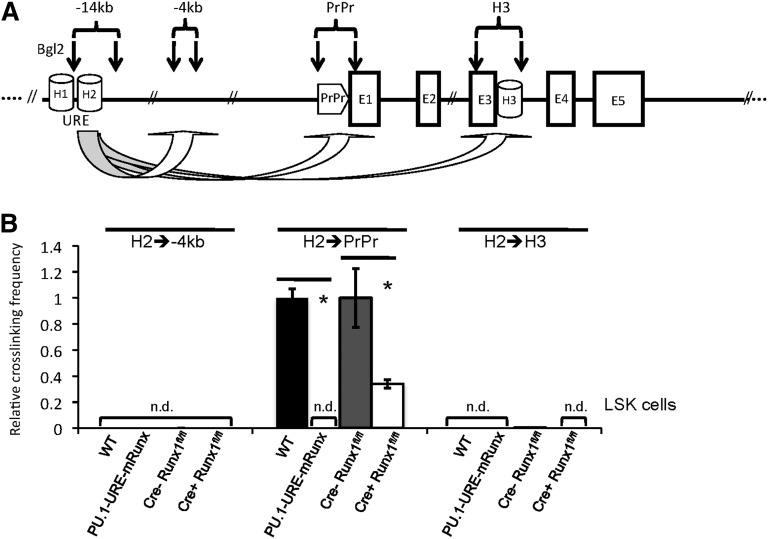
Figure 2.
Runx site mutation leads to loss of URE-proximal promoter interaction in HSCs and/or progenitors. (A) Simplified scheme of the PU.1 gene indicating the positions of homology regions (H1-H3), the −14kb URE, proximal promoter (PrPr), exon 1 through exon 5 (E1-E5), and Bgl2 restriction sites (vertical arrows) for quantitative 3C. The genomic region at −4kb was used as control. (B) Quantitative 3C demonstrates a loss of URE-PrPr interaction in mice with disrupted Runx binding sites (PU.1-URE-mRunx) and a significant loss after induced Runx1 deletion in LSK cells. After crosslinking and Bgl2 digestion, ligated DNA was purified, and interactions of the H2 region with indicated genetic locations was measured by TaqMan PCR and calibrated with an intergenetic DNA amplicon. Graphs represent the results of 4 independent quantitative TaqMan PCR experiments (average + SD; *P < .05). Crosslinking efficiencies are shown as relative values to H2-PrPr interactions of WT and Cre-Runx1fl/fl, respectively. n.d., not detectable.