Summary
A new degradable PEG-diester-dinorbornene/PEG-triester-trithiol hydrogel was evaluated for protein release. The hydrogel polymerized rapidly with seconds of UV irradiation and subsequently hydrolytically degraded in aqueous buffer over the course of approximately 3 weeks. Further, the hydrogel enabled the encapsulation and release of a model protein, bovine serum albumin (BSA), over 7 days with ~ 90% released at 48 h. This study serves as a proof-of-concept for the creation of hydrolytically degradable, PEG-ester-thiol-based hydrogels by a photoinitiated step growth mechanism for protein release. With this approach, degradation and release rates could be tuned by varying the monomer molecular weight and functionality in future studies.
Keywords: hydrogels, degradation, drug delivery systems, photopolymerization, thiol–ene
Introduction
Hydrogels increasingly are used for the controlled delivery of proteins, such as growth factors, cytokines, and chemoattractants, whose spatiotemporal presentation regulate tissue homeostasis, regeneration, or disease progression.[1] Proteins are loaded within hydrogel matrices by a number of mechanisms aimed at tuning the release while maintaining protein activity. For example, proteins loaded by physical entrapment are released by matrix swelling, degradation, and erosion, whereas the release of proteins loaded by sequestration is mediated by protein-matrix interactions.[2] In particular, hydrogels formed from degradable, macromolecular monomers are attractive owing to the ease of protein physical entrapment (loading) and release modulation (delivery). Monomers are designed with i) reactive functionalities conjugated to biocompatible polymers, such as poly(ethylene glycol) (PEG), to form hydrogels that entrap the protein of interest and ii) cleavable moieties to release the entrapped protein as the resulting hydrogel backbone degrades. The hydrogel degradation rate, and thus protein release, can be tuned by selecting appropriate cleavable groups, including hydrolytically degradable esters,[3] enzymatically degradable peptide sequences,[4] or photolytically degradable nitrobenzyl ethers.[5]
Hydrolytically degradable PEG-based hydrogels have a rich history in controlled protein release.[6] Traditionally, poly(lactic acid) (PLA) of varied length, or a similar polyester, is attached to PEG, terminally functionalized with (meth)acrylate end groups and polymerized by free-radical chain polymerization to form a hydrolytically degradable hydrogel with a heterogeneous network structure. Degradation is dictated by initial network properties and chemistry, where increasing the number of lactic acid repeat units increases the degradation and release rates; however, increased PLA content also increases the network's hydrophobicity, affecting protein loading, and increases the acidity of the local microenvironment with degradation as lactic acid byproducts are generated, eliciting an inflammatory response.[7, 8] Zustiak and Leach offered an alternative approach using a Michael-type polymerization of a four-arm PEG-vinyl sulfone (PEG-VS) with PEG-diester-dithiols to form hydrolytically degradable hydrogels with homogenous network structures while avoiding the acidic byproducts of PLA degradation.[8] Pritchard et al. recently examined Michael-type polymerization of PEG-triester-trithiols and PEG-diacrylates as an injectable, in situ forming hydrolytically degradable hydrogel for small molecule therapeutic release.[9] Further, Baldwin and Kiick have developed multifunctional PEG-ester-thiol and heparin-maleimide hydrogels that form by Michael-type polymerization and degrade by a retro-Michael-type addition reaction under reducing conditions and by ester hydrolysis over longer times. These PEG-ester-thiol hydrogels are promising for controlled drug release applications.[10] However, Michael-type reactions do not afford spatial and temporal control of hydrogel formation, and gel formation commences upon mixing. In contrast, photopolymerization reactions allow control over where and when polymerization occurs, offering facile methods such as micromolding and photolithography to form well-defined hydrogel geometries.[11] For example, Rolland et al. have used photolithographic techniques to form PEG-based nano- to micron-scale hydrogel particles with encapsulated proteins, small molecule pharmaceuticals, and oligonucleotides, where particle shape has been observed to influence cell uptake.[12] Furthermore, the added control of polymerization could allow polymer-protein solutions to be pre-mixed and stored in liquid form, if desired.
