Abstract
Background
Conserved proteins preferentially expressed in synaptic terminals of the nervous system are likely to play a significant role in brain function. We have previously identified and molecularly characterized the Sap47 gene which codes for a novel synapse associated protein of 47 kDa in Drosophila. Sequence comparison identifies homologous proteins in numerous species including C. elegans, fish, mouse and human. First hints as to the function of this novel protein family can be obtained by generating mutants for the Sap47 gene in Drosophila.
Results
Attempts to eliminate the Sap47 gene through targeted mutagenesis by homologous recombination were unsuccessful. However, several mutants were generated by transposon remobilization after an appropriate insertion line had become available from the Drosophila P-element screen of the Bellen/Hoskins/Rubin/Spradling labs. Characterization of various deletions in the Sap47 gene due to imprecise excision of the P-element identified three null mutants and three hypomorphic mutants. Null mutants are viable and fertile and show no gross structural or obvious behavioural deficits. For cell-specific over-expression and "rescue" of the knock-out flies a transgenic line was generated which expresses the most abundant transcript under the control of the yeast enhancer UAS. In addition, knock-down of the Sap47 gene was achieved by generating 31 transgenic lines expressing Sap47 RNAi constructs, again under UAS control. When driven by a ubiquitously expressed yeast transcription factor (GAL4), Sap47 gene suppression in several of these lines is highly efficient resulting in residual SAP47 protein concentrations in heads as low as 6% of wild type levels.
Conclusion
The conserved synaptic protein SAP47 of Drosophila is not essential for basic synaptic function. The Sap47 gene region may be refractory to targeted mutagenesis by homologous recombination. RNAi using a construct linking genomic DNA to anti-sense cDNA in our hands is not more effective than using a cDNA-anti-sense cDNA construct. The tools developed in this study will now allow a detailed analysis of the molecular, cellular and systemic function of the SAP47 protein in Drosophila.
Background
The "synapse-associated protein of 47 kDa" (SAP47) of Drosophila melanogaster was discovered as the first member of a novel conserved protein family of unknown function [1]. In-situ hybridization and immunostaining using a monoclonal antibody (MAB nc46) showed prominent expression of the Sap47 gene in the nervous system and the specific localisation of the gene product in synaptic terminals. Cross-reacting proteins of similar size were found in several insect species [1]. The SAP47 protein does not contain any domains defined by Prosite patterns that could be indicative of a specific function or molecular interaction. However, by an iterative search of the protein database a novel domain named BSD was identified due to weak but significant amino acid similarity between numerous proteins including BTF2-like transcription factors, synapse associated proteins (SAP47), and DOS2-like proteins [2]. As a first step towards a comprehensive functional analysis of SAP47 we have now generated gene knock-out flies that lack the protein. Unexpectedly, these flies fail to show an obvious phenotype. The high conservation of the protein throughout evolution demonstrates, however, that SAP47 must serve a fitness related function. In order to identify such a function, diverse behavioural assays have to be employed. Quantitative behavioural characterization is notoriously affected by genetic background phenomena, such that extensive out-crossing and comparison with transgenic "rescue" strains will be mandatory. Furthermore, functional defects of a nervous system as reflected by behavioural deficits can often be associated with specific brain structures. To test such a possibility it is necessary to selectively control gene expression in defined populations of neurons. In the present work we have therefore constructed and describe here a set of transgenic tools for cell-specific gene suppression and over-expression that will be decisive for the detailed characterization of SAP47 function in the nervous system of Drosophila at the molecular, cellular and systemic level.
Results
The Drosophila Sap47 gene codes for at least nine protein isoforms
Reichmuth et al. [1] described two independent cDNAs (Sap47-I and Sap47-II) of the Sap47 gene encoding 347 and 351 amino acids, respectively. The corresponding proteins have a calculated molecular mass of 35.06 and 35.50 kDa, respectively, but Western blots show an apparent molecular weight of 47 kDa [1]. The combined information from available cDNA and EST sequences as well as putative transcripts suggested by computer analysis (available at http://fly.ebi.ac.uk), suggests the existence of additional Sap47 splice variants that code for at least three further derived proteins of calculated molecular mass 37.89, 55.31, and 56.98 kDa (Figure 1A). The updated gene structure consists of at least 11 exons and 10 introns. The exon-intron boundaries all conform to the canonical consensus sequences [3]. Six of the exons (2, 8, 9, 10 and 11, and the 3'-region of the exon 5) are used alternatively to generate the different variants. The open reading frames all begin in exon 1 but are terminated alternatively in exons 9, 10, or 11 by in-frame stop codons.
Figure 1.
