Summary:
To ensure the stable transmission of the genome during vertebrate cell division, the mitotic spindle must attach to a single locus on each chromosome, termed the centromere. The fundamental requirement for faithful centromere inheritance is the controlled deposition of the centromere-specifying histone, CENP-A. However, the regulatory mechanisms that ensure the precise control of CENP-A deposition have proved elusive. Here, we identify Polo-like kinase 1 (Plk1) as a centromere-localized regulator required to initiate CENP-A deposition in human cells. We demonstrate that faithful CENP-A deposition requires integrated signals from Plk1 and cyclin-dependent kinase (CDK), with Plk1 promoting the localization of the key CENP-A assembly factor, the Mis18 complex, and CDK inhibiting Mis18 complex assembly. By bypassing these regulated steps, we uncoupled CENP-A deposition from cell cycle progression, resulting in mitotic defects. Thus, CENP-A deposition is controlled by a two-step regulatory paradigm comprised of Plk1 and CDK that is crucial for genomic integrity.
Introduction
During cell division, the genome must be segregated equally between the daughter cells. To accomplish this, the mitotic spindle must attach to each chromosome at a single locus, termed the centromere. Chromosomes lacking a functional centromere are unable to attach to the segregation apparatus, resulting in chromosome loss. In contrast, chromosomes with multiple centromeres can attach simultaneously to opposing spindle poles, resulting in chromosome mis-segregation and DNA damage. Indeed, chromosomes with multiple centromeres are frequently observed in cancers and can promote genomic instability and characteristics of tumorigenesis (Gisselsson et al., 2000; Gascoigne and Cheeseman, 2013).
In most eukaryotes, centromeres are specified epigenetically by the presence of the histone H3 variant, CENP-A (Black et al., 2010). Thus, centromere inheritance depends on the maintenance of CENP-A-containing nucleosomes at a single site on each chromosome. During DNA replication, existing CENP-A-containing nucleosomes are distributed to the replicated sister chromatids. Subsequently, CENP-A-containing nucleosomes must be replenished at centromeres. CENP-A deposition is restricted both spatially, to existing centromeres, and temporally, to G1 phase in human cells (Jansen et al., 2007). Current models suggest that this temporal restriction is crucial for faithful centromere inheritance and function (Gómez-Rodríguez and Jansen, 2013). However, the regulatory paradigms that control the propagation of this crucial epigenetic mark remain poorly understood.
The restriction of CENP-A deposition is accomplished at least in part through the regulated recruitment and function of its dedicated deposition machinery. In human cells, CENP-A incorporation is carried out by at least two sets of assembly factors: the Mis18 complex, which assembles from Mis18α, Mis18β, and M18BP1/KNL2 (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007), and the CENP-A chaperone, HJURP (Dunleavy et al., 2009; Foltz et al., 2009). The full Mis18 complex localizes to centromeres beginning at anaphase onset (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007) (Fig. 1A). HJURP recruitment and new CENP-A deposition then occur during G1 (Jansen et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009) (Fig. 1A). Recent work demonstrated that cyclin-dependent kinase 1 and 2 (CDK1 and CDK2) negatively regulate CENP-A deposition to restrict this process to G1 (Silva et al., 2012). However, thus far it has not been possible to uncouple CENP-A deposition from its temporal regulation without also disrupting cell cycle progression (Silva et al., 2012). This suggests that key mechanistic steps or regulatory paradigms for the control of CENP-A deposition remain to be defined.
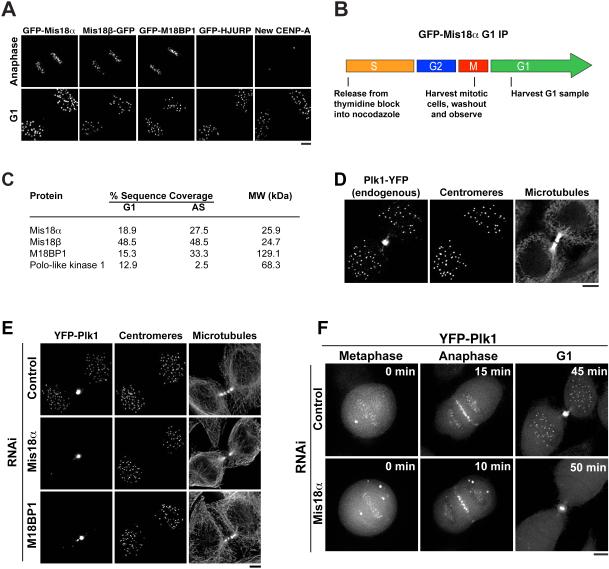
Figure 1. Plk1 localizes to G1 centromeres in a Mis18 complex-dependent manner.
A) Images showing the localization of components of the CENP-A deposition pathway in anaphase and G1. Time-lapse images of single cells are shown for Mis18α, Mis18β, M18BP1 and HJURP. New CENP-A-SNAP was labeled using a quench-pulse strategy (Jansen et al., 2007) in fixed cells. B) Schematic describing the isolation of G1 samples of GFP-Mis18α cells for analysis by mass spectrometry. C) Summary of mass spectrometry results following immunoprecipitation of GFP-Mis18α. Proteins shown are those identified in the GFP-Mis18α immunoprecipitation, but not in unrelated immunoprecipitations of other GFP-tagged proteins. AS: asynchronous sample, generated from cells that failed to arrest in nocodazole. D) G1 localization of Plk1 tagged with YFP at the endogenous locus. Centromeres are marked with anti-centromere antibodies. E) Immunofluorescence images showing YFP-Plk1 localization in Mis18 complex-depleted cells (with γ adjustment). Centromeres are identified using anti-centromere antibodies. F) Time lapse images of YFP-Plk1 in Mis18α-depleted cells. Numbers represent minutes after the metaphase image. Panels are not scaled equivalently, but are scaled (with γ adjustment) to show the full range of data. Scale bars = 5 μm. See also Fig. S1.
We sought to determine the molecular basis for the regulation of CENP-A deposition. Our data establish a regulatory paradigm for CENP-A deposition that combines global regulation by CDK and a centromere-localized initiation signal provided by Polo-like kinase 1 (Plk1). Defining the mechanisms by which Plk1 and CDK control CENP-A deposition allowed us to bypass the cell cycle regulation of CENP-A deposition, resulting in severe mitotic defects. Thus, the regulation of CENP-A deposition downstream of Plk1 and CDK is critical to protect the integrity of the genome.
Results
Plk1 displays Mis18 complex-dependent localization to G1 centromeres
To identify potential factors that regulate CENP-A deposition, we began by isolating GFP-Mis18α by affinity purification from HeLa cells that were synchronized by mitotic shake-off and then allowed to progress into G1 (Fig. 1B). Mass spectrometry analysis identified the established components of the Mis18 complex – Mis18α, Mis18β, and M18BP1 (Fig. 1C). In addition, we found that Plk1 co-purified with the Mis18 complex (Fig. 1C). The isolation of Plk1 with the Mis18 complex from G1 cells was unexpected, as Plk1 has been described predominantly as an M-phase kinase (Barr et al., 2004). To assess the relevance of the association between the Mis18 complex and Plk1, we analyzed HeLa cells stably expressing YFP-Plk1. Prior work focused on the localization of Plk1 to centrosomes, mitotic kinetochores, the spindle midzone, and the midbody (Archambault and Glover, 2009). In addition, as reported by others (Arnaud et al., 1998; Kishi et al., 2009), we found that YFP-Plk1 localized to centromeres in G1, concurrent with Mis18α localization (Fig. S1A). We observed identical localization when we tagged the endogenous Plk1 locus with YFP using CRISPR/Cas-mediated genome editing (Plk1-YFP) (Fig. 1D). In contrast, the related Polo-like kinases, Plk2 and Plk3, did not localize to G1 centromeres (Fig. S1B). Depletion of Mis18α or M18BP1 by RNAi abolished Plk1 localization to G1 centromeres (Fig. 1E, F; Fig. S1C, D). In contrast, Plk1 localization to the spindle midzone, midbody, and mitotic kinetochores was unaffected by Mis18α or M18BP1 depletion (Fig. 1E, F; Fig. S1C; data not shown). These observations indicate that Plk1 localizes to G1 centromeres in a Mis18 complex-dependent manner.
