Abstract
During an influenza A(H7N7) virus outbreak among poultry in Italy during August–September 2013, infection with a highly pathogenic A(H7N7) avian influenza virus was diagnosed for 3 poultry workers with conjunctivitis. Genetic analyses revealed that the viruses from the humans were closely related to those from chickens on affected farms.
Keywords: Influenza, avian influenza A(H7N7) virus, zoonoses, transmission, viruses, Italy
In Europe, avian influenza viruses of subtype H7 have been responsible for several disease outbreaks among poultry, which resulted in human infections (1,2). Notably, since 2000, outbreaks of avian influenza caused by high and low pathogenicity influenza A(H7N1) viruses and low pathogenicity A(H7N3) viruses occurred on poultry farms located mainly in northeastern Italy (3). On August 14, 2013, infection caused by a highly pathogenic avian influenza A(H7N7) virus was initially detected on a layer farm in Ostellato, Ferrara Province, Italy, representing the start of an epizootic that affected another 5 poultry farms in Ferrara and Bologna Provinces (Emilia-Romagna Region) during the next 3 weeks. Nearly 1 million chickens on the 6 farms were culled (4). All workers (≈200) who participated in depopulating infected premises applied strict infection prevention procedures and were monitored for symptoms. Among the workers, infection with highly pathogenic A(H7N7) avian influenza virus was confirmed for 3 who had conjunctivitis but no respiratory symptoms. We describe the clinical and virologic findings of the investigation conducted with regard to these 3 human cases of influenza A(H7N7) virus infection.
The Study
On August 28, 2013, a previously healthy 51-year-old poultry worker (patient 1) noted unilateral conjunctivitis. After the worker was examined by a physician, a conjunctival swab sample was collected and tested at St. Orsola Hospital in Bologna; it was positive for influenza A virus subtype H7. Three days later, a 46-year-old poultry worker (patient 2) sought care for bilateral conjunctivitis and other symptoms, such as chills and muscle aches. On September 4, a 49-year-old man (patient 3) sought care for bilateral conjunctivitis. Conjunctival swab samples collected from patients 2 and 3, tested at the same laboratory, also produced positive results for influenza A virus subtype H7.
Patients 1 and 2 worked with breeding and cleaning on a farm in Mordano, Bologna Province; they had not used personal protective equipment (PPE) until August 21, when influenza A(H7N7) virus infection in poultry was diagnosed. Thereafter, they were involved in culling and wore PPE, including face masks with eye protection. Patient 3 had not previously worked with animals, but he participated in depopulation procedures, while wearing PPE, during the 3-week outbreaks on the farms in Ostellato and Mordano. Because the 3 patients had worked for the same poultry company on the affected farms, located inside a 1.5-km–radius area, the date of exposure for each patient is difficult to infer (4). All 3 patients were isolated at home; without specific antiviral treatment, symptoms resolved in a few days. Six family contacts were placed under clinical surveillance for 10 days.
Conjunctival swab samples were collected from each patient, and aliquots of these samples were sent to the Istituto Superiore di Sanità, Rome, Italy, where they were confirmed as influenza virus subtype H7N7 by 2 real-time reverse transcription PCR (rRT-PCR) assays (5,6). A traditional RT-PCR assay was conducted by using specific primers (available upon request), and PCR products were sequenced by using a BigDye Terminator version 3.1 kit (Life Technologies, Austin, TX, USA). Full-genome sequencing was also performed by using the Ion Torrent PGM apparatus (Life Technologies, Carlsbad, CA, USA). Virus isolation in MDCK cells was successful for patient 3 only, whose conjunctival swab sample showed the highest virus load as revealed by a low cycle threshold (18.3) obtained by rRT-PCR (5). Full-length gene sequencing was conducted on the clinical sample from patient 3 (GenBank accession nos. KF918334–KF918341), whereas the sequence analysis of the other 2 samples was focused on hemagglutinin and neuraminidase genes and a few regions of the other genes (nt 159–525 and nt 994–1107 of polymerase basic protein [PB] 2, nt 115–453 and nt 1726–2259 of PB1, nt 19–297 of nucleocapsid protein, and nt 715–981 of matrix protein) to trace the avian virus responsible for the human infections. In particular, the virus from the chicken with the index case (A/chicken/Italy/13VIR4527–11/2013) was isolated on August 13 in Ostellato, and 6 other strains were isolated from chickens during the 6 outbreaks that occurred among poultry until September 3, 2013 (7) (Table 1). Antigenic characterization of the subtype H7N7 virus isolated from a human was conducted by hemagglutination inhibition assay with turkey erythrocytes (8) and H7 reference antiserum. To define virus susceptibility to neuraminidase inhibitors (oseltamivir and zanamivir), we performed the fluorescent MUNANA (2′-[4-methylumbelliferyl]-α-D-N-acetylneuraminic acid)–based assay (8).
