of s-(2-succino) cysteine (2sc), a biomarker of mitochondrial stress in obesity and diabetes. Obesity (Silver Spring) 2012;20:263–269. doi: 10.1038/oby.2011.340. [DOI] [PubMed] [Google Scholar]
- 10.Frizzell N., Lima M., Baynes J.W. Succination of proteins in diabetes. Free Radic. Res. 2011;45:101–109. doi: 10.3109/10715762.2010.524643. [DOI] [PubMed] [Google Scholar]
- 11.Bardella C., el-Bahrawy M., Frizzell N., Adam J., Ternette N., Hatipoglu E., Howarth K., O’Flaherty L., Roberts I., Turner G., et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and hlrcc patients is a robust biomarker of mutation status. J. Pathol. 2011;225:4–11. doi: 10.1002/path.2932. [DOI] [PubMed] [Google Scholar]
- 12.Alderson N.L., Wang Y., Blatnik M., Frizzell N., Walla M.D., Lyons T.J., Alt N., Carson J.A., Nagai R., Thorpe S.R., et al. S-(2-succinyl)cysteine: A novel chemical modification of tissue proteins by a krebs cycle intermediate. Arch. Biochem. Biophys. 2006;450:1–8. doi: 10.1016/j.abb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Merkley E.D., Metz T.O., Smith R.D., Baynes J.W., Frizzell N. The succinated proteome. Mass Spectrom. Rev. 2014;33:98–109. doi: 10.1002/mas.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan L.B., Martinez-Garcia E., Nguyen H., Mullen A.R., Dufour E., Sudarshan S., Licht J.D., Deberardinis R.J., Chandel N.S. The proto-oncometabolite fumarate binds glutathione to amplify ros-dependent signaling. Mol. Cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ternette N., Yang M., Laroyia M., Kitagawa M., O'Flaherty L., Wolhulter K., Igarashi K., Saito K., Kato K., Fischer R., et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013;3:689–700. doi: 10.1016/j.celrep.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blatnik M., Frizzell N., Thorpe S.R., Baynes J.W. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by fumarate in diabetes: Formation of S-(2-succinyl)cysteine, a novel chemical modification of protein and possible biomarker of mitochondrial stress. Diabetes. 2008;57:41–49. doi: 10.2337/db07-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Valera V.A., Padilla-Nash H.M., Sourbier C., Vocke C.D., Vira M.A., Abu-Asab M.S., Bratslavsky G., Tsokos M., Merino M.J., et al. Uok 262 cell line, fumarate hydratase deficient (fh-/fh-) hereditary leiomyomatosis renal cell carcinoma: In vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet. Cytogenet. 2010;196:45–55. doi: 10.1016/j.cancergencyto.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai R., Brock J.W., Blatnik M., Baatz J.E., Bethard J., Walla M.D., Thorpe S.R., Baynes J.W., Frizzell N. Succination of protein thiols during adipocyte maturation: A biomarker of mitochondrial stress. J. Biol. Chem. 2007;282:34219–34228. doi: 10.1074/jbc.M703551200. [DOI] [PubMed] [Google Scholar]
- 19.Racusen L.C., Monteil C., Sgrignoli A., Lucskay M., Marouillat S., Rhim J.G., Morin J.P. Cell lines with extended in vitro growth potential from human renal proximal tubule: Characterization, response to inducers, and comparison with established cell lines. J. Lab. Clin. Med. 1997;129:318–329. doi: 10.1016/S0022-2143(97)90180-3. [DOI] [PubMed] [Google Scholar]
- 20.Parker E., Newby L.J., Sharpe C.C., Rossetti S., Streets A.J., Harris P.C., O’Hare M.J., Ong A.C. Hyperproliferation of pkd1 cystic cells is induced by insulin-like growth factor-1 activation of the ras/raf signalling system. Kidney Int. 2007;72:157–165. doi: 10.1038/sj.ki.5002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam J., Yang M., Soga T., Pollard P.J. Rare insights into cancer biology. Oncogene. 2014;33:2547–2556. doi: 10.1038/onc.2013.222. [DOI] [PubMed] [Google Scholar]
- 22.Yang M., Soga T., Pollard P.J., Adam J. The emerging role of fumarate as an oncometabolite. Front. Oncol. 2012;2:85. doi: 10.3389/fonc.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uniprot. [(accessed on 21 March 2014)]. Available online: http://www.uniprot.org.