Plk1 activity is required for new CENP-A deposition
The co-purification of Plk1 with the Mis18 complex and the localization of Plk1 to G1 centromeres suggested that Plk1 might contribute to CENP-A deposition. To test this, we inhibited Plk1 kinase activity using the small molecule BI2536 (Lénárt et al., 2007; Steegmaier et al., 2007) and assessed the incorporation of new CENP-A using a CENP-A-SNAP quench-pulse assay (Jansen et al., 2007) (Fig. 2A). We observed a dramatic defect in the deposition of new CENP-A following BI2536 treatment (Fig. 2A; Fig. S2A). We also observed a reduction in new CENP-A incorporation following treatment with the bulky ATP analogue 3MB-PP1 in an RPE1 cell line expressing an analogue-sensitive allele of Plk1 (Plk1as) (Burkard et al., 2007) (Fig. S2B). These data indicate that Plk1 activity is required for CENP-A deposition.
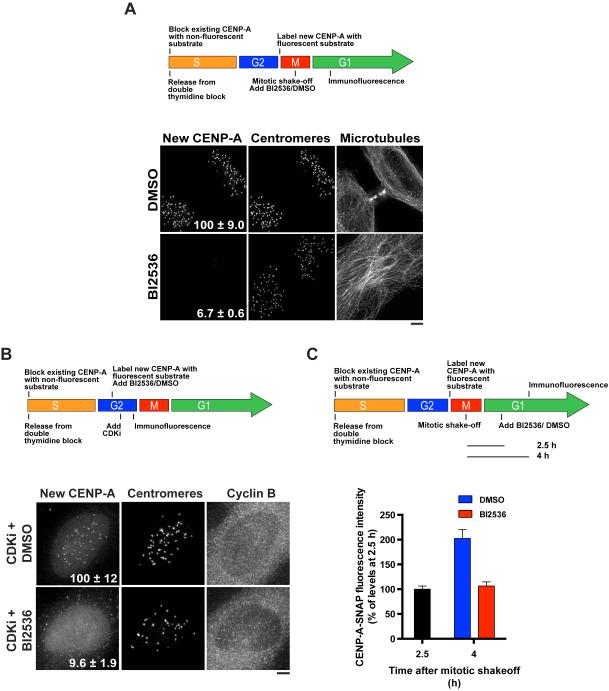
Figure 2. Plk1 activity is required for new CENP-A deposition.
A) Top: Schematic of the cell synchronization and SNAP-CENP-A labeling strategy to detect the deposition of newly synthesized CENP-A using a fluorescent quench-pulse strategy (Jansen et al., 2007). Mitotic cells were harvested and allowed to progress through G1 in the presence of BI2536 or DMSO for 2.5 h before staining. Bottom: Immunofluorescence images showing incorporation of new CENP-A-SNAP (labeled with SNAP-Cell Oregon Green) following treatment with BI2536 or DMSO. Centromeres are identified using anti-centromere antibodies. The microtubule morphology observed following BI2536 treatment is characteristic of failed cytokinesis due to Plk1 inhibition. Numbers represent CENP-A-SNAP centromeric fluorescence intensity as percent of control, ± s.e.m, p < 0.001 (Student’s t-test), n = 20 G1 cell pairs. B) Top: Schematic of the cell synchronization and SNAP-CENP-A labeling strategy used to test the Plk1 dependence of new CENP-A deposition in G2 phase following inhibition of CDK by flavopiridol (CDKi). Bottom: Immunofluorescence images showing incorporation of new CENP-A-SNAP (labeled with SNAP-Cell TMR-Star) following treatment with BI2536 or DMSO, and CDK inhibition. Centromeres are identified using anti-centromere antibodies. Numbers represent CENP-A-SNAP centromeric fluorescence intensity as percent of control, ± s.e.m, p < 0.001 (Student’s t-test), n = 20 cyclin Bhigh cells. C) Top: Schematic of the cell synchronization and SNAP-CENP-A labeling strategy used to test whether the maintenance of newly deposited CENP-A depends on Plk1. Bottom: Quantification of centromeric fluorescence intensity of new CENP-A-SNAP (labeled with SNAP-Cell Oregon Green) as percent of levels at 2.5 h, ± s.e.m, n = 20 G1 cell pairs per condition per time point. Scale bars = 5 μm. See also Fig. S2.
In human cells, CENP-A deposition is restricted to the G1 phase of the cell cycle (Jansen et al., 2007). As Plk1 plays an established role in cell cycle progression (Barr et al., 2004), we sought to test whether the observed defects in CENP-A deposition were due to global effects of Plk1 inhibition on cell state. However, the BI2536-treated cells with a G1-like morphology that we analyzed for our experiments displayed cell cycle markers consistent with an unperturbed G1 state, including increasing levels of nuclear Cdt1-RFP, minimal levels of geminin-GFP (Sakaue-Sawano et al., 2008), diffuse PCNA staining, and low cyclin B1 levels (Fig. S2C, D). This suggests that Plk1 regulates CENP-A deposition independently of its previously reported effects on cell cycle progression.
Recent work demonstrated that CENP-A deposition can be induced in S and G2 cells following the inhibition of CDK (Silva et al., 2012). As we also observed Plk1 localization to centromeres during S phase and G2 (Fig. S2E), we tested whether CENP-A incorporation following CDK inhibition depended on Plk1. Treatment with BI2536 severely disrupted CENP-A deposition in G2 cells following CDK inhibition (Fig. 2B). Together, these data indicate that Plk1 plays a critical role in CENP-A deposition, and that Plk1-dependent regulation of new CENP-A deposition does not depend on residual CDK activity, or regulatory circuits and events that are specific to mitotic exit, such as cytokinesis (Petronczki et al., 2007).
Previous work found that newly deposited CENP-A must be actively maintained by a process involving MgcRacGAP (Lagana et al., 2010), which is also a substrate of Plk1 (Wolfe et al., 2009). Therefore, we sought to test whether Plk1 was required to maintain new CENP-A at centromeres. To this end, we allowed cells with fluorescently labeled new CENP-A-SNAP to progress through G1 for 2.5 h before the addition of BI2536. In this assay, we found that newly deposited CENP-A remained intact following BI2536 treatment (Fig. 2C). In contrast, ongoing CENP-A deposition was halted following Plk1 inhibition (Fig. 2C). This suggests that Plk1 is continuously required to direct CENP-A deposition, but that it is not required to maintain newly incorporated CENP-A.
The Mis18 complex and HJURP, but not CENP-C, require Plk1 activity for proper localization to G1 centromeres
We next sought to determine the mechanisms by which Plk1 promotes CENP-A incorporation. As a first step, we assessed the functional contributions of Plk1 to each step in the CENP-A deposition process. Previous work implicated the constitutive centromere protein CENP-C as a centromere-localized binding partner for the Mis18 complex (Moree et al., 2011; Dambacher et al., 2012). However, the functional contribution of CENP-C to Mis18 complex recruitment in mammalian cells has remained unclear (Dambacher et al., 2012). Therefore, we analyzed Mis18 complex localization following depletion of CENP-C by RNAi. As CENP-C depletion causes a mitotic arrest, we drove cells into G1 using an inhibitor of the checkpoint kinase Mps1. We found that depletion of CENP-C, but not the constitutive centromere protein CENP-T, strongly reduced Mis18 complex localization to G1 centromeres (Fig. 3A), indicating that CENP-C is required for Mis18 complex recruitment. However, treatment with the Plk1 inhibitor BI2536 did not affect CENP-C localization (Fig. 3B), indicating that Plk1 inhibition does not result in the global destabilization of interphase centromeres.