Table 1. Comparison of the nucleotide sequences of highly pathogenic avian influenza A(H7N7) virus isolate from chickens and humans, Italy, August–September 2013*.
| Virus, location (date)† | PB2 |
PB1 |
PA |
HA |
NP |
NA |
M |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 232‡ | 279 | 333 | 1044 | 183‡ | 1882 | 2133 | 1251 | 471 | 1018‡ | 1410 | 72 | 515‡ | 541‡ | 1040‡ | 1347 | 818‡ | 884‡ | |||||||
| Chicken | ||||||||||||||||||||||||
| A/CK/4527, Ostellato (Aug 13)§ | C | T | T | C | C | C | C | G | A | G | G | G | A | T | C | A | A | G | ||||||
| A/CK/4541, Ostellato (Aug 13) | T | • | • | • | • | • | T | • | G | A | • | • | • | A | • | • | G | • | ||||||
| A/CK/4603, Mordano (Aug 19) | T | • | • | • | • | • | T | • | • | • | • | • | • | A | • | G | G | • | ||||||
| A/CK/4678, Portom (Aug 21) | T | • | • | T | • | • | T | • | • | • | • | A | • | A | • | G | G | • | ||||||
| A/CK/4774, Mordano (Aug 27) | T | • | C | • | • | T | T | • | • | • | A | • | • | A | • | • | • | • | ||||||
| A/CK/5091, Bondeno (Sep 2) | T | C | • | T | T | • | T | • | • | • | • | A | • | A | • | G | G | A | ||||||
| A/CK/5051,
Mordano
(Sep 3) |
T |
• |
• |
– |
|
• |
• |
T |
|
A |
|
• |
• |
• |
|
• |
|
• |
A |
• |
• |
|
• |
• |
| Human¶ | ||||||||||||||||||||||||
| 1, Mordano (Aug 29) | T | • | • | • | • | • | T | – | • | • | • | • | • | A | • | – | G | • | ||||||
| 2, Mordano (Sep 2) | T | • | • | • | • | • | T | – | • | • | • | • | • | A | • | • | • | • | ||||||
| 3, Mordano /Ostellato (Sep 7) | T | • | • | • | • | • | T | A | • | • | • | • | G# | A | A# | • | • | • | ||||||
*Dots indicate no nucleotide changes; –, notavailable. HA, hemagglutinin; NA, neuraminidase; NP, nucleocapsid protein; M, matrix; PA, polymerase acidic; PB, polymerase basic; Portom, Portomaggiore. †Date indicates date collected. A/CK/4527, A/chicken/Italy/13VIR4527–11/2013; A/CK/4541, A/chicken/Italy/13VIR4541–34/2013; A/CK/4603, A/chicken/Italy/13VIR4603/2013; A/CK/4678, A/chicken/Italy/13VIR4678–1/2013; A/CK/4774, A/chicken/Italy/13VIR4774/2013; A/CK/5091, A/chicken/Italy/13VIR5091–1/2013; A/CK/5051, A/chicken/Italy/13VIR5051–3/2013. ‡Nonsynonymous substitution. §First isolate from chicken (index case). ¶Clinical samples. #Nucleotide changes found only in the specimen from patient 3 and responsible for the D172G and P347Q amino acid changes.