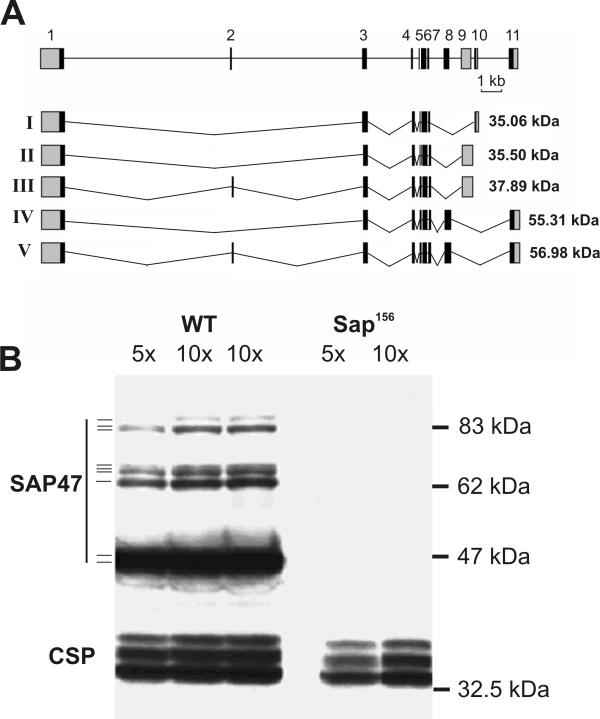
Sap47 gene structure and protein isoforms. (A). Top: Exon-intron structure of the Sap47 gene of Drosophila melanogaster. Boxes represent exons 1 to 11, coding sequences are black. I-V: Five proposed splice isoforms of the Sap47 gene. The corresponding calculated molecular masses are given on the right. (B) Western blot analysis of head homogenates from wild type (WT, w1118) and Sap47 null mutant flies (control for specificity of signals). On the original blot 9 SAP47 bands can be identified as indicated on the left. Staining of cysteine string proteins (CSP) with monoclonal antibody ab49 serves as loading control.
Using modified Western blot conditions (I. Schwenkert, unpublished) we are now able to identify with the anti-SAP47 monoclonal antibody nc46 a total of 9 SAP47 isoforms in adult heads (ca. 46, 47, 62, 65, 66, 67, 86, 87, 90 kDa). Comparison of the blots from wild type with those of Sap47 null mutants demonstrates the specificity of the signals (Figure 1B). In an attempt to associate the molecular weights of the novel SAP47 isoforms with the transcripts we have expressed cDNAs Sap47-I and Sap47-V in E. coli. In Western blots of bacterial lysate the corresponding proteins migrate at 46/47 and 86/87 kDa (data not shown). Thus the protein isoforms of 46, 47, 86, and 87 kDa can presumably be accounted for by the proposed cDNAs I – V. Whether the isoforms between 62 and 67 kDa represent post-translational modifications or are produced by additional splice variants remains to be investigated.
Sequence comparison of homologous proteins in fly and human
Figure 2 illustrates the homologies between SAP47 of Drosophila (DM) and proteins found by BLASTP in the Anopheles (AG), human (HS), and mouse (MM) Proteome. Amino acid (aa) identity between DM and AG up to amino acid 378 is 55%. Using TBLASTN detects highly significant identities between DM and AG up to the very end of SAP47, indicating conservation among diptera also of the C-terminus. Identity between HS and MM is low in 103 aas following a conserved stretch of 11 aas at the N-terminus, but over-all high (85%). Divergence between mammals and diptera is large at the N-terminus which is ca. 150 aas shorter in mammals. Aa identity is high in a central domain, with an over-all value of 23%.
Figure 2.

SAP47 homologues from diptera and mammals. Sequence comparison of the largest SAP47 isoform of Drosophila melanogaster (DM) with its homologues of Anopheles gambiae (AG), human (HS), and mouse (MM) reveals high homology in a central domain. Homologies between Drosophila and human are shown in red.
Mutagenesis
Homologous recombination
Gene targeting by homologous recombination is a powerful method to knock out genes in yeast and mice. Rong and Golic reported a variant applicable in Drosophila and successfully applied it to knock out several genes [4-6]. In the present work we have attempted to mutate the Sap47 gene using the techniques and the materials described by Rong and Golic [4]. Two different large "donor" constructs (ca. 5 kb and ca. 7.6 kb donor-target homology) were created and transformed into the fly germ line. We isolated five independent transgenic lines: four lines for the 5 kb and one for the 7.6 kb donor construct. By crossing these transgenic flies with FLP/SceI lines kindly provided by Dr. K.G. Golic we generated animals which contain all three components of the "gene targeting" system (donor, FLP recombinase, and I-SceI meganuclease transgenes). The action of FLP recombinase and I-SceI produces a linear extra-chromosomal recombinogenic donor DNA molecule that can recombine with and destroy the Sap47 gene. The progeny of these flies were selected for eye colour and the position of the donor construct on the third chromosome. By screening of ca. 280,000 animals containing the 5 kb large "donor" construct and ca. 125,000 animals containing the 7.6 kb construct one single fly with a non-targeted recombination event was isolated, but no targeted recombinants were detected. It thus seems that the Sap47 gene is refractory to this method of mutagenesis.