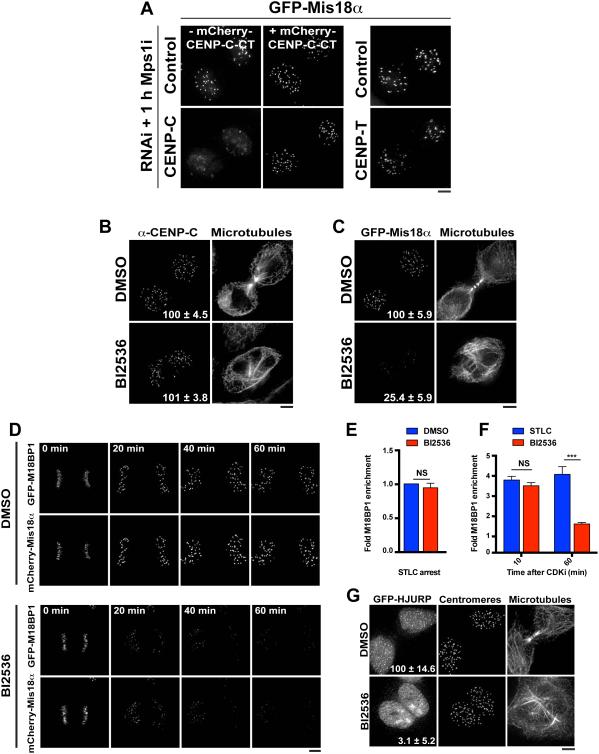
Figure 3. Plk1 is required for the localization of the Mis18 complex and HJURP to G1 centromeres.
A) Live cell images of GFP-Mis18α cells following 48 h treatment with siRNAs against the indicated targets. Penetrant RNAi was confirmed by observation of a disorganized metaphase plate before cells were driven into G1 using an Mps1 inhibitor (Mps1i; AZ3146) for 1 h. The Mis18α recruitment defect observed in CENP-C RNAi can be rescued by expression of the RNAi-resistant CENP-C C-terminus (CT: aa 510-934). Right: depletion of the constitutive centromere protein CENP-T does not affect Mis18α localization. B) Immunofluorescence images showing localization of CENP-C following treatment with DMSO or BI2536. Numbers represent centromeric fluorescence intensity as percent of control, ± s.e.m, n = 20 G1 cell pairs, p > 0.05 (Student’s t-test). C) Immunofluorescence images showing GFP-Mis18α localization following treatment with BI2536 or DMSO. Numbers represent centromeric fluorescence intensity as percent of control, ± s.e.m, n = 20 G1 cell pairs for each condition, p < 0.001 (Student’s t-test). D) Time-lapse images of live cells co-expressing GFP-M18BP1 and mCherry-Mis18α following treatment with BI2536 or DMSO. BI2536 was added in early anaphase. Numbers indicate minutes after BI2536 addition. E) Quantification of centromeric GFP-M18BP1 levels in live prometaphase-like cells following treatment with BI2536. Cells were treated with the Eg5 inhibitor STLC for 4 h to induce a mitotic arrest before addition of BI2536 or DMSO for 1 h. Numbers are presented as fold enrichment over initial centromeric fluorescence, ± s.e.m, n = 10 cells. NS: not significant, p > 0.05 (Student’s t-test). F) Quantification of centromeric GFP-M18BP1 levels in live cells following treatment with BI2536. To avoid confounding effects due to delays in uptake and function of the drug, cells were pre-treated with BI2536 for 1h. As a control, cells were treated with STLC, which, like BI2536, induces a mitotic arrest with monopolar spindle. To bypass the arrest, mitotic exit was induced with the CDK inhibitor flavopiridol for both BI2536-and STLC-treated cells. Times after CDK inhibition were chosen to correspond to anaphase (10 min) and G1 (60 min). Numbers are presented as fold increase over centromeric fluorescence at t = 0, ± s.e.m, n = 10 cells. ***: p < 0.001; NS: not significant, p > 0.05 (Student’s t-test). G) Immunofluorescence images showing localization of GFP-HJURP following treatment with BI2536 or DMSO. Centromeres are identified using anti-centromere antibodies. The images are not scaled equivalently, but are scaled to show the full range of the data. Numbers represent centromeric fluorescent intensity as percent of control, ± s.e.m, n = 20 G1 cell pairs, p < 0.001 (Student’s t-test). Scale bars = 5 μm. See also Fig. S3.
We next tested the contribution of Plk1 to Mis18 complex localization. BI2536-treated cells displayed a substantial decrease in GFP-M18BP1 and mCherry- or GFP-Mis18α localization to G1 centromeres (Fig. 3C, D), indicating that Plk1 activity is required for robust Mis18 complex localization. In addition to localizing to G1 centromeres, we found that GFP-M18BP1 localized to centromeres throughout mitosis (Fig. S3A, B), consistent with previous reports for Xenopus laevis M18BP1 localization (Moree et al., 2011). Therefore, we also tested the effects of Plk1 inhibition on GFP-M18BP1 localization in both prometaphase and an anaphase-like state induced by CDK inhibition. In contrast to the defects observed in G1 cells (Fig. 3C; Fig. 3F, t = 60 min after CDK inhibition), the prometaphase (Fig. 3E) and anaphase-like (Fig. 3F, t = 10 min after CDK inhibition) localization of M18BP1 was unaffected by Plk1 inhibition. Taken together, these data indicate that Plk1 is required to maintain the localization of the Mis18 complex at centromeres specifically during G1, the period when CENP-A deposition occurs.
Finally, we analyzed the effect of Plk1 inhibition on the centromere localization of the CENP-A chaperone, HJURP. Consistent with the defects in CENP-A deposition and Mis18 complex localization described above, BI2536-treated cells exhibited striking defects in the centromere localization of GFP-HJURP (Fig. 3G). These data indicate that Plk1 activity is required for multiple aspects of the CENP-A deposition process.
The Mis18 complex is a key target of Plk1 during CENP-A deposition
To define the direct targets of Plk1, we performed in vitro kinase assays using recombinant components of the CENP-A deposition machinery. For these assays, we reconstituted the full Mis18 complex by co-expression of its subunits in bacteria (Fig. S4A). Plk1 directly phosphorylated the Mis18 complex based on radioactive kinase assays (Fig. 4A; Fig. S4B) and mass spectrometry analysis of in vitro phosphorylated samples (Fig. S4C). In contrast, Plk1 did not efficiently phosphorylate HJURP, or a C-terminal region of CENP-C containing the M18BP1-binding region (Moree et al., 2011; Dambacher et al., 2012) that we found to be necessary and sufficient for Mis18 complex recruitment (Fig. 4A; Fig. 3A). These data suggest that the Mis18 complex is a major target of Plk1 in the CENP-A deposition pathway.
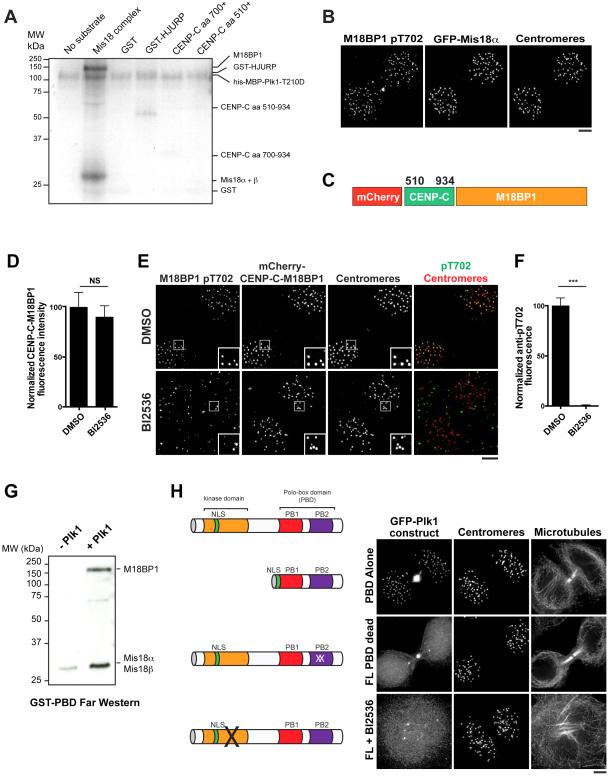
Figure 4. Plk1 binds to and phosphorylates the Mis18 complex.
