Abstract
MicroRNAs (miRNAs) are involved in the pathogenesis of a number of cardiovascular diseases. In this review article, we have summarized the role of miRNAs in regulating lipid metabolism and how their therapeutical inhibition may lead to new approaches to treat cardiometabolic diseases, including atherosclerosis and metabolic syndrome. Specific miRNAs, such as miR-33a and -33b, represent one of the most interesting and attractive targets for metabolic-related disorders and anti-miR33 approaches are under intensive investigation. In addition to miR-33, other miRNAs, including miR-122, are also emerging as key players in lipid metabolism. More recently miRNAs were shown to exert their activities in a paracrine manner and also systemically. The latter is possible due to lipid-carriers, including lipoproteins, that transport and protect miRNAs from degradation. The emerging strong connection between miRNAs, lipoproteins and lipid metabolism indicates the existence of a reciprocal modulation that might go beyond atherosclerosis.
Introduction
MiRNAs are short non-coding RNAs involved in the regulation of gene expression at the post-transcriptional level [1]. MiRNAs are implicated in the pathogenesis of various cardiovascular diseases and have also been considered as potential targets for therapeutic intervention [2]. To date, several miRNAs have been described to finely regulate lipid metabolism and the progression and regression of atherosclerosis including miR-122, miR-33, miR-106, miR-758, miR-26, miR-370, miR-378/378*, let-7, miR-27, miR-103, miR-107, miR34a, miR-143 and miR-335 [3–20]. More recently, miRNAs were shown to exert their activities in cells different from those were they have been generated and also systemically. The latter activity is possible thanks to lipid-carriers, including lipoproteins that transport and protect miRNAs from degradation but could also modulate their delivery.
In this review we aim to to: i) present the complex mechanism by which miRNAs regulate lipid metabolism, ii) illustrate how their therapeutical modulation may lead to new treatments for cardiometabolic diseases, and iii) discuss how the transport of miRNAs in the circulation by lipoproteins and other lipid carriers might represent a new aspect in biology further connecting lipid lipoproteins and miRNAs.
miRNAs modulating lipid metabolism
miR-33 and miR-122 represent the miRNAs that have been extensively investigated for their role in modulating genes involved in lipid metabolism, however several other miRNAs are emerging.
miR-33 family
The sterol-regulatory element-binding proteins (SREBPs) are key regulators of lipid metabolism [23]. They bind to the promoter region and activate the transcription of many genes involved in cholesterol and fatty acid biosynthesis and uptake and in the production of TG and phospholipids. The Srebp1 gene encodes for two transcripts, Srebp1a and 1c, which primarily regulate lipogenic genes such us acetyl-CoA carboxylase (Acc), Scd and Fasn. On the other hand, SREBP2 preferentially controls cholesterol metabolism by regulating genes involved in cholesterol biosynthesis, such as Hmgcr and most of the mevalonate-pathway genes, and cholesterol uptake, including the low-density lipoprotein receptor (Ldlr).
Interestingly, several groups have recently reported the intriguing localization of the miR-33 family members within the introns of the Srebp genes [8, 12, 13, 19]. miR-33a, which is conserved in organisms from Drosophila to humans, is located in intron 16 of the Srebp2 gene, whereas miR-33b is encoded in intron 17 of the Srebp1 gene. Both mature miRNAs have similar seed sequences and therefore are predicted to target similar genes. Of note, miR-33b is not conserved in rodents, which may explain some of the differences between the lipid/lipoprotein metabolism in rodents versus humans. Both miRNAs appear to be co-transcribed with their host genes and regulate physiological processes related to the genes where they are encoded [13, 19]. For instance, like many intronic miRNAs, miR-33a is co-transcribed when Srebp2 is activated and targets genes involved in cholesterol export, such as the adenosine tri-phosphate binding cassette (ABC) transporters, ABCA1 and ABCG1, and the endolysosomal transport protein, Niemann-Pick C1 (NPC1) [8, 12, 13, 19]. This regulatory function of miR-33a may function as a gatekeeper to ensure that the cell is protected under low sterol conditions from additional sterol loss. ABCG1 is not a conserved target in rodents, pointing out that the regulation of cholesterol efflux between species may be differently regulated at the post-transcriptional level. The identification of ABCA1 as a main miR-33 target gene leads to the hypothesis that miR-33 may be regulating high-density lipoproteins (HDL) levels in vivo. Indeed, three independent studies have demonstrated that endogenous inhibition of miR-33 using a variety of strategies leads to a significant increase in hepatic ABCA1 expression and plasma HDL levels [8, 12, 13, 19]. These findings were later confirmed in the miR-33 deficient mice [24]. Most importantly, anti-miR-33 therapy also resulted in increased plasma HDL levels in non-human primates.