In this work, we aimed to create a hydrolytically degradable hydrogel that incorporates multiple ester-containing functional groups towards tuning degradation profiles and has a homogeneous network structure affording spatiotemporal control of hydrogel formation and protein encapsulation. PEG-based hydrogels formed by thiol-norbornene photopolymerization form rapidly and offer spatiotemporal control of the polymerization.[13] Hydrogels based on this scheme have been studied for release of several therapeutics, and uniquely, thiol-norbornene hydrogels have been observed to have a protective effect on encapsulated protein bio-efficacy as compared to hydrogels formed by traditional free radical chain polymerization of acrylate-functionalized monomers.[14] For controlled protein release, Aimetti et al. examined thiol-ene gels that release proteins when exposed to the enzyme human neutrophil elastase towards combating inflammatory processes.[15] For small molecule release, Yang et al. conjugated dexamethasone to matrix metalloproteinase (MMP)-sensitive peptide tethers that were incorporated as pendant groups within thiol-ene gels and demonstrated the release of dexamethasone upon exposure of the hydrogel to MMP-secreting cells.[16] Both of these systems rely on MMP-degradable peptide linkers, which can be expensive. Shih and Lin recently observed that PEG-norbonene hydrogels linked with either simple thiol-containing peptides (noncleavable CGGGC and chymotrypsin cleavable CGGY↓C) or small molecules (dithiothreitol) that are not MMP-labile have appreciable hydrolytic degradation at physiological or slightly basic pH (pH of 7.4 to 8), owing to cleavage of the ester-linked norbornene.[17] These formative works demonstrate the promise of peptide-linked norbornene-based hydrogels for controlled therapeutic release or their potential for hydrolytic degradation, respectively. Building upon these prior works, PEG-based linkers are often less expensive and more hydrophilic than peptide crosslinkers, and accordingly, there is interest in understanding and controlling the degradation of various labile PEG-linked systems for protein release.[8] Furthermore, water-mediated degradation of networks could be advantageous for applications where specific enzymes are not readily available or vary in concentration. In this work, we employ two ester-containing monomers, a PEG-diester-dinorbornene and a PEG-triester-trithiol, which allow hydrolytic degradation at six sites per crosslink, adding to the toolbox of network chemistries towards tuning degradation profiles.
We synthesized a PEG-diester-dinorbornene (Mn ~ 3350 g/mol) and selected a commercially available PEG-triester-trithiol (Mn ~ 1300 g/mol) for hydrogel formation and protein encapsulation by radical-mediated step-growth thiol–ene photopolymerization. Hydrogel formation and degradation were characterized with rheometry, and degradation was compared with similar PEG-diacrylate-based hydrogels formed by Michael-type polymerization. Last, release of a model protein, bovine serum albumin (BSA), was evaluated. Owing to their simple chemistry and facile, rapid light-triggered formation, these hydrolytically degradable thiol—ene hydrogels are promising for the in situ formation of protein-releasing hydrogel depots and microparticles.
Materials and Methods
Materials
The tri-thiol crosslinker, ethoxylated-trimethylolpropane tri-3-mercaptopropionate (PEG-triester-trithiol or ETTMP, Mn = 1300 g/mol) was generously donated by Evans Chemetics. N,N'-Dicyclohexylcarbodiimide (DCC), glycine, sodium chloride, and 4-(dimethylamino)pyridine (DMAP) were purchased from Alfa Aesar. Chloroform and sodium hydroxide were purchased from Fisher Scientific. Dichloromethane (DCM, extra dry) was purchased from Acros Organics. Phosphate buffered saline (PBS, pH 7.4) was purchased from Invitrogen. 4-(2-hydroxyethoxy)phenyl-(2-hydroxy-2-propyl)ketone (I2959) was generously donated by BASF. All other chemicals were purchased from Sigma-Aldrich.
Synthesis of poly(ethylene glycol) dinorbornene
PEG-dinorbornene was synthesized based on a modified version of a published protocol.[13] Briefly, DCC (5x molar excess relative to hydroxyl groups on PEG) and 5-norbornene-2-carboxylic acid (10x) were dissolved in 50 mL DCM. The DCC and norbornene solution was stirred while venting for 3 minutes and then purged with argon for 15 minutes. In a separate flask, PEG (10 g, Mn = 3350 g/mol) and DMAP (0.5x) were dissolved in 50 mL DCM and purged with argon for 15 minutes. After purging, pyridine (5x) was added to the PEG and DMAP, and the resulting PEG solution was added to the flask containing DCC and norbornene. The reaction was allowed to proceed overnight under argon.