A) Autoradiogram showing Plk1 phosphorylation of recombinant proteins in the CENP-A deposition pathway in the presence of 32P-ATP. The approximate migration of each protein is indicated on the right based on GelCode Blue staining (see Fig. S4B). aa: amino acid. B) Immunofluorescence images of G1 cells expressing GFP-Mis18α, co-stained with α-M18BP1 pT702. Centromeres are marked with α-CENP-A antibodies. C) Schematic of the CENP-C-M18BP1 fusion used to bypass regulated M18BP1 localization. Numbers represent amino acid positions within CENP-C. D) Quantification of mCherry-CENP-C-M18BP1 levels following treatment with BI2536 or DMSO as percent of DMSO levels. Both DMSO and BI2536-treated populations were depleted for endogenous M18BP1. Error bars represent s.e.m, n = 20 G1 cell pairs. NS: not significant, p > 0.05. E) Immunofluorescence images showing CENP-C-M18BP1-expressing cells stained for α-M1B81P pT702 following treatment with BI2536. Centromeres are identified with α-CENP-A antibody. F) Quantification of pT702 centromeric fluorescence in CENP-C-M18BP1-expressing cells following treatment with BI25356 (quantification of Fig. 4E). Error bars represent s.e.m, n = 20 G1 cell pairs, *** p < 0.001. G) Far-Western analysis of recombinant GST-PBD binding to the recombinant Mis18 complex in the presence or absence of Plk1. H) Left: schematic of modified GFP-Plk1 constructs. Right: Immunofluorescence images showing localization of modified GFP-Plk1 constructs: PBD alone (PBD truncation + nuclear localization signal), FL PBD dead (full length protein with mutations rendering the Polo-Box unable to bind to its substrates (Elia et al., 2003b)), FL + BI2536 (full-length protein after treatment with the Plk1 inhibitor BI2536). Images are scaled with γ adjustment. Scale bars = 5 μm. See also Fig. S4.
A subset of the phosphorylation sites in the Mis18 complex that we mapped in vitro has also been identified by mass spectrometry analysis of endogenous samples (Dephoure et al., 2008; Shiromizu et al., 2013) (Fig. S4C). To directly test whether the Mis18 complex is a substrate of Plk1 in vivo, we generated an antibody specific to phospho-T702 on M18BP1 (Fig. S4D). This antibody detected centromeres by immunofluorescence in control cells, but not following M18BP1 RNAi (Fig. 4B; Fig. S4E). Treatment with BI2536 abolished this signal (data not shown). However, it remained possible that the signal was eliminated because BI2536 treatment also causes Mis18 complex delocalization (Fig. 3). To overcome this, we uncoupled Mis18 complex localization from Plk1 activity by generating an in-frame fusion between M18BP1 and the C terminal domain of CENP-C described above (CENP-C-M18BP1) (Fig. 4C). Localization of the CENP-C-M18BP1 fusion was unaffected by Plk1 inhibition (Fig. 4D), consistent with the Plk1-independent localization of CENP-C (Fig. 3B). Despite the continued localization of the CENP-C-M18BP1 fusion, the pT702 signal at centromeres was eliminated following BI2536 treatment (Fig. 4E, F). Collectively, these data suggest that Plk1 directly phosphorylates the Mis18 complex in vitro and in cells.
Plk1 binds to many of its substrates via a phosphopeptide-binding module termed the Polo-Box Domain (PBD) (Elia et al., 2003a). Therefore, we sought to determine if the Mis18 complex and the Plk1 PBD interact directly. Substrates are primed to interact with the PBD by kinases including CDK (Elia et al., 2003a) and Plk1 itself (known as self-priming) (Burkard et al., 2007; Neef et al., 2007). We found that GST-PBD bound robustly to the recombinant Mis18 complex by Far-Western analysis, but only when the Mis18 complex had been previously phosphorylated with Plk1 (Fig. 4G). Consistent with a Plk1 phosphorylation-dependent interaction between the Mis18 complex and the Plk1 PBD, we found that Plk1 localization to G1 centromeres required both a functional PBD and Plk1 kinase activity (Fig. 4H). Therefore, Plk1 can phosphorylate and bind to the Mis18 complex directly via its PBD.
Plk1 phosphorylation of the Mis18 complex promotes new CENP-A deposition
To test the consequences of Mis18 complex phosphorylation by Plk1, we generated cell lines expressing RNAi-resistant versions of M18BP1, Mis18α, or Mis18β. Wild type versions of these constructs were functional to carry out CENP-A deposition in the absence of the corresponding endogenous proteins (Fig. 5A, B; Fig. S5A). We next generated mutants in which the mapped Plk1 phosphorylation sites were mutated to alanine to prevent their phosphorylation (Plk1-A mutants; see Table S2). In the presence of the endogenous proteins, these mutants displayed wild type localization (data not shown), suggesting that these mutations do not substantially disrupt the structural integrity of these proteins.
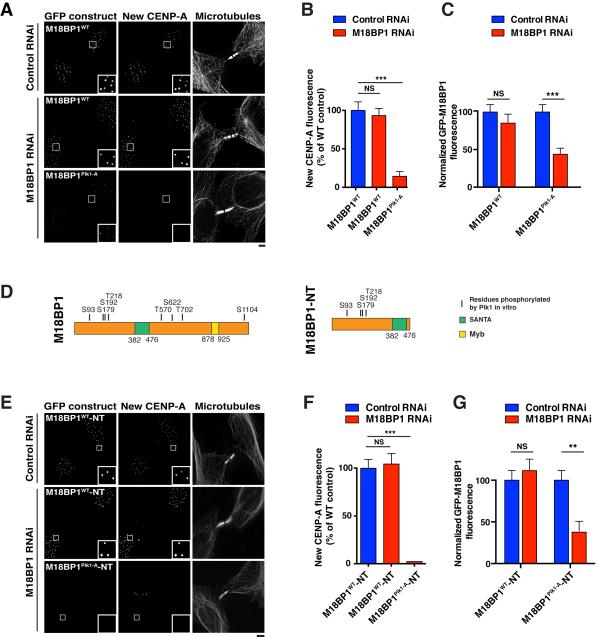
Figure 5. Plk1 phosphorylation of the Mis18 complex is required for CENP-A deposition.