HDL is believed to play a key role in reverse cholesterol transport (RCT), which mediates the clearance of excess cholesterol from cells to the liver for excretion to bile and feces [25]. This process is particularly relevant for the removal of cholesterol from lipid-loaded macrophages that accumulate in atherosclerotic lesions and is thought to contribute predominantly to the atheroprotective effects of HDL. Interestingly, anti-miR-33 therapy promotes RCT and regresses atherosclerosis in LDLr knockout mice [26]. The effect of miR-33 on RCT appears to be more complex and not only mediated by ABCA1. In this regard, Allen RM et al has recently reported that miR-33 also targets two canalicular transporters, ABCB11 and ATP8B1, that regulate bile secretion, which is essential for controlling whole body sterol homeostasis [27]. This study also suggests that anti-miR-33 therapy may be useful for treating recurrent cholestasis. Indeed, pretreatment of mice with anti-miR-33 oligonucleotides rescues the hepatotoxic effect of diet and statins [27].
In addition to the important role of miR-33 in regulating cholesterol metabolism, the conservation of miR-33 in the Drosophila melanogaster genome, an organism that does not synthesize sterols, and the genomic localization of miR-33b in the intron of the Srebp1 gene, led several groups to study the contribution of miR-33 in controlling other metabolic pathways. Importantly, miR-33a and miR-33b contribute to the regulation of fatty acid metabolism, modulating the expression of carnitine O-octanyl transferase (Crot), carnitine palmitoyltransferase 1A (Cpt1A), and hydroxyacyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase-enoyl-CoA hydratase (trifunctional protein) β-subunit (Hadhb) [8, 28]. CROT and CPT1A regulate the transport of fatty acids to the mitochondria for their degradation and HADHB is directly involved in mitochondrial fatty acid β-oxidation. Over-expression of miR-33 in hepatic cells significantly reduces the degradation of fatty acids leading to an accumulation of neutral lipids [8, 28]. Conversely, endogenous inhibition of miR-33 increases the degradation of fatty acids [8, 28]. These observations suggest that anti-miR-33 therapy may be useful for treating hepatic steatosis by increasing the degradation rate of fatty acids in the liver. Accordingly, Rayner and colleagues also demonstrated that non-human primates treated with anti-mir-33 oligonucleotides have a significant reduction of plasma VLDL levels [16]. This effect on plasma lipoproteins could be explained by a reduced lipidation and secretion of ApoB-containing lipoproteins from the liver owing to the increased fatty acid oxidation in the liver of non-human primates treated with anti-miR-33 oligonucleotides.
miR-33 also controls the expression of sirtuin 6 (Sirt6) and AMP-activated protein kinase (Ampkα1), which is involved in regulating lipid and glucose metabolism [28]. SIRT6 is a NAD+-dependent histone deacetylase that plays an important role in regulating glucose metabolism [29, 30]. Indeed, SIRT6-deficient mice develop normally but succumb to lethal hypoglycemia early in life. Moreover, hepatic-specific disruption of SIRT6 results in fatty liver formation because of the enhanced glycolisis and triglyceride synthesis [31]. SIRT6 is regulated by miR-33 in humans and non-human primates but not in other species, suggesting that the regulation of fatty acid and glucose metabolism by miR-33 may be different between species. AMPKα1, a master regulator of cellular energy, is also an important target of miR-33 [28]. AMPKα1 phosphorylates and inhibits key lipogenic enzymes including HMGCR and ACC. Thus, the inhibition of AMPKα1 by miR-33 may increase HMGCR and ACC activity to boost intracellular levels of cholesterol and fatty acids. Altogether, these results suggest a paradigm in which miR-33a and miR-33b act in concert with their host genes, Srebp2 and Srebp1, to increase intracellular cholesterol and fatty acid levels by balancing the transcriptional induction and posttranscriptional repression of lipid metabolism genes.