The next day, the mixture was filtered to remove dicyclohexylurea. The PEG was concentrated by rotary evaporation, precipitated in 9x volume ethyl ether, centrifuged, and the ethyl ether supernatant was discarded. The solid PEG was re-dissolved in chloroform. The PEG/chloroform solution was washed twice sequentially with aqueous 0.05 M glycine, 0.05 M sodium hydroxide, and 0.05 M sodium chloride (the PEG product remained in the chloroform layer) and once with aqueous 5.1 M sodium chloride. Finally, the PEG/chloroform mixture was precipitated in 9x volume ethyl ether and centrifuged three times. Product purity was confirmed by 1H-NMR in DMSO-d6: 400 mHz, δ 6.20-5.86 (m, 4H), δ 3.65-3.40 (m, 304H).
Modulus evolution during hydrogel formation
Rheological measurements were performed on a TA Instruments AR-G2 rheometer with UV-visible light attachment that allows sample irradiation by a filtered Exfo Omnicure Series 2000 light source. Hydrogel formation studies were performed at a strain of 2% and a frequency of 1 Hz, which was determined to be in the linear viscoelastic regime for these hydrogels. Samples were irradiated with 365 nm light at 10 mW/cm2.
For measurements of modulus evolution during hydrogel formation, hydrogels were formed directly on the rheometer. Solutions consisting of 18% w/w total monomer and 0.5% w/w I2959 in citric acid buffer (51 mM citric acid and 56 mM sodium citrate in water, adjusted to pH 5) were placed on the rheometer and exposed to 365 nm UV light. The citric acid buffer was used to ensure acidic conditions during polymerization, minimizing disulfide formation. The ratio of monomers was such that the thiol concentration was equal to the norbornene concentration.
Hydrogel degradation
For degradation studies, PEG-diester-dinorbornene/PEG-triester-trithiol hydrogels were formed in molds consisting of two 75 mm by 25 mm glass slides treated with an anti-adhesion agent (e.g., Rain-X) and separated by a 1 mm rubber spacer. The monomer solutions were prepared at the same concentrations as for the measurements of modulus evolution and pipetted into the molds. The hydrogels were formed by irradiating the molds with 365 nm UV light at an intensity of 10 mW/cm2 for 30 seconds. Hydrogel discs were then punched with an 8-mm diameter biopsy punch and washed with PBS to remove any citric acid and unreacted monomers.
For comparison, PEG-diacrylate/PEG-triester-trithiol hydrogels were formed in the same molds as described above. PEG-diacrylate (Mn = 1000 g/mol) and PEG-triester-trithiol were mixed to a final monomer composition of 18% w/w, using equal molar concentrations of thiol and acrylate functionalities. Triethanolamine was added at 12.4 mM, and the solution was allowed to gel for one hour. Samples were again punched with an 8-mm biopsy punch and washed with PBS.
Degradation was monitored by measuring mass swelling and shear modulus over time. Hydrogels were allowed to swell to equilibrium for 24 hours before any measurements were made. To determine mass swelling, samples were removed from PBS and weighed to record their wet mass. The samples were then frozen at -20°C, lyophilized, and weighed again to record their dry mass. Mass swelling was calculated as wet weight divided by dry weight.
For the shear modulus measurements, strain sweeps from 0.1% to 10% strain at 0.4 Hz were performed on the AR-G2 rheometer at each timepoint. Shear moduli were measured at strains in the linear viscoelastic regime for each sample.