A) Immunofluorescence images showing new CENP-A deposition in cells expressing GFP fusions of either M18BP1WT or M18BP1Plk1-A following treatment with the indicated siRNAs. New CENP-A is labeled using SNAP-Cell TMR-Star. B) Quantification of centromeric fluorescence intensity of new CENP-A-SNAP following replacement of endogenous M18BP1 with RNAi-resistant GFP-M18BP1WT or GFP-M18BP1Plk1-A. Numbers are presented as a percentage of the intensity in M18BP1WT cells + control RNAi. Error bars represent s.e.m, n = 20 G1 cell pairs. NS: not significant, p > 0.05; ***: p < 0.001. Wild type and mutant cell lines were generated from the same parental CENP-A-SNAP cell line (see Table S1) and after generation continue to have equivalent levels of total CENP-A-SNAP protein (data not shown). C) Quantification of centromeric GFP-M18BP1WT or GFP-M18BP1Plk1-A fluorescence intensity in cells in which M18BP1 has been depleted. Error bars represent s.e.m, n = 20 G1 cell pairs. ***: p < 0.001; NS: not significant, p > 0.05 (Student’s t-test). D) Left: Schematic of M18BP1 showing residues phosphorylated by Plk1 in vitro. Right: Schematic of an N Terminal domain of M18BP1 (M18BP1-NT) that is sufficient for M18BP1 centromere localization, Mis18α recruitment and CENP-A deposition. SANTA: SANT-associated domain; Myb: Myb DNA-binding domain. E) Immunofluorescence images showing new CENP-A deposition in cells expressing GFP fusions of either M18BP1WT-NT or M18BP1Plk1-A-NT following treatment with the described siRNAs. New CENP-A is labeled using SNAP-Cell TMR-Star. F) Quantification of centromeric fluorescence intensity of new CENP-A-SNAP following replacement of endogenous M18BP1 with RNAi-resistant GFP-M18BP1WT-NT or GFP-M18BP1Plk1-A-NT. Numbers are presented as a percentage of the intensity in M18BP1WT-NT cells + control RNAi. Error bars represent s.e.m, n = 20 G1 cell pairs. NS: not significant, p > 0.05; ***: p < 0.001. G) Quantification of centromeric GFP-M18BP1WT-NT or GFP-M18BP1Plk1-A-NT fluorescence intensity in cells in which M18BP1 has been depleted. Localization of GFP-M18BP1Plk1-A-NT is weak even in the presence of the endogenous protein (Fig. S5G). Error bars represent s.e.m, n = 20 G1 cell pairs. **: p < 0.005; NS: not significant, p > 0.05 (Student’s t-test). Scale bars = 5 μm. See also Fig. S5.
To determine the importance of these phosphorylated residues for CENP-A deposition, we tested CENP-A incorporation in the mutant cell lines following depletion of the endogenous proteins by RNAi. Cells expressing mCherry-Mis18αPlk1-A, Mis18βPlk1-A-GFP, or co-expressing Mis18αPlk1-A and Mis18βPlk1-A did not display defects in new CENP-A deposition following depletion of their endogenous counterparts (Fig. S5A). In contrast, cells expressing GFP-M18BP1Plk1-A displayed severe defects in new CENP-A-SNAP incorporation following depletion of endogenous M18BP1 (Fig. 5A, B). We attempted to mimic Plk1 phosphorylation by mutating the Plk1 phosphorylation sites to aspartate (GFP-M18BP1Plk-D). However, GFP-M18BP1Plk-D displayed similar defects in CENP-A deposition as GFP-M18BP1Plk1-A (Fig. S5B). We speculate that aspartate does not effectively mimic the phosphate group in this context and thus renders the mutant non-functional. These data indicate that Plk1 phosphorylation of M18BP1 is required for CENP-A deposition.
In the absence of endogenous M18BP1, we also observed a significant reduction in the levels of GFP-M18BP1Plk1-A at centromeres (Fig. 5A, C). To test whether the defect in M18BP1Plk1-A localization was caused by a global decrease in the levels of kinetochore proteins due to defective CENP-A deposition, we tested the effect of directly depleting CENP-A by RNAi on GFP-M18BP1Plk1-A localization (Fig. S5C, D). CENP-A depletion had a minimal effect on GFP-M18BP1Plk1-A levels (Fig. S5D), consistent with previous reports demonstrating a limited reduction in CENP-C levels at a similar time point following induction of a conditional CENP-A knockout (Fachinetti et al., 2013). This suggests that the observed reduction in M18BP1Plk1-A localization is due to a defect intrinsic to the mutant. Collectively, these data demonstrate that direct phosphorylation of the Mis18 complex by Plk1 promotes M18BP1 localization and new CENP-A deposition.
The identified Plk1 phosphorylation sites in M18BP1 are present throughout the protein (Fig. 5D). We found that an N-terminal region (amino acids 1-490) was sufficient for M18BP1 centromere localization and Mis18α recruitment (Fig. S5E, F), and is functional to restore CENP-A deposition to M18BP1-depleted cells (Fig. 5E, F). Therefore, we tested the requirements for the Plk1 phosphorylation sites that we identified in this N terminal (NT) region (GFP-M18BP1Plk1-A-NT) (Fig. 5D). GFP-M18BP1-NT showed robust centromere localization in the presence and absence of endogenous M18BP1 (Fig. 5E, 5G). In contrast, GFP-M18BP1Plk1-A-NT localized weakly to centromeres in the presence of the endogenous protein (Fig. S5G), and this localization was further reduced upon depletion of endogenous M18BP1 (Fig. 5E, G). In addition, CENP-A deposition was severely defective in cells expressing GFP-M18BP1Plk1-A-NT following M18BP1 RNAi (Fig. 5E, F). These data indicate that Plk1 phosphorylation of the N-terminus of M18BP1 is critical for M18BP1 localization and function.
The phenotypes observed in the M18BP1Plk1-A mutant suggest that Plk1 phosphorylation of M18BP1 promotes its localization and new CENP-A incorporation. To distinguish whether the primary function of Plk1 during CENP-A deposition is to regulate M18BP1 localization, we bypassed the regulated M18BP1 localization using the CENP-C-M18BP1 fusion. As described above, in the absence of the CENP-C fusion partner, M18BP1Plk1-A displayed severely defective CENP-A deposition following M18BP1 depletion (Fig. 5A, B). In contrast, CENP-C-M18BP1Plk1-A partially restored CENP-A deposition to M18BP1-depleted cells (Fig. S5H), although to a lesser extent than wild type CENP-C-M18BP1. This suggests that Plk1 phosphorylation of M18BP1 primarily affects CENP-A deposition by modulating M18BP1 localization.
Cyclin-dependent kinase regulates Mis18 complex assembly
The combination of these data demonstrates that Plk1 acts as a key regulator for CENP-A deposition, and functions at least in part through modulating M18BP1 localization. Previous work demonstrated that CDK1/2 activity inhibits CENP-A deposition (Silva et al., 2012). We therefore sought to define the mechanisms by which these two kinases coordinately regulate the CENP-A deposition process. CDK has been proposed to act by restricting M18BP1 localization to anaphase and G1 (Silva et al., 2012). However, when we mutated the full complement of serine and threonine residues corresponding to CDK consensus phosphorylation sites to alanine (GFP-M18BP1CDK-A; Table S2), we found that this mutant displayed similar temporal localization to wild type GFP-M18BP1, localizing to centromeres from mitotic entry through G1 (data not shown). In contrast, Mis18α and Mis18β did not localize until anaphase onset (Fig. S3A, Fig. 1A), suggesting that assembly of the Mis18 complex is cell cycle regulated.
To test whether CDK controls the recruitment of Mis18α and Mis18β, we next used the CENP-C-M18BP1 fusion, which localizes to centromeres constitutively (Fig. S6A). Despite the constitutive localization of CENP-C-M18BP1, Mis18α localization remained restricted to anaphase/G1 in cells expressing this fusion (Fig. S6A). In contrast, expression of a fusion between the CENP-C C-terminal domain and M18BP1CDK-A (CENP-C-M18BP1CDK-A) resulted in premature GFP-Mis18α recruitment (Fig. 6A). These data indicate that the CDK-dependent inhibition of CENP-A deposition occurs at least in part through preventing assembly of the Mis18 complex at centromeres.