Jump-out mutagenesis of the Sap47 gene
Recently, a line carrying an EPgy2 P element insertion in the 5' untranslated region of the Sap47 gene (EY07944 line kindly provided by Dr. H. Bellen) became available. This line was used for jump-out mutagenesis because the insertion itself did not visibly influence Sap47 gene expression (data not shown). Screening of ca. 230 white-eyed jump-out "candidates" by Western blots lead to the identification of three lines with a significant reduction in SAP47 expression and three null mutants for the Sap47 gene (Figure 3A). The loss of SAP47 expression in the jump-out null mutants was confirmed by immunohistochemical staining of frozen head sections (Figure 3B). The genomic EcoRI-digested DNA of the isolated Sap47 null mutants was analyzed by Southern blots using ca. 7.1 kb EcoRI-EcoRI genomic fragment of the gene as a probe. The analysis shows that the Sap47156, Sap47201 and Sap47208 null alleles of the gene suffered ca. 2.1 kb, 5.8 kb and 1.3 kb deletions in the Sap47 locus respectively.
Figure 3.
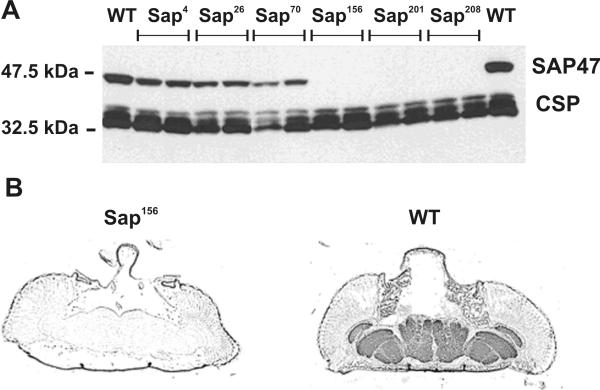
SAP47 expression in the Sap47 mutants generated by P-element jump-out. (A). Western blot analysis of head extracts from jump-out lines (Sap4 – Sap208) and wild type (WT) control. The jump-out lines Sap4, Sap26, and Sap70 show a reduction in SAP47 expression, the lines Sap156, Sap201, and Sap208 lack any detectable SAP47 expression. CSP: loading control. (B). Immunohistochemical staining of frozen head sections. WT: wild type control; Sap156: Sap47 jump-out null mutant line.
Transgenic cDNA RNAi constructs
In a first approach, we used a cDNA-cDNA RNAi construct to target the Sap47 gene. Figure 4A shows the design of the cDNA RNAi vector. We fused a ca. 1.1 kb fragment of the Sap47-I cDNA containing exons 1 and 3 through 7 to the same fragment in opposite orientation. The construct is under UAS control in the pUAST vector and transcription can be induced by Gal4 driver lines. 20 independent transgenic lines for this cDNA RNAi construct were isolated by transformation into the w1118 wild type. The RNAi transgene was activated by crossing the RNAi transgenic flies with an actin-Gal4 line. Figure 5A shows a Western blot containing head extracts from 5 independent lines expressing the Sap47 cDNA RNAi transgene in comparison to wild type. The 47 kDa SAP47 protein is strongly reduced in the heads of flies from the lines V and VII.5. The reduction of SAP47 in the RNAi-expressing flies was confirmed by immunohistochemical staining of frozen head sections and nerve-muscle preparations of third instar larvae (data not shown). To quantitatively determine the suppression of the SAP47 protein, we analyzed head extracts from the "best" RNAi-expressing line (VII.10) and serial dilutions of wild-type head extracts on Western blots (Figure 5B). The analysis shows that 1 head of the Sap47-RNAi line actin-Gal4/VII.10 expresses an amount of SAP47 protein equivalent to that found in 1/16 of a wild-type head.
Figure 4.