Protein release
For protein release studies, hydrogels were formed the same way as in the hydrogel degradation studies except with the addition of 2% w/w bovine serum albumin (BSA) to the monomer solution. After the hydrogel discs were punched, they were submerged in 15 mL PBS to wash. The PBS was removed, and the hydrogels were each placed in 15 mL of fresh PBS and gently rocked. Residual citric acid in the hydrogel decreased the pH of the PBS to 7.07, so the pH remained between 7.0 and 7.4 for the course of the release experiment. At each timepoint, a 1-mL aliquot was taken from the PBS/BSA solution and frozen, and 1 mL of fresh PBS was added back to the solution. Once all timepoints were collected, the BSA concentration in each aliquot was measured with a Bio-Rad Protein Assay Kit II using the manufacturer's microassay procedure. If appropriate, samples were diluted to ensure the measurements were taken in the linear range of the assay. The release solution concentration and mass of released BSA thus were measured, taking into account the cumulative sample dilution that occurred with removal and addition of PBS aliquots at each timepoint.
Statistics
Data are presented as mean ± standard error unless otherwise specified. The number of samples for each study is as follows: the modulus evolution studies, n = 3; the hydrogel degradation studies, n = 5; and the protein release studies, n = 6.
Results and Discussion
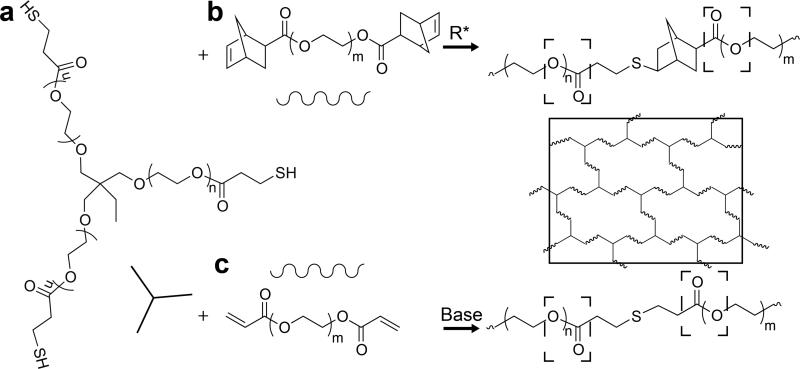
Hydrolytically degradable PEG hydrogels were formed by free-radical step growth polymerization of a commericially available PEG-triester-trithiol (Mn ~ 1300 g/mol) (Figure 1a) with a linear PEG-dinorbornene (Mn ~ 3350 g/mol) in aqueous solution with a balanced stoichiometric ratio of norbornene to thiol (Figure 1b). This simple approach results in a water-swollen polymer network that contains six ester bonds per crosslink. While only one composition was examined here, the crosslink density, modulus, and degradation profiles of this base system can be facilely tuned by adjusting the PEG-dinorbornene molecular weight or stoichiometric ratio of vinyl to thiol.[3, 18] Additionally, PEG-triester-trithiol and PEG-diacrylate (Mn ~ 1000 g/mol) were polymerized by base-catalyzed Michael-type addition (Figure 1c). This formulation has been previously published as a hydrolytically degradable, in situ forming hydrogel for controlled therapeutic release[9] and was used as a control to which the degradation profile of the PEG-dinorbornene hydrogel was compared.
Figure 1. Hydrolytically degradable thiol–ene hydrogel chemistries.
a) PEG-triester trithiol was polymerized with b) PEG-dinorbornene to form a polymer network (solid box) by free radical step growth polymerization. Each thiol-norbonene bond is adjacent to two hydrolytically degradable ester bonds (dashed boxes). Degradation of the resulting PEG-dinorbornene hydrogels was compared to hydrogels formed with the PEG-triester-trithiol and c) PEG-diacrylate via base-catalyzed Michael-type polymerization.
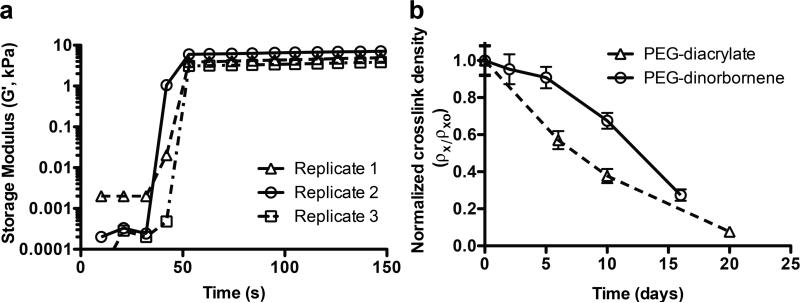
PEG-triester-trithiol and PEG-dinorbornene rapidly polymerized by free radical step growth polymerization (0.5 wt% photoinitiator, 365 nm at 10 mW/cm2) (Figure 2a).