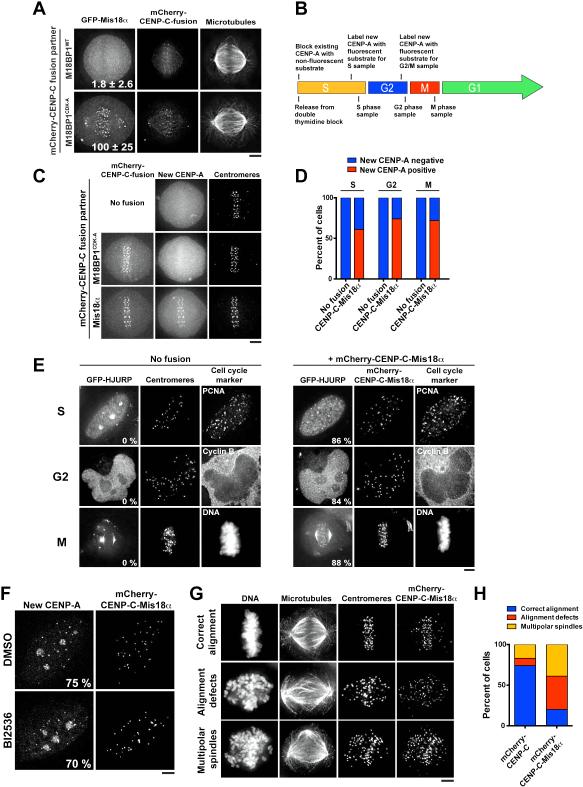
Figure 6. Bypassing CDK and Plk1 regulation induces cell-cycle-uncoupled CENP-A deposition.
A) Immunofluorescence images showing GFP-Mis18α localization in cells transiently transfected with either mCherry-CENP-C-M18BP1WT or mCherry-CENP-C-M18BP1CDK-A. Numbers represent centromeric fluorescence intensity as percent of cells transfected with CENP-C-M18BP1CDK-A ± s.e.m, n = 20 cells, p < 0.001 (Student’s t-test). B) Schematic of the cell synchronization and SNAP-CENP-A labeling strategy to detect the deposition of newly synthesized CENP-A in S, G2 and M phases. C) Immunofluorescence images showing the presence or absence of new CENP-A-SNAP (labeled with SNAP-Cell Oregon Green) at mitotic centromeres in cells expressing either mCherry-CENP-C-M18BP1CDK-A or mCherry-CENP-C-Mis18α after 24 h of induction of the fusion. Centromeres are marked with α-CENP-A. D) Quantification of the percent of cells observed with new CENP-A in each cell cycle stage, 24 h after induction of the fusion. n ≥ 100 cells per stage. E) Immunofluorescence images showing the recruitment of HJURP to centromeres throughout the cell cycle, indicative of ongoing CENP-A deposition. Centromeres are marked with anti-centromere antibodies. Numbers represent percentage of transfected cells in which GFP-HJURP was observed at centromeres, n = 50 cells. F) Immunofluorescence images showing deposition of new CENP-A-SNAP (labeled with SNAP-Cell Oregon Green) in S phase following treatment of CENP-C-Mis18α-expressing cells with BI2536. S phase cells are identified by punctate PCNA foci (not shown). Numbers represent percent of cells showing robust centromeric new CENP-A-SNAP, n = 100 cells per condition. G) Immunofluorescence images summarizing mitotic defects observed in CENP-C-Mis18α-expressing cells after 48 h of induction of the fusion. Centromeres are marked with anti-centromere antibodies. H) Quantification of the percent of cells observed with the chromosome alignment phenotypes depicted in part G. n = 100 cells. Scale bars = 5 μm. See also Fig. S6.
We next sought to determine whether Plk1 and CDK regulate separate aspects of the CENP-A deposition pathway. Premature mitotic recruitment of Mis18α in cells expressing the CENP-C-M18BP1CDK-A was not affected by BI2536 treatment or mutation of the Plk1 phosphorylation sites in M18BP1 alongside the CDK sites (CENP-C-M18BP1CDK-A-Plk1-A; Table S2) (Fig. S6B, C). Thus, Plk1 is not required for the assembly of the Mis18 complex. Overall, our data suggest that Plk1 and CDK control distinct steps in the CENP-A deposition process, with CDK regulating Mis18 complex assembly and Plk1 regulating M18BP1 localization.
The cell cycle restriction of CENP-A deposition is crucial for genomic integrity
The results above define key regulatory steps for the control of the CENP-A deposition process downstream of Plk1 and CDK. To determine if additional regulatory steps are required to promote CENP-A deposition, we sought to bypass these steps and uncouple CENP-A deposition from its normal cell cycle restriction. Although cells expressing the CENP-C-M18BP1CDK-A fusion recruit Mis18α to mitotic centromeres, we did not observe new CENP-A deposition during mitosis at the expression levels tested (Fig. 6B, C). As our work suggested that Plk1 and CDK both regulate steps upstream of Mis18α recruitment, we directly targeted Mis18α to centromeres. Strikingly, in cells expressing a fusion between the CENP-C C-terminal domain and Mis18α (CENP-C-Mis18α), we observed newly deposited CENP-A-SNAP at centromeres in S, G2, and M phase cells (Fig. 6B, C, D; Fig. S6D).
To determine whether CENP-A deposition is actively occurring during G2 and M phase in cells expressing the CENP-C-Mis18α fusion, we analyzed the localization of the CENP-A chaperone, HJURP. In wild type cells, HJURP recruitment is restricted to G1 phase, concurrent with CENP-A deposition (Fig. 1A; Fig. 6E). In contrast, we observed HJURP localization to centromeres in S, G2 and M phase cells expressing the CENP-C-Mis18α fusion (Fig. 6E). This suggests that CENP-C-Mis18α expressing cells incorporate new CENP-A throughout the cell cycle. To test whether CENP-C-Mis18α expression bypasses the requirement for Plk1, we treated cells with BI2536 immediately following the quenching of existing CENP-A-SNAP with non-fluorescent substrate, and allowed cells to progress through S phase in the presence of the inhibitor. In CENP-C-Mis18α-expressing cells, CENP-A deposition continued following BI2536 treatment (Fig. 6F), indicating that Plk1 acts upstream of Mis18α localization to control CENP-A deposition.
To test the consequences of uncoupling CENP-A incorporation from its cell cycle regulation, we analyzed the behavior of mitotic cells expressing the CENP-C-Mis18α fusion. Intriguingly, cells expressing the CENP-C-Mis18α fusion exhibited severe mitotic defects including dramatically misaligned chromosomes and multipolar spindles (Fig. 6G, H). These phenotypes are consistent with defective centromere and kinetochore function. In contrast, cells expressing the CENP-C C-terminal fragment alone displayed infrequent mitotic defects (Fig. 6H). These data suggest that the precise control of CENP-A deposition downstream of CDK and Plk1 is critical for proper chromosome segregation and genomic integrity.
Discussion
A key goal of the ongoing research in centromere biology has been to define the epigenetic mechanisms that direct the sequence-independent propagation of centromeres. However, the regulatory mechanisms that ensure proper centromere inheritance have remained elusive. Recent work demonstrated that CDK contributes to the cell-cycle restriction of centromere inheritance by globally inhibiting CENP-A deposition (Silva et al., 2012). Here, we defined the requirement for a positive, centromere-localized regulatory signal provided by Plk1 to initiate CENP-A deposition (Fig. 7). This dual control of CENP-A deposition – combining global CDK regulation with a site-specific licensing kinase – is analogous to the paradigms that ensure the fidelity of other key cell-cycle events. For example, although cellular changes in CDK activity are required to restrict DNA replication and centriole duplication to specific windows of the cell cycle, the initiation of these processes requires the licensing kinase Dbf4-dependent kinase (DDK) to act at replication origins (Bell and Dutta, 2002) or Plk4 to act at centrioles (Nigg, 2007).
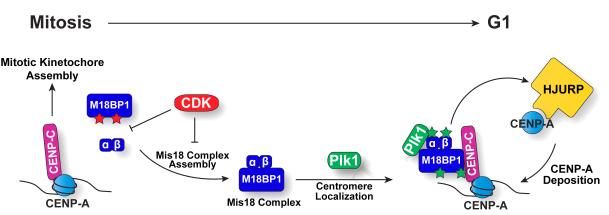
Figure 7. Model for the control of CENP-A deposition by Plk1 and CDK.