Finally, the expression of insulin receptor substrate 2 (IRS2) is also regulated by miR-33 [28]. IRS2 is an adaptor protein that controls insulin signaling in the liver. Overexpression of miR-33 in human hepatic cells significantly inhibits insulin signaling by reducing IRS2 expression, protein kinase B (PKB; also known as AKT) phosphorylation and FOXO1 cytoplasmic localization [28]. These data indicate that miR-33a and –b impact pathways influencing three of the primary risk factors in metabolic syndrome, namely insulin resistance, low HDL and high VLDL and suggest that anti-miR-33 therapies may be an attractive approach for treating this global health concern.
miR-122
miR-122 is the most abundant miRNA in the liver where it accounts for ~ 75% of total miRNA expression. The role of miR-122 in the liver is multifaceted; it is often down-regulated in hepatocellular carcinoma and is associated with the regulation of hepatitis C infection [21]. miR-122 is also the first miRNA identified to play a role in regulating lipid metabolism [3–5]. Two studies in mouse and non-human primates demonstrate that anti-miR-122 therapy results in a significant reduction of plasma cholesterol levels [4, 5]. These results have been recently recapitulated in miR-122 deficient mouse models [21, 22]. miR-122 liver-specific knockout and miR-122 germline knockout mice showed a significant reduction (~30%) of total serum cholesterol and triglyceride (TG) levels. Even though the effect of anti-miR-122 therapy and the phenotype of the miR-122 null mice on lipid metabolism have been replicated in several studies, the molecular mechanics by which the inhibition or absence of miR-122 results in a significant decrease of total cholesterol remains poorly understood. Stoffel and colleagues observed that in mice treated with miR-122 antagomirs, at least 11 genes involved in cholesterol biosynthesis, including 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr), 7-Dehydrocholesterol reductase (Dhcr7), mevalonate kinase (Mvk), 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (Hmgcs 1), farnesyl diphosphate synthase (Fdps) and squalene epoxidase (Sqle), were significantly down-regulated [3]. Similarly, miR-122 knockout mice showed a significant inhibition of genes involved in cholesterol biosynthesis, including Hmgcr [21, 22]. Moreover, Tsai and colleagues also found a significant down-regulation of the microsomal TG transfer protein (MTTP), which is essential for the assembly of lipoproteins, in miR-122 null mice [21]. Indeed, the TG secretion rate was significantly reduced in miR-122 deficient mice and rescued after over-expressing MTTP in the liver, suggesting that the phenotype in the miR-122 knockout mice is primarily due to effects on MTTP expression. Intriguingly, Mttp is not a direct target of miR-122 and the mechanism by which miR-122 regulates its expression is still unknown. Altogether, these results demonstrate that miR-122 plays a key role in regulating serum cholesterol and TG levels by controlling cholesterol biosynthesis and very-low density lipoprotein (VLDL) secretion in the liver.