The 3 patients were infected with an avian-origin influenza A(H7N7) virus. In particular, molecular analyses showed that the hemagglutinin and neuraminidase gene sequences of A/Italy/3/2013 virus isolate (GenBank accession nos. KF712391, KJ136817) were identical to those of the clinical specimens. The hemagglutinin amino acid sequence of this virus, and those from patients 1 and 2, showed complete homology to most of the isolates from chickens, including the hemagglutinin cleavage site containing multiple basic amino acids (PKRRERR*GL) responsible for the highly pathogenic phenotype (9). The neuraminidase sequence from the isolate from patient 3 differed from those of clinical specimens from the other 2 patients and most isolates from chickens by 2 amino acids (D172G and P347Q) (Table 1). Neither neuraminidase stalk deletions nor neuraminidase inhibitor sequence-based resistance were detected in the influenza viruses from the humans (10,11); the drug sensitivity of the viruses was further confirmed by phenotypic neuraminidase inhibitor susceptibility assays performed on A/Italy/3/2013 virus (data not shown).
Phylogenetic analyses of the hemagglutinin and neuraminidase genes (Figures 1, 2) confirmed that the H7N7 isolates from the human patient in Italy were closely related to H7 low pathogenicity avian influenza viruses that had been circulating among wild birds and poultry in Europe during the past 3 years. Antigenic characterization of the A/Italy/3/2013 virus by hemagglutination inhibition assay (Table 2) showed that the virus was recognized poorly by antiserum raised against A/Anhui/1/2013 (H7N9), A/turkey/Italy/214845/2002 (H7N3), and A/turkey/England/647/1977 (H7N7) and recognized well by antiserum raised against A/goose/Czech Republic/1848/2009 (H7N9), A/African starling/England/983/79 (H7N1), and A/turkey/Italy/3889/99 (H7N1).
Figure 1.
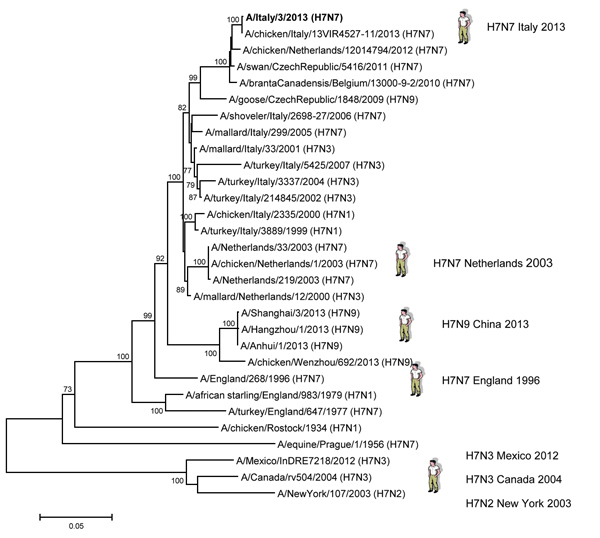
Phylogenetic analysis of the hemagglutinin gene of the influenza A(H7N7) virus, Italy, isolated from humans during August–September 2013. The phylogenetic tree was constructed by using the neighbor-joining method and MEGA 5 software (http://www.megasoftware.net) with 1,000 bootstrap replicates (bootstrap values >70% are shown next to nodes). The influenza A(H7N7) virus isolated from a human in 2013 is shown in boldface. Scale bar indicate nucleotide substitutions per site.
Figure 2.