Figure 2. Hydrogel formation and degradation.
a) Formation of the PEG-triester-trithiol and PEG-dinorbornene hydrogels was monitored using photorheometry (18% w/w total monomer with 0.5% w/w photoinitiator, 3 replicates shown). Modulus of the monomer solution was monitored for ~ 40 s, upon which time the irradiation was commenced (365 nm at 10 mW/cm2). Polymerization and crosslinking were rapid, as indirectly measured with modulus, where little change in modulus was observed after ~ 10 s of irradiation (G′o,av = 8.7 ± 1.45 kPa). b) Hydrogel degradation was monitored with rheometry and swelling as measures of crosslink density ρx, which was calculated from these and normalized to its initial value ρxo. Degradation of the PEG-dinorbornene hydrogel (circles, solid line) initially is slower than the control PEG-diacrylate hydrogel (triangles, dashed line); however, both degrade on the same timescale with complete degradation after ~ 25 days.
Photorheometry was utilized to assess mechanical property evolution to indirectly monitor the progression of the polymerization. The resulting hydrogel modulus is relatively constant after approximately 10 seconds of irradiation, indicating that the polymerization is complete. We hypothesized that this short free-radical polymerization time may be appropriate for protein encapsulation, as recent studies show that radical-mediated thiol–ene polymerizations over short times maintain protein bioactivity.[14] To ensure that each sample was completely polymerized, all replicates for hydrogel degradation and protein release experiments were irradiated for 30 seconds.
Hydrogel degradation was examined by monitoring the temporal decrease in storage modulus (G′) and increase in mass swelling (q) that occur with crosslink cleavage. Temporally evolving crosslink density (ρx) then was calculated using rubber elasticity theory: ρx = G′Q⅓(RT)-1, where R is the universal gas constant, T is absolute temperature, G′ was measured by rheometry, and Q is volumetric swelling calculated from measured q.[19] ρx at each time point was normalized to the initial crosslink density (ρxo) for each formulation.
Degradation of the PEG-dinorbornene hydrogels was initially slower than the PEG-diacrylate control, potentially owing to a stabilizing effect of the norbornene ring on ester(s) within the crosslinks relative the alkyl linker (Figure 2b). Over the 3-week study, the PEG-dinorbornene hydrogel degrades on a similar timescale to the PEG-diacrylate hydrogel (both fully degraded after ~ 25 days). Additionally, this degradation rate appears to be faster than that reported for a similarly crosslinked multifunctional PEG-norbornene polymerized with non-ester-containing dithiol (degradation over 50 days at pH ~ 7.4),[17] indicating that the esters within the PEG-triester-trithiol contribute to hydrogel degradation as previously observed by Pritchard et al.[9]
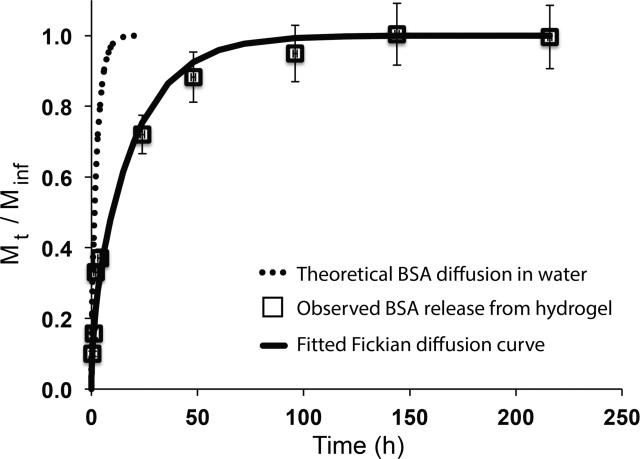
Protein release from this new hydrogel formulation was examined using BSA as a model protein. BSA was encapsulated during PEG-dinorbornene hydrogel formation (1 mg per sample). The mass of protein released over time was monitored using the Bio-Rad Protein Assay Kit II. Most of the protein is released over 2 days (Figure 3) (~ 90% released by 48 h). While a small amount of degradation occurs over this timeframe, the release profile appears to be largely diffusion-mediated. However, the BSA release profile is comparable to that achieved with multifunctional PEG-ester-thiols and PEG-vinyl sulfones polymerized by Michael-type polymerization.[8] Additionally, all of the BSA loaded is not observed to diffuse out of the hydrogel, possibly owing to chain transfer to BSA during hydrogel formation, which has been reported by others.[14, 20] Approximately 80% of the BSA is recovered, which is consistent with results published by McCall and Anseth for several model proteins in a similar hydrogel system.[13] Additionally, McCall and Ansethhave shown that the thiol-norbornene reaction has a protective effect on protein bioactivity during free radical encapsulation.[14]
Figure 3. Model protein release.