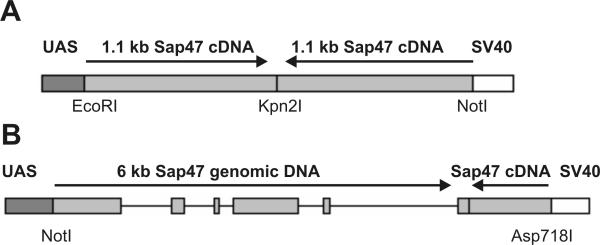
Structure of the two different RNAi constructs. (A). Schematic of the Sap47 cDNA RNAi construct regulated by the yeast UAS enhancer. The transgene produces dsRNA as the construct carries the same fragment of the Sap47-I cDNA arranged as a dimer with dyad symmetry without a heterologous spacer. (B). Genomic-cDNA RNAi construct for the Sap47 gene. Exons 3 to 7 and part of exon 8 with their intervening sequences (introns 3 to 7) of the Sap47 gene were fused to an inverted corresponding fragment of the Sap47-I cDNA and cloned into the pUAST plasmid.
Figure 5.
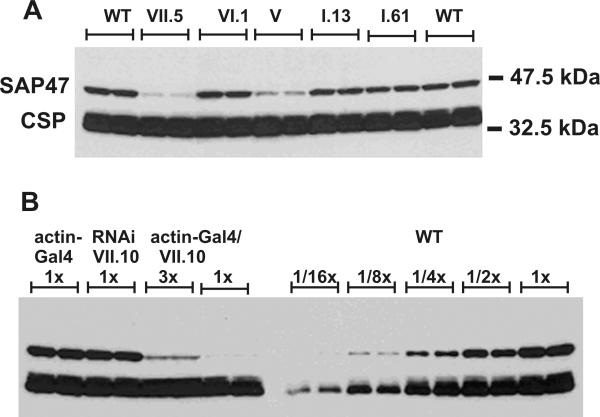
SAP47 expression in the Sap47 cDNA RNAi lines. (A). Western blot of head extracts (one head/lane) from control (WT: wild type (w1118)) and five independent RNAi transgenic lines (VII.5, VI.1, V, I.13, I.61). SAP47 protein levels in VII.5 and V RNAi transgenic lines is drastically reduced. CSP: loading control. (B). Semi-quantitative Western blot of head extracts from Sap47 cDNA RNAi expressing flies (line VII.10) and wild type as control. WT: wild type (w1118). x-values indicate the number of heads loaded per lane. Actin-Gal4/VII.10: animals expressing RNAi under the control of actin-Gal4. RNAi reduces SAP47 protein levels in these flies by an approximate factor of 16. Normal SAP47 expression is observed in RNAi VII.10 (RNAi line without a Gal4 driver) and in actin-Gal4 (driver line alone). CSP: loading control.
Inspection of the Western blot in Fig. 5A reveals a dramatic difference in residual SAP47 expression in different RNAi lines. A few RNAi lines show a very stringent suppression with only ca. 6% residual expression, other lines did not show any visible reduction in SAP47 expression. This difference in suppression could be caused by a position effect of the transgene insertion or by instability of the palindromic sequences of the construct. To check whether the difference was due to structural instability of the RNAi constructs, we examined the genomic DNA of 12 randomly selected RNAi transgenic lines by Southern blot. For each line we analyzed EcoRI/NotI-digested DNA samples, each derived from a pool of 100 flies, by hybridization using the complete ca. 2.2 kb EcoRI/NotI fragment of the Sap47 cDNA RNAi construct as a probe. This probe detects bands of 11.5, 7.0, and 1.8 kb, derived from the endogenous Sap47 allele, and a band of ca. 2.2 kb, derived from the full length RNAi transposon (data not shown). In fact, the 2.2 kb transposon band was detectible only in 2/3 of the RNAi transgenic flies, 1/3 of the lines did not show any visible signal of 2.2 kb. These results indicate that the RNAi transgene was lost in a substantial number of transgenic lines although the red eye colour demonstrated successful germ-line transformation by the pUAST vector.
Transgenic genomic-cDNA RNAi construct
Two groups have described the genomic-cDNA RNAi approach as a simple and very effective strategy to knock out gene function in living organisms [7,8]. Therefore, we used this method to suppress the Sap47 gene activity in Drosophila. Figure 4B shows the design of the Sap47 genomic-cDNA RNAi construct. We fused genomic DNA containing exons 3 through 8 with the intervening introns to a corresponding inverted cDNA fragment of the Sap47 locus. The construct was inserted into the pUAST vector to allow its expression under the control of transgenic Gal4. The P-element mediated germ line transformation of w1118 embryos lead to the isolation of at least 11 independent transgenic lines. By crossing the flies with an actin-Gal4 line animals heterozygous for both transgenes were generated. Western blot of head homogenates from these lines expressing the genomic-cDNA RNAi transgene under actin-Gal4 control demonstrate a strong reduction of SAP47 expression in several transgenic flies containing a single copy of genomic-cDNA RNAi construct. A corresponding reduction was detected by immunohistochemical staining of frozen head sections and nerve-muscle preparations (data not shown). For the line with the strongest reduction we semi-quantitatively determined the suppression of SAP47 by Western blotting and showed, that protein extracts from one adult RNAi-expressing fly head of the genomic-cDNA RNAi line actin-Gal4/2.4 contains an amount of SAP47 protein equivalent to that found in 1/8 of Drosophila wild type head (data not shown).