- 24.Forman H.J., Ursini F., Maiorino M. An overview of mechanisms of redox signaling. J. Mol. Cell Cardiol. 2014;73:2–9. doi: 10.1016/j.yjmcc.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooi A., Wong J.C., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W., Tan M.H., Zhang Z., Yang X.J., et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Adam J., Hatipoglu E., O’Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., et al. Renal cyst formation in fh1-deficient mice is independent of the hif/phd pathway: Roles for fumarate in keap1 succination and nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abou-Sleiman P.M., Muqit M.M., Wood N.W. Expanding insights of mitochondrial dysfunction in parkinson’s disease. Nat. Rev. Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 28.Vasseur S., Afzal S., Tardivel-Lacombe J., Park D.S., Iovanna J.L., Mak T.W. Dj-1/park7 is an important mediator of hypoxia-induced cellular responses. Proc. Natl. Acad. Sci. USA. 2009;106:1111–1116. doi: 10.1073/pnas.0812745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumunk D., Reichelt U., Hildebrandt J., Krause H., Ebbing J., Cash H., Miller K., Schostak M., Weikert S. Expression parameters of the metabolic pathway genes pyruvate dehydrogenase kinase-1 (pdk-1) and dj-1/park7 in renal cell carcinoma (rcc) World J. Urol. 2013;31:1191–1196. doi: 10.1007/s00345-012-0874-5. [DOI] [PubMed] [Google Scholar]
- 30.Milani P., Ambrosi G., Gammoh O., Blandini F., Cereda C. Sod1 and dj-1 converge at nrf2 pathway: A clue for antioxidant therapeutic potential in neurodegeneration. Oxid. Med. Cell. Longev. 2013;2013:836760. doi: 10.1155/2013/836760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merikallio H., Paakko P., Kinnula V.L., Harju T., Soini Y. Nuclear factor erythroid-derived 2-like 2 (nrf2) and dj1 are prognostic factors in lung cancer. Hum. Pathol. 2012;43:577–584. doi: 10.1016/j.humpath.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Van Horssen J., Drexhage J.A., Flor T., Gerritsen W., van der Valk P., de Vries H.E. Nrf2 and dj1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic. Biol. Med. 2010;49:1283–1289. doi: 10.1016/j.freeradbiomed.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Liu S.Q., Bhatnagar A., Ansari N.H., Srivastava S.K. Identification of the reactive cysteine residue in human placenta aldose reductase. Biochim. Biophys. Acta. 1993;1164:268–272. doi: 10.1016/0167-4838(93)90258-s. [DOI] [PubMed] [Google Scholar]
- 34.Wood Z.A., Schroder E., Robin Harris J., Poole L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 35.Poynton R.A., Hampton M.B. Peroxiredoxins as biomarkers of oxidative stress. Biochim. Biophys. Acta. 2014;1840:906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell D.A., Marletta M.A. Thioredoxin catalyzes the s-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 37.Rouault T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Model. Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., Casey P.J. Site-specific analysis of protein s-acylation by resin-assisted capture. J. Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adam J., Yang M., Bauerschmidt C., Kitagawa M., O’Flaherty L., Maheswaran P., Ozkan G., Sahgal N., Baban D., Kato K., et al. A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell Rep. 2013;3:1440–1448. doi: 10.1016/j.celrep.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DAVID Bioinformatics Resources. [(accessed on 21 March 2014)]; Available online: http://david.abcc.ncifcrf.gov/
- 41.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Huang da W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., et al. David bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Protein Index. [(accessed on 21 March 2014)]. Available online: http://www.ebi.ac.uk/IPI.