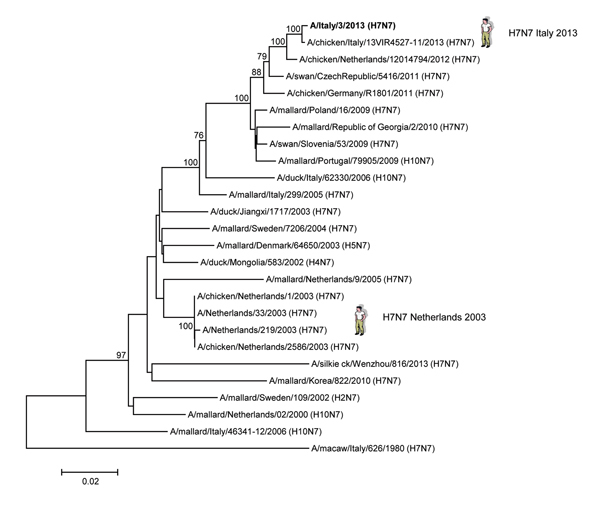
Phylogenetic analysis of the neuraminidase gene of the influenza A(H7N7) virus, Italy, isolated from humans during August–September 2013. The phylogenetic tree was constructed by using the neighbor-joining method and MEGA 5 software (http://www.megasoftware.net) with 1,000 bootstrap replicates (bootstrap values >70% are shown next to nodes). The influenza A(H7N7) virus isolated from a human in 2013 is shown in boldface. Scale bar indicate nucleotide substitution per site.
Table 2. Antigenic analyses of influenza A/H7 viruses*.
| Viruses | Antiserum |
|||||
|---|---|---|---|---|---|---|
| A/Anhui/ 1/13 (ferret) | A/TK/It/ 3889/99 (ferret)† | A/TK/It/ 214845/02 (ferret) | A/GS/CZ/ 1848/09 (chicken)‡ | A/TK/Eng/ 647/77 (chicken)‡ | A/AS/Eng/ 983/79 (chicken)‡ | |
| Reference | ||||||
| A/Anhui/1/2013 (H7N9) | 320 | 80 | 80 | 80 | 40 | 80 |
| A/turkey/Italy/3889/1999 (H7N1) | 80 | 80 | 80 | 40 | <40 | <40 |
| A/turkey/Italy/214845/2002 (H7N3) | 40 | 80 | 80 | <40 | <40 | 40 |
| A/goose/Czech Republic /1848/2009 (H7N9)§ | 40 | 40 | 40 | 320 | 40 | 40 |
| A/turkey/England/647/1977 (H7N7) | 40 | <40 | 40 | 40 | 160 | 40 |
| A/African starling/England/983/1979 (H7N1)§ |
40 |
<40 |
40 |
40 |
40 |
160
|
| Test | ||||||
| A/Italy/3/2013 (H7N7) | 40 | 40 | <40 | 160 | 40 | 80 |
*Data are hemagglutination inhibition titers. Homologous titers of reference viruses to serum samples are indicated in boldface. AS, African starling; GS, goose; TK, turkey. †Antiserum from National Institute for Biologic Standards and Control, UK. ‡Antiserum from Animal Health and Veterinary Laboratories Agency, UK. §Inactivated virus.
Genome sequences of the internal proteins of A/Italy/3/2013 virus and from the clinical samples revealed high identity to the circulating H7N7 strains from chickens in the area. Although deducing the exact exposure for each patient is difficult, partial sequence analysis showed that the virus from patient 1 was more related to A/chicken/Italy/13VIR4603/2013 and that the viruses from patients 2 and 3 were more related to A/chicken/Italy/13VIR5051–3/2013 (Table 1). These 2 viruses from chickens had been isolated during different outbreaks in Mordano (7) and differed somewhat from that from the chicken with the index case (A/chicken/Italy/13VIR4527–11/2013). Neither mammalian host adaptation markers, including the E627K mutation in PB2, nor the common mutations associated with adamantane resistance (L26F, V27A/G, A30S/T/V, S31N, G34E) in matrix protein 2 were found in the H7N7 strain from humans (12–14).
Conclusions
This study provides further evidence of H7 subtype–specific ocular tropism (1). Our molecular findings suggest direct transmission of the virus from chickens to humans; the lack of known host adaptation markers does not support human-to-human transmission. The presence of 2 mutations in neuraminidase from the specimen of patient 3, which contained the highest viral load, might suggest a correlation with the efficiency of infection and replication in the conjunctiva. Indeed, specific neuraminidase mutations have been observed in H7N7 viruses from the Netherlands and have been associated with enhanced replication (15). However, further studies are needed to determine their exact role in the pathogenesis of the infection. Clinical surveillance was immediately applied to all exposed workers and cohabiting contacts, and no further human cases of H7N7 infection were identified. For the purpose of investigating human subclinical infections by H7 viruses, a serologic surveillance program is ongoing in the affected areas.