Rescue of null mutants and over-expression of SAP47
In order to be able to directly relate an observed phenotype of the null mutant to the lack of the encoded protein one has to demonstrate that ectopic expression of the protein in the null mutant reverts the phenotype. In addition, ectopic expression in the wild type leads to over-expression of the protein which may by itself cause a phenotype indicative of the protein's function. For these reasons the Sap47-I cDNA was cloned into the pUAST vector and transformed into the germ line of Drosophila. Of 11 transgenic lines 4 were selected for further analysis. After crossing these flies with an elav-Gal4 driver line, Western blots of head homogenates demonstrate a two to five fold increase in SAP47 expression compared to the wild type (Fig. 7).
Figure 7.
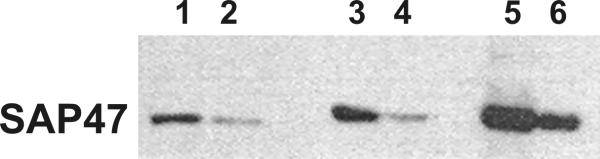
Over-expression of SAP47 by a UAS-Sap47-I cDNA construct. The transgene was driven by neuronal Gal4 expression of the elav-Gal4 line. The following head equivalents were loaded per lane: 1: 1 WT; 2: 1/2 WT, 3: 1/2 Sap47-I1/elav-Gal4; 4: 1/4 Sap47-I1/elav-Gal4; 5: 1/2 Sap47-I3.1/elav-Gal4; 6: 1/4 Sap47-I3.1/elav-Gal4.
Discussion
Using the available sequence information from our own lab [1] and from the Drosophila genome project we updated the putative exon-intron structure of the Sap47 gene. The gene consists of at least 11 exons and 10 introns, producing different splice variants that presumably code for at least five different protein isoforms. In Western blots 9 isoforms can be discerned. By comparison with proteins translated from bacterially expressed cDNAs Sap47-I and Sap47-V it is concluded that the 347 and 551 amino acid proteins migrate anomalously at ca. 46/47 and 85/86 kDa apparent molecular weight, respectively. The Western blot signals that cannot be accounted for by known cDNAs may either represent products of additional splice variants or post-translationally modified isoforms.
Four strategies were used in this work to manipulate Sap47 expression in Drosophila melanogaster: "gene targeting" by homologous recombination, RNA interference (RNAi), transgenic over-expression, and "jump-out" mutagenesis.
For homologous recombination, a DNA fragment containing incomplete 5' and 3' sequences of the target gene interrupted by the 18 bp recognition site for the endonuclease I-SceI was cloned between two target sequences for the site-specific FLP recombinase. In this approach transgenic expression of the recombinase induces the excision of the transgene forming a ring of extra-chromosomal DNA. Subsequent transgenic expression of the endonuclease linearizes this DNA ring. Two events of homologous recombination of the linear DNA then could result in the disruption of the endogenous gene sequence. In previous work, successful targeting of several genes using this technique varied in the range of about one in 500 to one in 30,000 gametes [4,6]. Plausible explanations for detecting in the present experiments zero targeting events and only 1 non-targeted insertion among 405,000 gametes by this procedure are difficult to provide, but rapid degradation of the cut donor fragment, rapid intramolecular repair of the double strand break, or an unfavourable chromatin structure at the Sap47 locus all may influence success rates of the technique [6].