- 44.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Trudgian D.C., Thomas B., McGowan S.J., Kessler B.M., Salek M., Acuto O. Cpfp: A central proteomics facilities pipeline. Bioinformatics. 2010;26:1131–1132. doi: 10.1093/bioinformatics/btq081. [DOI] [PubMed] [Google Scholar]
- 46.O’Flaherty L., Adam J., Heather L.C., Zhdanov A.V., Chung Y.L., Miranda M.X., Croft J., Olpin S., Clarke K., Pugh C.W., et al. Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Hum. Mol. Genet. 2010;19:3844–3851. doi: 10.1093/hmg/ddq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.STRINGv9.1. [(accessed on 21 March 2014)]. Available online: http://string-db.org/
- 48.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. String 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. Dj-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekito A., Koide-Yoshida S., Niki T., Taira T., Iguchi-Ariga S.M., Ariga H. Dj-1 interacts with hipk1 and affects h2o2-induced cell death. Free Radic. Res. 2006;40:155–165. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- 51.Junn E., Jang W.H., Zhao X., Jeong B.S., Mouradian M.M. Mitochondrial localization of dj-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009;87:123–129. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J., Li L., Chin L.S. Parkinson disease protein dj-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum. Mol. Genet. 2010;19:2395–2408. doi: 10.1093/hmg/ddq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally R.S., Davis B.K., Clements C.M., Accavitti-Loper M.A., Mak T.W., Ting J.P. Dj-1 enhances cell survival through the binding of cezanne, a negative regulator of nf-kappab. J. Biol. Chem. 2011;286:4098–4106. doi: 10.1074/jbc.M110.147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K.S., Kim J.S., Park J.Y., Suh Y.H., Jou I., Joe E.H., Park S.M. Dj-1 associates with lipid rafts by palmitoylation and regulates lipid rafts-dependent endocytosis in astrocytes. Hum. Mol. Genet. 2013;22:4805–4817. doi: 10.1093/hmg/ddt332. [DOI] [PubMed] [Google Scholar]
- 55.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The parkinson’s disease protein dj-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirota K., Matsui M., Iwata S., Nishiyama A., Mori K., Yodoi J. Ap-1 transcriptional activity is regulated by a direct association between thioredoxin and ref-1. Proc. Natl. Acad. Sci. USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell D.A., Morton S.U., Fernhoff N.B., Marletta M.A. Thioredoxin is required for s-nitrosation of procaspase-3 and the inhibition of apoptosis in jurkat cells. Proc. Natl. Acad. Sci. USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen C.N., Price J.S., Wilkinson K.D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 59.Boudreaux D.A., Maiti T.K., Davies C.W., Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase uchl1 into productive conformation. Proc. Natl. Acad. Sci. USA. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishikawa K., Li H., Kawamura R., Osaka H., Wang Y.L., Hara Y., Hirokawa T., Manago Y., Amano T., Noda M., et al. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase l1 variants. Biochem. Biophys. Res. Commun. 2003;304:176–183. doi: 10.1016/S0006-291X(03)00555-2. [DOI] [PubMed] [Google Scholar]
- 61.Ooi A., Dykema K., Ansari A., Petillo D., Snider J., Kahnoski R., Anema J., Craig D., Carpten J., Teh B.T., et al. Cul3 and nrf2 mutations confer an nrf2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013;73:2044–2051. doi: 10.1158/0008-5472.CAN-12-3227. [DOI] [PubMed] [Google Scholar]
- 62.Yap T.A., Gerlinger M., Futreal P.A., Pusztai L., Swanton C. Intratumor heterogeneity: Seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps110. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 63.Fisher R., Larkin J., Swanton C. Inter and intratumour heterogeneity: A barrier to individualized medical therapy in renal cell carcinoma? Front. Oncol. 2012;2:49. doi: 10.3389/fonc.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frizzell N., Thomas S.A., Carson J.A., Baynes J.W. Mitochondrial stress causes increased succination of proteins in adipocytes in response to glucotoxicity. Biochem. J. 2012;445:247–254. doi: 10.1042/BJ20112142. [DOI] [PubMed] [Google Scholar]
- 65.Piroli G.G., Manuel A.M., Walla M.D., Jepson M.J., Brock J.W., Rajesh M.P., Tanis R.M., Cotham W.E., Frizzell N. Identification of protein succination as a novel modification of tubulin. Biochem. J. 2014 doi: 10.1042/BJ20131581. [DOI] [PMC free article] [PubMed] [Google Scholar]