Acknowledgments
We are grateful to Ruth Manvell, Ian Brown, and Bob Newman for supplying antiserum for hemagglutination inhibition testing and to Tiziana Grisetti for editing the manuscript.
This work was funded by the Italian Ministry of Health project “Virological surveillance of epidemic and pandemic influenza” (grant no. 4M13) and by the Emilia-Romagna Region. Work at the Medical Research Council National Institute for Medical Research was funded by Medical Research Council program no. U117512723.
Biography
Dr Puzelli is a researcher in the Department of Infectious, Parasitic and Immune-mediated Diseases at the National Institute of Health, Rome. Her primary research interests include molecular mechanisms of genetic variability of influenza viruses and antiviral susceptibility.
Footnotes
Suggested citation for this article: Puzelli S, Rossini G, Facchini M, Vaccari G, Di Trani L, Di Martino A, et al. Human infection with highly pathogenic A(H7N7) avian influenza virus, Italy, 2013. Emerg Infect Dis [Internet]. 2014 Oct [date cited]. http://dx.doi.org/10.3201/eid2010.140512
Influenza Task Force members: Laura Calzoletti, Concetta Fabiani, Monica Meola, Annapina Palmieri, Arianna Boni, Guendalina Zaccaria (Istituto Superiore di Sanità, Rome); Marisa Cova, Valerio Parmeggiani, Claudio Po, Roberto Cagarelli, Gabriele Squintani, Emanuela Bedeschi (Emilia-Romagna Region, Bologna); and Maria Grazia Pompa (Ministry of Health, Rome).
References
- 1.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93. 10.1016/S0140-6736(04)15589-X [DOI] [PubMed] [Google Scholar]
- 2.Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–65. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, et al. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology. 2004;323:24–36. 10.1016/j.virol.2004.02.015 [DOI] [PubMed] [Google Scholar]
- 4.Bonfanti L, Monne I, Tamba M, Santucci U, Massi P, Patregnani T, et al. Highly pathogenic H7N7 avian influenza in Italy. Vet Rec. 2014;174:382. 10.1136/vr.102202 [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. CDC human influenza virus real-time RT-PCR diagnostic panel–-influenza A/H7 (Eurasian Lineage) assay [cited 2014 Jun 20]. http://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM349065.pdf
- 6.Monne I, Ormelli S, Salviato A, De Battisti C, Bettini F, Salomoni A, et al. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J Clin Microbiol. 2008;46:1769–73. 10.1128/JCM.02204-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Istituto Zooprofilattico Sperimentale delle Venezie. Highly pathogenic avian influenza subtype H7N7, Italy, August 2013. Laboratory and molecular findings [cited 2014 Jun 20]. http://www.izsvenezie.it/images/stories/Pdf/Temi/Aviaria/situazione_epidemiologica2013/presentazione_def_09_2013.pdf
- 8.World Health Organization. Manual for the laboratory diagnosis and virological surveillance of influenza [cited 2014 Apr 14]. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf?ua=1
- 9.Wood GW, McCauley JW, Bashiruddin JB, Alexander DJ. Deduced amino acid sequences at the haemagglutinin cleavage site of avian influenza A viruses of H5 and H7 subtypes. Arch Virol. 1993;130:209–17. 10.1007/BF01319010 [DOI] [PubMed] [Google Scholar]
- 10.Matsuoka Y, Swayne DE, Thomas C, Rameix-welti MA, Naffakh N, Warmes C, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol. 2009;83:4704–8. 10.1128/JVI.01987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir. Antivir Ther. 2007;12:603–16 . [PubMed] [Google Scholar]
- 12.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–2. 10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- 13.Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, et al. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J Infect Dis. 2007;196:258–65. 10.1086/518792 [DOI] [PubMed] [Google Scholar]
- 14.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, et al. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol. 2010;84:1597–606. 10.1128/JVI.01783-09 [DOI] [PMC free article] [PubMed] [Google Scholar]