Double-stranded RNA molecules can suppress gene activity in a sequence-specific manner [9-12]. Several groups have reported that expression of stabile transgenic inverted repeats in Drosophila can reduce gene expression [13-15], but the suppression is incomplete. Kalidas and Smith [7] noted that RNAi transgenes composed of genomic-cDNA fusions can efficiently suppress target genes in adult flies. These authors suggest that the level of suppression will be significantly greater and more uniform than reported for transgenic RNAi constructs composed of simple inverted repeats. In the present work we created and compared two different RNAi constructs for the Sap47 gene of Drosophila. The first construct consist of two cDNA fragments cloned into the pUAST as inverted repeats. The second one is a fusion product of a Sap47 genomic fragment and the corresponding cDNA sequences. After crossing the transgenic RNAi flies with an actin-Gal4 line, offspring were analysed for SAP47 expression by Western blot and immunohistochemistry. In most cases, we observed a visible reduction in the expression of SAP47 protein. About 54% of the RNAi cDNA transgenic lines heterozygote for both, RNAi and actin-Gal4 transgenes, show more than 75% reduction in protein level, a few lines show a very stringent suppression with only ca. 6% residual expression, but ca. 35% of the analysed lines did not show any visible reduction of SAP47 expression in Western blot. The same dramatic differences in residual SAP47 expression levels were observed for the genomic RNAi construct. 8 of 11 independent transgenic lines show less than 25% residual SAP47 expression, one less than 12%, 2 lines show a wild type like SAP47 signal in Western blots.
We have presented evidence that this difference in the SAP47 levels in RNAi expressing lines can be caused by structural instability of the transgenes. The Sap47 cDNA RNAi is a perfect palindromic construct. Giardano et al. [14] described somatic rearrangements of the inverted repeat in Drosophila, Collick et al. [16] reported instability of the inverted repeats in somatic or germline cells of mice. In our experiments, 1/3 of the RNAi lines analysed by Southern blots did not show any visible transgene bands of 2.2 kb, although they had red eyes demonstrating successful transformation of the transgene. The loss of transgenic sequences might represent a rearrangement of the perfect palindromic fragments of the cDNA RNAi construct in the germ line of the transgenic flies. These rearrangements can lead to the inactivation of the transgene and result in wild-type-like protein expression in these animals.
To compare the silencing effects of cDNA-cDNA RNAi vs. genomic-cDNA RNAi constructs, we tested head extracts from the "best" lines from the each group in the same Western blot under the same conditions (Figure 6). In this experiment, we did not find a dramatic difference between the two constructs.
Figure 6.
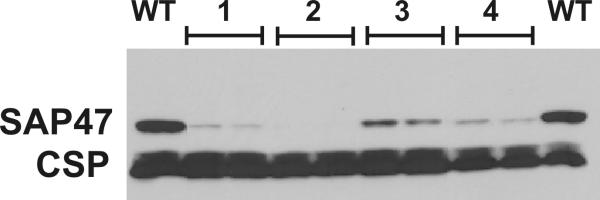
Sap47 gene suppression by the two RNAi techniques used is similar. The Western blot shows SAP47 protein levels in three independent cDNA-cDNA RNAi lines (1: line VII.5 (2 transgene insertions), 2: line VII.10 (2 transgene insertions), 3: line X.2) and in one genomic-cDNA RNAi line (4: line 2.4) WT: wild-type control; one head/lane. All RNAi constructs are under actin-Gal4 control. CSP: loading control.
The mobilisation of a EPgy2 P element inserted in the 5' untranslated region of the Sap47 gene and screening of ca. 230 "candidates" lead to the isolation of three independent knock-out strains (null mutants), containing deletions in the first exon and first intron of the Sap47 gene. In order to exclude the possibility that an N-terminally truncated protein was still expressed and possibly sufficient for SAP47 function, we repeated the Western blots with a monoclonal antibody (nb200) whose epitope is known to lie in the portion of the protein that is not affected by the deletion. Since again no signal was obtained (data not shown) we conclude that the deletions prevent production even of SAP47 fragments. From all three mutagenesis techniques that we used in this study, "gene targeting" by homologous recombination, RNA interference, and jump-out mutagenesis, only the last was an effective method to create Sap47 null mutants.
Conclusions
Surprisingly, neither knock-down, nor knock-out, nor over-expression of the Sap47 gene resulted in flies showing an obvious, immediately noticeable phenotype. Thus it will be necessary, first to exchange the genetic background of the mutants to that of the "standard" wild type used for behavioural assays, and then analyse complex behaviour of mutants and wild-type controls in detail under identical conditions. An elaborate collection of behavioural paradigms are available for Drosophila to test e.g. locomotor activity, olfactory or visual performance, courtship, sensory habituation, ethanol tolerance, and learning and memory. As mentioned above, the evolutionary conservation of the Sap47 gene assures us that the encoded protein has a fitness related function. This function now can be searched for.
Methods
Drosophila strains and transgenic flies
Balancer chromosomes and genetic markers are described in Lindsley and Zimm [17]. w1118 wild-type stock was used for generation of transgenic flies and as controls in these studies. Most of the flies employed for gene targeting by homologous recombination were kindly provided by Yikang Rong and Kent G. Golic. In the actin-Gal4 strain, the P [actin-Gal4, w+] transposon was inserted on the second chromosome (causing homozygous lethality). The transgenic flies were generated by standard P-element mediated transformation [18].