In addition to controlling cholesterol metabolism, miR-122 has been shown to regulate fatty acid synthesis and oxidation. In this regard, the inhibition of miR-122 using 2′-O-methoxyethyl (2′-MOE) phosphothionate-modified antisense oligonucleotides (ASO) in mice reduced the expression of multiple genes mainly involved in fatty acid synthesis, such as the sterol regulatory binding protein 1 (Srebp1), Fatty acid synthase (Fasn), Acetyl-CoA-Carboxylase (Acc) 1 and 2, and stearoyl-CoA desaturase (Scd), leading to a reduction of fatty acid content in the liver [5]. Of note, pharmacological inhibition of miR-122 improved liver steatosis in a high-fat-fed mouse model by increasing fatty acid oxidation and reducing fatty acid synthesis. In contrast, two independent studies have recently shown that mice lacking miR-122 in the liver or in the germline develop steatohepatitis by increasing the expression of genes involved in TG synthesis, including 1-acyl-glycerol-3-phosphate acyltransferase alpha (Agpat1), Agpat3, Agpat9, monoacylglycerol O-acyltransferase 1 (Mogat1), phosphatidic acid phosphatase 2A (Ppap2a) and Ppap2c [21, 22]. Additionally, miR-122 deficient mice also showed an increased expression of Cidec, also known as FSP27, which is a protein that localizes to lipid droplets and negatively regulates lipolysis. Perhaps this effect associated with the reduced MTTP expression observed in the miR-122 knockout mice may also lead to steatohepatits as a consequence of reduced VLDL synthesis and secretion from the liver and increased TG accumulation in hepatocytes. Based on these reports, miR-122 appears to play an important role in TG and cholesterol metabolism. The different results obtained using antisense oligonucleotide therapies and the miR-122 knockout models, together with the lack of understanding of the effects of miR-122 on lipid homeostasis and the possibilities of other adverse effects, such as hepatocellular carcinoma, has dampened the enthusiasm for developing anti-miR-122 therapies for treating lipid disorders.
Other miRNAs that regulate lipid metabolism
In addition to miR-122 and miR-33, other miRNAs regulate lipid metabolism. Similarly to miR-33, miR-758, miR-26 and miR-106b regulate ABCA1 expression and cellular cholesterol efflux [10, 17, 32]. miR-758 is down-regulated after cholesterol loading in macrophages and in the liver of mice fed a high fat diet [32]. Over-expression of miR-758, miR-26 and miR-106b reduces ABCA1 expression in macrophage, hepatic and neuronal cell lines. Thus, the post-transcriptional regulation of ABCA1 expression by miRNAs appears to be complex and mediated by multiple miRNAs and further work is needed to dissect out their specific, or may be redundant, role..
miR-370 targets directly Cpt1a, thereby reducing fatty acid β-oxidation [9]. Interestingly, over-expression of miR-370 in human hepatic cells increases the levels of miR-122 leading to an increase in lipogenic genes, including Srebp1 and Dgat2. These data suggest that the effect of miR-370 in regulating lipid metabolism may be mostly mediated by miR-122. miR-378/378* also regulates fatty acid metabolism [7]. Intriguingly, this miRNA is located within the peroxisome proliferator-activated receptor gamma coactivator-1 alpha (Pgc1α) gene and regulates triacylglycerol synthesis and storage during adipogenesis. Transfection of miR-378/378* in ST2 cells increases the expression of fatty acid binding protein 4 (FABP4), FASN, SCD1, Kruppel-like factor 15 (KLF15), and resistin [7]. However, the miR-378/378* targets that regulate the expression of these genes are unknown and warrant further investigation. Another interesting finding is that both strands of the miR-378/miR-378* duplex regulate TG synthesis in 3T3-L1 adipocytes, suggesting that miR-378 and miR-378* may cooperate to regulate lipid accumulation during adipogenesis. Other miRNAs have also been linked to adipocyte differentiation, such as let-7, miR-143, miR-335, miR-27 and miR-103/107 [6, 11, 14, 33, 34].
In addition, miRNAs have also been involved in the genetic regulation mediated by nuclear receptors. An early study reported that miR-34 is induced by the nuclear bile acid receptor farnesoid X receptor (FXR) and targets Sirt1 [35]. Importantly, miR-34a levels are elevated and SIRT1 is reduced in diet-induced obesty in mice. This expression was reverted upon FXR activation, suggesting that the FXR pathway controls SIRT1 levels via miR-34a inhibition. miR-26 is also activated by nuclear receptors. In this case, the liver X receptor (LXR) positively regulates miR-26 expression to fine-tune cellular cholesterol efflux in macrophages [17].