CENP-A deposition is accomplished by a two-step regulatory mechanism integrating critical signals from Plk1 and CDK. During S, G2 and M phases, CDK inhibits Mis18 complex assembly. In G1, Plk1 at centromeres binds to and phosphorylates the Mis18 complex to promote its localization and license CENP-A deposition.
To precisely define the roles for Plk1 and CDK in CENP-A deposition, we dissected the regulation of each step in the CENP-A deposition process. In particular, we analyzed 3 key points of regulation: 1) M18BP1 localization to centromeres, 2) Mis18 complex assembly, and 3) new CENP-A deposition by the CENP-A chaperone HJURP. We found that Plk1 is required for CENP-A deposition downstream of CENP-C localization, but upstream of Mis18α recruitment. Further, we defined the Mis18 complex as a direct substrate of Plk1, and demonstrated that Plk1 phosphorylation of M18BP1 promotes its localization and CENP-A deposition. These data indicate that M18BP1 is a key functional target of Plk1. We cannot exclude the possibility that Plk1 phosphorylation of other components of the CENP-A deposition pathway contributes to CENP-A incorporation. However, as the CENP-C-Mis18α fusion bypasses the requirement for Plk1 to promote CENP-A deposition, Plk1 phosphorylation of HJURP or MgcRacGAP, which function downstream of Mis18α localization (Lagana et al., 2010; Barnhart et al., 2011), is unlikely to play a critical role. In addition, although our data suggest that Plk1 phosphorylation controls CENP-A deposition primarily by regulating M18BP1 localization, artificial targeting of M18BP1 to centromeres as a CENP-C fusion does not fully bypass the requirement for Plk1 activity. In particular, we find that the CENP-C-M18BP1Plk1-A fusion does not fully restore CENP-A deposition in the absence of endogenous M18BP1. Thus, the phosphorylation sites in M18BP1 may regulate aspects of M18BP1 function in addition to controlling its localization.
In addition to identifying Plk1 as a regulator of M18BP1 localization, we also demonstrated that the assembly of the Mis18 complex is regulated by CDK. However, our data indicate that Plk1 and CDK act independently to control distinct steps during this process. For example, the regulation of M18BP1 localization by Plk1 does not require CDK activity, whereas regulation of Mis18 complex assembly by CDK does not require Plk1 phosphorylation. Thus, CENP-A deposition is accomplished by a two-step regulatory mechanism integrating critical signals from Plk1 and CDK. Together, these regulators provide the temporal and spatial cues to precisely control CENP-A deposition (Fig. 7).
Underlying the efforts to define the mechanisms that regulate CENP-A deposition is the assumption that the observed cell cycle restriction of this process is functionally important for the propagation or function of this epigenetic mark. By defining the key molecular events required for CENP-A deposition and their regulation, we developed a strategy to bypass the cell cycle regulation of this process. We found that bypassing Plk1 and CDK regulation by expression of a CENP-C-Mis18α fusion induced CENP-A deposition throughout the cell cycle, resulting in severe mitotic defects. This indicates that the precise regulation of CENP-A deposition by Plk1 and CDK is crucial for proper chromosome segregation. These data raise exciting new questions regarding the molecular consequences of uncoupling CENP-A deposition from cell cycle progression. For example, CENP-A deposition during S phase alongside canonical H3 may disrupt centromere integrity, or mitotic CENP-A deposition could destabilize chromosome condensation at centromeres. Ongoing CENP-A deposition during mitosis may also affect kinetochore assembly, either by preventing the recruitment of key kinetochore components, or by generating additional sites for kinetochore formation and thereby disrupting the higher order organization of the kinetochore. Together, our data define key roles for Plk1 and CDK in regulating CENP-A deposition, and establish the vital importance of this regulation for ensuring genomic integrity.
Experimental Procedures
Cell culture
HeLa cell lines were cultured as described previously in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin and 2 mM L-glutamine. hTERT-RPE1 Plk1as cells were maintained in DMEM:F12 with 10% fetal bovine serum (FBS), penicillin/streptomycin and 2 mM L-glutamine. For time-lapse imaging, cells were maintained in CO2-independent media (Invitrogen) with 10% FBS.
Unless otherwise indicated, cells were incubated in 10 μM BI2536 (Thermo Fisher Scientific) for 2.5 h, although severely defective CENP-A deposition was observed at concentrations down to at least 10 nM (Fig. S2A). Where indicated, cells were incubated with 5 μM flavopiridol (Sigma), 2 μM AZ3146 (Tocris), 10 μM 3MB-PP1 (Merck), or 10 μM STLC (Sigma) for 1-2.5 h. HeLa cells were synchronized by double thymidine block using 2 mM thymidine (Sigma) for all immunofluorescence and live cell imaging experiments unless otherwise stated.
Cell line generation and transfection
The cell lines used in this study are described in Table S1. Clonal cell lines stably expressing GFPLAP or mCherryLAP fusions were generated in HeLa cells as described previously (Cheeseman et al., 2004). Tetracycline-inducible cell lines were generated using the Flp-In T-Rex Expression system (Invitrogen) in a HeLa cell line (a gift from Stephen Taylor) and induced using 1 μg/ml tetracycline (Sigma) approximately every 12 h. Due to heterogeneity within inducible cell lines, cells were matched for expression levels based on similar fluorescence where appropriate and cells that lacked detectable fluorescence were disregarded. The wild type M18BP1 cDNA (Silva et al., 2012) was a gift from Lars Jansen (Gulbenkian Institute for Science). E. coli-optimized and RNAi-resistant M18BP1, Mis18α, Mis18β, CENP-C, and corresponding phosphomutants were synthesized by Genewiz or generated using Quikchange (Agilent). Point mutants are described in Table S2.
The Plk1 locus was tagged with eYFP at the C-terminus using CRISPR/Cas-mediated genome engineering in HeLa cells. The oligonucleotide sequences introduced for the targeting site, and to amplify the 5’ and 3’ homology arms are listed in Table S3. Cas9 and sgRNA were expressed in pX330-BFP ((Cong et al., 2013); a gift from Chikdu Shivalila and Rudolf Jaenisch, Whitehead Institute/MIT) as described (Wang et al., 2013). The YFP donor plasmid derived from pL452 (Liu et al., 2003) was a gift from Paul Fields and Laurie Boyer (MIT). pX330 and the donor were co-transfected into HeLa cells at 2.5 μg each and selected after 48 h with 800 μg/ml G418 (Life Technologies) for two weeks.
siRNAs (Table S4) and a non-targeting control were obtained from Dharmacon. RNAi experiments were conducted using Lipofectamine RNAi MAX and serum-free OptiMEM (Invitrogen). DMEM + 10% FBS was added 5–6 h after incubation. Cells were assayed 48 h after transfection. Transient transfections were performed using Lipofectamine 2000 and OptiMEM (Invitrogen) according to manufacturer’s instructions. The Premo FUCCI Cell Cycle Sensor BacMam 2.0 (Invitrogen) was used according to manufacturer’s instructions.
Immunofluorescence and microscopy
Immunofluorescence was conducted using the antibodies listed in Table S5. The pT702 phospho-specific antibody was generated against a synthesized phosphopeptide with the following amino acid sequence: GTLEN(pT)FEGHKSC (New England Peptide LLC, Covance). Serum from the immunized rabbit was depleted against the unphosphorylated peptide and affinity purified against the phosphorylated peptide. For immunofluorescence using the phospho-specific antibody, cells were pre-extracted for 8-10 min in phosphate buffered saline (PBS) + 0.5% Triton-X 100 before fixation in 4% formaldehyde in PBS. Cy2-, Cy3-, and Cy5-conjugated secondary antibodies were obtained from Jackson Laboratories. DNA was visualized using 10 μg/ml Hoechst. G1 cells were identified by nuclear morphology (decondensed chromosomes) and microtubule staining: either two daughter cells connected by a midbody, or two daughter cells connected by a microtubule pattern that is characteristic of cytokinesis failure due to Plk1 inhibition (e.g. Fig. 2A).