The EY07944 line used for jump-out mutagenesis was kindly provided by the Bellen laboratory http://flypush.imgen.bcm.tmc.edu. Virgin w1118;;EY07944(w+) were crossed with w1118;;P{Δ2-3}(99B), Sb/TM2, Ubx, e males. The F1 generation was selected for Sb to obtain w1118;;EY07944(w+)/P{Δ2-3}(99B), Sb animals. These were crossed with w1118;;TM3, Sb, e/TM6, Tb, e double balancer and the white Tb-marked F2 "jump-out" animals were crossed with Δblp,ΔSap47/TM3, Sb, e double mutants [19]. The progeny was characterized by Western blot using monoclonal antibodies nc46 and, as a loading control, ab49 (anti-CSP).
Western blotting and Immunohistochemistry
Drosophila heads were homogenized in Laemmli sample buffer. After separation by SDS-PAGE [20] proteins were transferred to a nitrocellulose membrane [21] and the membrane blocked in 5% milk-powder solution. Blots were immunostained with monoclonal antibodies and with horseradish peroxidase conjugated second antibody (Bio-Rad Laboratories GmbH, Muenchen, Germany) followed by ECL detection (Amersham Buchler GmbH, Braunschweig). For staining of motorneuronal synaptic boutons third instar larvae were fixed after preparation for 30 min at room temperature in 4% paraformaldehyde and incubated overnight at 4°C with primary antibody. Biotin-avidin-peroxidase system (Vector Laboratories Inc., Burlingame, USA) and diamino benzidine were used for visualizing the staining. The procedure used for immunostaining frozen sections of Drosophila brain has been described elsewhere [22]. Briefly, flies were fixed for 3 h in 4% paraformaldehyde and washed overnight in 25% sucrose. Frozen sections were incubated overnight at 4°C with primary antibody (monoclonal antibody nc46) and stained using again the biotin-avidin peroxidase system.
RNAi constructs
To prepare the Sap47-cDNA RNAi construct, a ca. 1.1 kb coding fragment of the Sap47-I cDNA was amplified by PCR with primers containing unique restriction sites (5'-GGCGTGAATTCAACATGTTTTCGGGCCTAAC-3' and 5'-CATGATCCGGAATCTTCATCTTCGCCGC-3' primer pair for sense cDNA fragment and 5'-CTGACATCCGGACAATCTTCATCTTCGCCG-3' and 5'-AAATAGCGGCCGCTTTCGGGCCTAACAAATC-3' primer pair for anti-sense cDNA fragment, respectively). The PCR-amplified fragments were digested with EcoRI/Kpn2I and Kpn2I/NotI respectively, subcloned into EcoRI/NotI-cut pBluescript KSII (Stratagene, La Jolla, USA) and sequenced. The resulting inverted-repeat sequence was excised as an EcoRI/NotI fragment, ligated into EcoRI/NotI-cut pUAST [23] and transformed into recombination-deficient SURE2 cells (Stratagene, La Jolla, USA).
For the Sap47-genomic RNAi construct, a 5025 bp genomic fragment of the Sap47 gene was obtained by cutting the Sap47 genomic lambda clone IV/4 [24] with MspA1I/Bsp68I and subcloning of the fragment into a Bsp68I-cut modified pBluescript KSII vector. A ca. 5 kb fragment was then excised using the NotI restriction site from pBluescript KSII poly-linker and the Bsp68I site. In parallel, the ca. 1 kb genomic fragment of the Sap47 gene (contains a portion of intron VII and the 5'-end of exon 8 with splice acceptor sequences) and a ca. 0.85 kb fragment of the Sap47-I cDNA (contains exons 3–8) were amplified using PCR with primers containing unique restriction sites (5'-GGGTCCAGAATTCTCGCGAATTTGGTTTTCC-3' and 5'-CCTGATCCGGAGTCTTTTGGTTTTAATCCATTCAAT-3' primer pair for genomic fragment and 5'-GGTCCTCCGGGATTTTGGTTTTAATCCATTCAATC-3' and 5'-GTTCAGGTACCGATGGCTGGGCAGTGC-3' primer pair for the cDNA, respectively), digested with EcoRI/Kpn2I and Asp718I/Kpn2I and subcloned into EcoRI/Asp718I-cut pBluescript KSII. A ca. 1.85 kb fusion-product was then isolated using the Asp718I site from pBluescript KSII and endogenous Bsp68I site. Finally, the ca. 5 kb NotI/Bsp68I genomic fragment and ca. 1.85 kb Asp718I/Bsp68I fusion-fragment were cloned into the NotI/Asp718I-cut pUAST. The structure of the cloned construct was verified by a combination of DNA sequencing and restriction mapping.