Altogether these findings summarize the importance of miRNAs in regulating lipid homeostasis and lipoprotein metabolism, opening new avenues for treating dyslipidemias and cardiometabolic disorders.
Lipoproteins mediate miRNAs transport and delivery
Although originally described to act as intracellular endogenous non-coding RNAs that control gene expression at post-translational level, many studies have demonstrated that miRNAs are present in the circulation (Mayr)(Mitchell PS Proc Natl Acad Sci U S A. 2008;105:10513–10518). Alterations in circulating miRNA profiles were associated not only with cancer (Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101:2087–2092) but also with atherosclerosis (Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol 2011; 31:2383–2390. McManus DD, Ambros V. Circulating MicroRNAs in cardiovascular disease. Circulation 2011; 124:1908–1910.), and nonalcoholic steatohepatitis (Cheung O, Puri P, Eicken C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008; 48:1810–1820.). While these observations clearly support the role for circulating miRNA signatures as novel disease biomarkers; recent evidence suggests that circulating extracellular miRNAs may also be biologically active and play a role in intercellular communication (Grange C, Tapparo M, Collino F, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 2011; 71:5346–5356). This mechanism has been described so far for several cell types including adipocytes, cancer and immune cells (Curr opini lipidol-Remaley), and the possibility that circulating miRNAs could act as hormones, favoring intercellular communication in several physiopathological conditions is emerging (Curr opini lipidol-Remaley).
While the chemical structure, suggests that miRNAs should be immediately degraded by nuclease in the circulation, they are relatively stable in plasma (El-Hefnawy T Clin Chem 2004;50:564–73) and the incubation with detergents [23] or sonication [29] favors their degradation. These observations indicate that miRNAs circulate in a protected form, probably complexed to lipid related molecules (Curr Opinin lipidol 2012). Extracellular miRNAs were found, by size-exclusion chromatography, to coelute with Argonaute 2 (AGO2), an intracellular structure–function protein found within the RNA-induced silencing complex (Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108:5003–5008), which suggests that proteins, in addition to lipid-carriers, could transport miRNAs. It has also been proposed that extracellular AGO2 bound to miRNAs is likely to be released into the circulation as a consequence of cell lysis/necrosis and may not participate in intercellular communication (Curr Opinin lipidol 2012), but this hypothesis has not been tested yet.
Among the lipid-based molecules, both microvescicles, from the smaller exosomes (40–100nm in diameter) (Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654–659) to the larger membrane-derived vesicles (50–4000nm in diameter) (Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470–1476), apoptotic bodies (Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010; 107:810–817) and lipoproteins contain miRNAs (Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13:423–433). While these molecules share a lipid related structure, they differ in several aspects, including their interaction with cells and their metabolism. For instance, vesicles (microvesicle/exosomes) containing miRNAs could be delivered to cells either by a process involving endocytosis (Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004; 104:3257–3266) or by membrane fusion and the release of content into the cytoplasm (Montecalvo A, Larregina AT, Shufesky WJ et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood in Press), with only the second mechanism able to preserve miRNA function. However, in vivo, microvescicle fusion with target cell membranes and the release of their genetic contents inside the recipient cells is mainly a nonselective process that could be prevalent in phagocytes (Feng D, Zhao WL, Ye YY, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010; 11:675–687), thus potentially limiting the cell specificity of these miRNA lipid carriers.