Immunofluorescence images were acquired on a DeltaVision Core deconvolution microscope (Applied Precision) equipped with a CoolSnap HQ2 CCD camera and deconvolved where appropriate. For immunofluorescence, approximately 10 Z-sections were acquired at 0.2 μm steps using a 100×, 1.4 NA Olympus U-PlanApo objective. In general, live cell imaging was performed on the DeltaVision microscope using a 60×/1.42 NA Olympus U-PlanApo objective. For the initial characterization of localization (Fig. 1A) and localization of M18BP1 following CDK inhibition (Fig. 3E, F), images were acquired on a Nikon Ti-E inverted microscope with Perfect Focus system as part of an Andor Revolution 500 XD laser system including a Yokogawa CSU-X1 spinning disk confocal and Andor iXon 897 EMCCD camera using a 100×/1.49 NA Apo TIRF objective. For live cell imaging, approximately 4 Z-sections were acquired at 1 μm steps at 5-10 min intervals for 1 h, with re-focusing using DIC before each time point. Images are scaled equivalently when shown for comparison, unless otherwise stated. Quantification of fluorescence intensity was conducted on unprocessed images using Metamorph (Molecular Devices).
SNAP-CENP-A labeling
SNAP quench-pulse labeling was performed as described (Jansen et al., 2007) using a quench of 10 μM SNAP-Cell Block and a pulse of either 3 μM SNAP-Cell TMR-Star or 5 μm SNAP-Cell Oregon Green (New England Biosciences). To assay CENP-A deposition in G1, cells were arrested in G1/S by double thymidine block and existing CENP-A-SNAP was saturated with non-fluorescent SNAP-Cell Block. Cells were released from the block for approximately 9 h before addition of the fluorescent SNAP substrate. SNAP-Cell Oregon Green and SNAP-Cell TMR-Star were used as indicated in the figure legends. For RNAi experiments, cells were fixed after approximately 11 h. For BI2536 treatment, mitotic cells were collected by mitotic shake-off, split into two pools, and plated on poly-lysine coated coverslips. BI2536 was immediately added to one pool, and an equivalent volume of DMSO was added to the other. The cells were allowed to progress through G1 for 2-2.5 h before fixation and immunofluorescence.
To assay CENP-A deposition in S phase, cells were quenched and released from G1/S as above and allowed to progress for 5 h before addition of the fluorescent SNAP substrate, and fixed at 6 h after release. S phase cells were then selected by punctate PCNA foci. To assay CENP-A deposition in G2 and M phase, cells were allowed to progress for 8 h before addition of the fluorescent SNAP substrate. Cells were fixed at 9 h (G2) or 10 h (M) after release. G2 cells were identified by high cytoplasmic cyclin-B. Mitotic cells were identified by DNA morphology or the presence of a mitotic spindle.
Protein expression and purification
GFPLAP-Mis18α was isolated from HeLa cells as described previously (Cheeseman and Desai, 2005). To obtain cells in G1, cells were arrested overnight in 20 nM nocodazole, collected by mitotic shake-off, released by washout and harvested when centromeric localization of Mis18α was observed for the majority of cells by microscopy. The asynchronous sample was comprised of the cells that failed to arrest in nocodazole. The immunoprecipitated proteins were identified by mass spectrometry of tryptic digests using an LTQ XL Ion Trap mass spectrometer (Thermo Fisher Scientific) coupled with a reverse phase gradient over C18 resin (Phenomenex). Data were analyzed using SEQUEST software.
For recombinant expression of the Mis18 complex, E. coli-optimized 6x-His-Mis18α, E. coli-optimized Mis18β, and human M18BP1 were cloned into pST39 and expressed in Rosetta 2 (DE3)pLysS competent cells (EMD Biosciences). The complex was bound to nickel NTA-agarose (Qiagen) in 50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.1% Tween-20 and washed in 50 mM sodium phosphate buffer pH 8.0, 500 mM NaCl, 40 mM imidazole and 0.1% Tween-20. The complex was eluted in 50 mM sodium phosphate buffer pH 7.0, 500 mM NaCl and 250 mM imidazole and exchanged into 20 mM HEPES pH 7.5, 150 mM KCl, 1 mM DTT. The complex was analyzed by mass spectrometry to confirm the presence of all three components. 6x-His-GST-PBD (residues 326–603) and the 6x-His-MBP-Plk1 T210D plasmid were gifts from Daniel Lim and Michael Yaffe (MIT).
In vitro phosphorylation and Far-Western analysis
Kinase assays were performed in 50 mM HEPES pH 7.5, 150 mM KCl, 10 mM MgCl2, 200 μM ATP, 1mM DTT at 33 °C for 45 min. For radioactive assays, 2 μCi γ32P-ATP was added to each reaction. 6x-His-MBP-Plk1-T210D purified from E. coli using Ni-NTA agarose (Qiagen) was used for radioactive assays; His-Plk1 (Invitrogen; generated by baculovirus expression) was used for phosphosite mapping and Far-Western analysis. Phosphorylation by both of these kinases was abrogated by the addition of BI2536 (data not shown). Far-Western analysis was performed as described using 6x-His-GST-PBD (Lowery et al., 2007).
Supplementary Material
Highlights.
Polo-like kinase 1 (Plk1) is required for CENP-A deposition in human cells.
Plk1 phosphorylates the Mis18 complex to promote its localization.
Cyclin-dependent kinase (CDK) inhibits Mis18 complex assembly.
Bypassing Plk1 and CDK regulation causes constitutive CENP-A deposition.
Acknowledgments
We thank members of the Cheeseman laboratory, Defne Yarar, David Sabatini, Peter Reddien, Steve Bell, Frank Solomon, Terry Orr-Weaver and Rick Young for discussions and critical reading of the manuscript. We are grateful to Daniel Lim and Michael Yaffe (MIT) for providing the GST-Plk1-PBD and advice on working with Plk1. We thank Aaron Straight (Stanford University) for the CENP-A-SNAP cell line, Prasad Jallepalli (Sloan Kettering) for the Rpe1-Plk1as cell line, and Lars Jansen (Gulbenkian Insitute) for the M18BP1 cDNA and CENP-A-SNAP plasmid. We thank Paul Fields and Laurie Boyer (MIT) and Chikdu Shivalila and Rudolf Jaenisch (Whitehead Institute and MIT) for advice and reagents for CRISPR/Cas9-mediated genome editing. This work was supported by a Scholar Award from the Leukemia & Lymphoma Society, a grant from the NIH/National Institute of General Medical Sciences (GM088313), and a Research Scholar Grant (121776) from the American Cancer Society to IMC. IMC is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LET, Foltz DR, Cleveland DW. Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb. Symp. Quant. Biol. 2010;75:403–418. doi: 10.1101/sqb.2010.75.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. U.S.a. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 20052005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Fröhlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.a. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003a;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Elia AEH, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003b;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosome Res. 2013;21:407–418. doi: 10.1007/s10577-013-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Pettersson L, Höglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. U.S.a. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Rodríguez M, Jansen LET. Basic properties of epigenetic systems: lessons from the centromere. Curr. Opin. Genet. Dev. 2013;23:219–227. doi: 10.1016/j.gde.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi KK, van Vugt MATMM, Okamoto K-IK, Hayashi YY, Yaffe MBM. Functional dynamics of Polo-like kinase 1 at the centrosome. CORD Conference Proceedings. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, Ladouceur A-M, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters J-M. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong S-E, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. Embo J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Glotzer M, Kraut N, Peters J-M. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, Tomonaga T. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J. Proteome Res. 2013;12:2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk Activity Couples Epigenetic Centromere Inheritance to Cell Cycle Progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BA, Takaki T, Petronczki M, Glotzer M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110. doi: 10.1371/journal.pbio.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.