Sap47 rescue construct
BglII and NotI restriction sites were attached by linker PCR (primer pair: 5'-CCTACTAGATCTCCAACATGTTTTCGGGCCTAA-3' and 5'-CTTAACGCGGCCCATTCAATCTTCATCTTCATCTTC-3') to the Sap47-I cDNA and inserted into the pUAST vector which was transformed into the germ line of Drosophila by standard methods.
Authors' contributions
NF conceived of the study, performed most of the work and wrote a first draft of the ms, SB cloned and analysed the Sap47 genomic region, SH generated the "rescue" transformants and analysed them, MB was involved in the sequence analysis, EB supervised the four theses, participated in the design and coordination of the study, and wrote the final ms.
Acknowledgments
Acknowledgements
We thank D. Dudaczek for excellent immunohistochemistry on frozen sections, Dr. K. G. Golic for provision of the fly strains for the "gene targeting" mutagenesis, Dr. H. Bellen for the EY07944 line. This work was financed by DFG Grants (Bu566, SFB581/B6, and SFB554/A2), the MD-PhD program and a scholarship from the University of Wuerzburg.
Contributor Information
Natalja Funk, Email: nfunk@biozentrum.uni-wuerzburg.de.
Sonja Becker, Email: sonja.becker@gsf.de.
Saskia Huber, Email: Saskia.Huber@gsf.de.
Marion Brunner, Email: marionbrunner@t-online.de.
Erich Buchner, Email: buchner@biozentrum.uni-wuerzburg.de.
References
- Reichmuth C, Becker S, Benz M, Debel K, Reisch D, Heimbeck G, Hofbauer A, Klagges B, Pflugfelder GO, Buchner E. The sap47 gene of Drosophila melanogaster codes for a novel conserved neuronal protein associated with synaptic terminals. Mol Brain Res. 1995;32:45–54. doi: 10.1016/0169-328X(95)00058-Z. [DOI] [PubMed] [Google Scholar]
- Doerks T, Huber S, Buchner E, Bork P. BSD: a novel domain in transcription factors and synapse-associated proteins. Trends Biochem Sci. 2002;27:168–170. doi: 10.1016/S0968-0004(01)02042-4. [DOI] [PubMed] [Google Scholar]
- Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2017. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/S0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Cerutti H. RNA interference: traveling in the cell and gaining functions? Trends Genet. 2003;19:39–46. doi: 10.1016/S0168-9525(02)00010-0. [DOI] [PubMed] [Google Scholar]
- Szweykowska-Kulinska Z, Jarmolowski A, Figlerowicz M. RNA interference and its role in the regulation of eukaryotic gene expression. Acta Biochim Pol. 2003;50:217–229. [PubMed] [Google Scholar]
- Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Matzke AJM, Kooter JM. RNAi: Guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccin A, Salameh A, Benna C, Sandrelli F, Mazzotta G, Zordan M, Rosato E, Kyriacou CP, Costa R. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 2001;29:E55–5. doi: 10.1093/nar/29.12.e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- Spradling AC. P-element mediated transformation. In: Roberts DB, editor. In Drosophila: A practical approach. Oxford: IRL Press; 1986. pp. 175–197. [Google Scholar]
- Becker S, Gehrsitz A, Bork P, Buchner S, Buchner E. The black-pearl gene of Drosophila defines a novel conserved protein family and is required for larval growth and survival. Gene. 2001;262:15–22. doi: 10.1016/S0378-1119(00)00548-5. [DOI] [PubMed] [Google Scholar]
- Bollag DM, Rozycki MD, Edelstein SJ. Protein methods. 2. New York: Wiley-Liss; 1996. [Google Scholar]
- Khyse-Anderson J. Electroblotting of multiple gels. A simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamid gels to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022X(84)90040-X. [DOI] [PubMed] [Google Scholar]
- Buchner E, Buchner S, Crawford G, Mason WT, Salvaterra PM, Sattelle DB. Choline acetyltransferase-like immunreactivity in the brain of Drosophila melanogaster. Cell Tissue Res. 1986;246:57–62. [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Becker S. PhD thesis. University of Wuerzburg; 1997. Sap47 und black-pearl. Molekulare Charakterisierung und Mutagenisierung zweier konservierter Genloci von Drosophila melanogaster. [Google Scholar]