On the contrary, lipoproteins represent a highly evolutionary conserved system to transport lipids, proteins and also miRNAs in circulation. The interaction between specific proteins of lipoproteins and their receptors on cell membranes ensures a selective interaction and exchange of lipoprotein-transported moieties. Although both LDL and HDL have been shown to transport miRNAs, they present a different miRNA signature, with LDL presenting a miRNA profile closer to that of plasma exosomes than that of HDL (Remaley; Nat Cell Biol 2011). HDL represents so far the only class of lipoproteins widely studied for its ability to retrieve, transport and deliver miRNAs to cells (Remaley; Nat Cell Biol 2011). HDL reconstituted with human apolipoprotein A-I (ApoA-I) and phosphatidylcholine(PC) (rHDL) is able to retrieve a large amount of miRNAs in vivo. Following injection of rHDL in mice (wild type or apolipoprotein E-deficient on chow or atherogenic diet), the majority of miRNAs retrieved by the lipoproteins were the same independently of mice genotype or the presence of atherogenic diet; however, the presence of the atherogenic diet was associated with a relatively different amount of miRNAs loaded on HDL (Remaley; Nat Cell Biol 2011). Although the exact mechanism of miRNA loading on rHDL is unknown, it is reasonable to speculate that ApoA-I might bridge the lipoprotein to the cells, and PC, that was shown to complex small RNAs (Lu D, Rhodes DG. BBA 2002), might bind the miRNAs. MiRNA export from macrophages to HDL is favored by the induction of ABCA1 expression but also by the inhibition of the ceramide pathway (Remaley; Nat Cell Biol 2011). Of note, the latter was shown to promote miRNA export to exosomes (Trajkovic K, Science 2008), therefore the possibility that miRNAs loading on HDL or exosomes could be related to the different bioavailability of cellular lipids should be investigated. Furthermore, as HDL undergos a maturation process consisting of continuous lipid and protein exchange not only with cells but mainly with the other lipoprotein classes, the possibility that HDL could transport miRNAs acquired also from other lipoproteins can not be excluded.
The delivery of miRNAs from HDL to cells is highly dependent on the scavenger receptor B1 (SR-BI) (Remaley; Nat Cell Biol 2011). SR-BI, a scavenger receptor with a mission to transport HDL lipids. Curr Opin Lipidol 2004;15:287–295), which may also lead to less degradation of miRNAs. Indeed rHDL-mir223 complexes when incubated with hepatocytes released miR-223 to the cells and reduced the expression on their targets into the cells (Remaley; Nat Cell Biol 2011).
Of note, none of the miRNAs described above to modulate lipid metabolism are among those prevalent on human HDL from healthy subjects or patients with familial hypercholesterolemia (Remaley; Nat Cell Biol 2011). This raises the possibility that miRNAs carried by HDL could influence pathways other than those strictly related to lipid metabolism. For instance, HDL is emerging as a new player in immunity [36, 37] and an in silico analysis predicted that the most abundant miRNAs present on human HDL might affect signaling pathways associated with innate and adaptive immunity (Remaley; Nat Cell Biol 2011). These observations support the concept of additional functions of lipoproteins in modulating intercellular communication.
Conclusion and perspectives
A deep connection between miRNAs, lipoproteins and lipid metabolism is emerging. Specific miRNAs, such as miR-33a and –b, represent one of the most interesting and attractive targets for metabolic-related disorders and anti-miR33 approaches are under intensive investigation. Nevertheless, future studies in non-human primates and rodents treated chronically with anti-miR-33 oligonucleotides are important to rule out potential adverse effects. miR-33 also targets multiple genes involved in glucose metabolism and inflammation; therefore the role of miR-33 in regulating these physiological processes needs to be clarified. Additional experiments that integrate proteomics, system biology and RNA-sequencing in different tissues will help us to elucidate how the modulation of gene networks by miR-33 contribute to the regulation of metabolic processes including lipid and glucose metabolism. Lipoproteins can also potentially affect miRNA availability and function, thus paving the road for new and yet unexplored lipoprotein functions. In this context, several open questions remain. Is the miRNOme of lipoprotein species (in addition to HDL) affected in different dyslipidemic conditions and could be a more specific marker than total miRNA signature of CVD progression? Can lipoprotein species exchange miRNAs? Is this process the consequence of lipid exchange? Is the HDL enrichment in miRNAs a mechanism to force their catabolism via the liver? Can pharmacological treatment targeting lipoprotein levels affect circulating miRNA-related responses? Answering these questions will be critical to fully establish a role for lipoproteins in influencing miRNA biology besides their role in lipid metabolism.
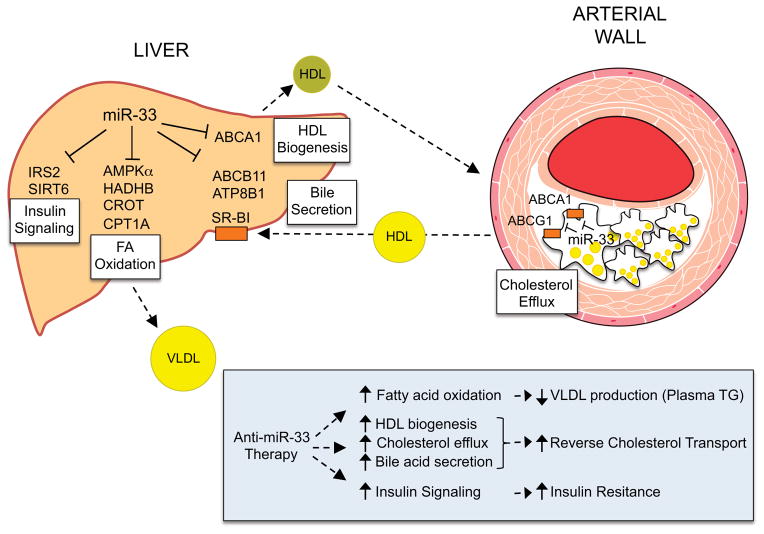
Figure 1. miR-33 regulates lipid metabolism and insulin signaling.
In the liver miR-33 regulates the expression of genes involved in HDL biogenesis (ABCA1), fatty acid oxidation (CPT1A, CROT, HADHB and AMPKα), bile secretion (ABCB11 and ATP8B1) and insulin signaling (IRS 2 and SIRT6). In the periphery, including the arterial wall, miR-33 regulates cellular cholesterol efflux by controlling the expression of ABCA1 and ABCG1. The bottom box summarizes the potential therapeutical effects of anti-miR-33 therapy, including increasing plasma HDL levels, reverse cholesterol transport and insulin signaling, and reducing VLDL production
Acknowledgments
GDN is supported by grants from Fondazione Cariplo (2010-0768) Società Italiana Studio Aterosclerosi Lombardia Chapter, PUR 2009 University of Milan and pump priming MCM 2012, and Blizard Institute QMUL. ALC is supported by PUR 2009 University of Milan and Fondazione SISA. CF-H is supported by grants from the National Institutes of Health (R01HL107953 and R01HL106063). AGGIUNGERE IL MIO CARIPLO, IL FONDO MINISTERO E LA SISA
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Condorelli G, Latronico MV, Dorn GW. 2nd, microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 4.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 7.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 299:E198–206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, Macdougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010 doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C. miR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276:2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, Yahagi N, Kobayashi K, Yatoh S, Takahashi A, Suzuki H, Urayama O, Yamada N, Shimano H. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385:492–496. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 15.Ou Z, Wada T, Gramignoli R, Li S, Strom SC, Huang M, Xie W. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Mol Endocrinol. 25:584–596. doi: 10.1210/me.2010-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 18.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G, Catapano AL. MicroRNA 143–145 deficiency impairs vascular function. Int J Immunopathol Pharmacol. 2012;25:467–474. doi: 10.1177/039463201202500216. [DOI] [PubMed] [Google Scholar]
- 21.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 24.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 26.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, Jou W, Gius D, Deng CX. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23:925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norata GD, Pirillo A, Ammirati E, Catapano AL. Emerging role of high density lipoproteins as a player in the immune system. Atherosclerosis. 2012;220:11–21. doi: 10.1016/j.atherosclerosis.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 37.Norata GD, Pirillo A, Catapano AL. HDLs, immunity, and atherosclerosis. Curr Opin Lipidol. 2011;22:410–416. doi: 10.1097/MOL.0b013e32834adac3. [DOI] [PubMed] [Google Scholar]