BSA release from the PEG-dinorbornene hydrogel was observed over approximately 10 days, where ~ 90% of the BSA is released within 48 hours. While 1 mg of BSA per sample was loaded, approximately 0.82 mg on average was recovered (Minf). BSA release was evalulated by fitting a Fickian diffusion coefficient to the release curve (Deff ~ 1.4 × 10-8 cm2/s; open squares data; solid line predicted release based on Fickian fit). This release profile is substantially slower than Fickian diffusion of BSA in aqueous solution (Do ~ 1 × 10-7 cm2/s; dashed line predicted Fickian diffusion in water).
The effective diffusion coefficient for BSA in the PEG-dinorbornene hydrogel was examined as a measure of controlled release. To fit an effective diffusion coefficient to the release data, the mass of protein released at any time (Mt) was divided by the total mass of BSA released (Minf),[21] and assuming Fickian diffusion, a numerical algorithm in MATLAB was used to fit Deff to the resulting release curve (Figure 3). Deff for this hydrogel formulation was ~ 1.4 × 10-8 cm2/s, approximately an order of magnitude smaller than the diffusion coefficient for BSA in water of ~ 1.0 × 10-7 cm2/s.[8] Slowed diffusion indicates that the hydrogel affords some level of controlled release, providing a local depot or reservoir for protein delivery over short timescales. With this base strategy, diffusion could be slowed further by decreasing the PEG molecular weight or total monomer concentration.
Conclusions
Hydrogels formed by photopolymerization of ester-containing norbornene- and thiolfunctionalized PEG show promise for the controlled release of therapeutics. These hydrogels polymerize rapidly upon irradiation, which is advantageous for protein encapsulation and in situ polymerization, and hydrolytically degrade over the timescale of weeks. BSA release from the current hydrogel formulation appears to be diffusion controlled, where the effective diffusion coefficient for BSA out of the hydrogel is an order of magnitude smaller than the diffusion coefficient in water. With the proof-of-concept demonstrated for this base approach, tuning of degradation rates and profiles can be explored further by varying PEG-dinorbornene molecular weight, total monomer concentration, and the ratio of norbornene to thiol.
Acknowledgements
The authors would like to thank the Molecular Design of Advanced Biomaterials NIH Center of Biomedical Research Excellence at the University of Delaware for funding (NIH COBRE P20-RR017716). MSR would like to thank the Chemistry-Biology Interface Program at the University of Delaware for additional support.
References
- 1.Tayalia P, Mooney DJ. Adv. Mater. 2009;21:3269. doi: 10.1002/adma.200900241. [DOI] [PubMed] [Google Scholar]; Lin CC, Anseth KS. Pharm. Res. 2009;26:631. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vermonden T, Censi R, Hennink WE. Chem. Rev. 2012;112:2853. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]
- 2.Censi R, Di Martino P, Vermonden T, Hennink WE, Control J. Release. 2012;161:680. doi: 10.1016/j.jconrel.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Metters A, Hubbell J. Biomacromolecules. 2005;6:290. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 4.Patterson J, Hubbell JA. Biomaterials. 2010;31:7836. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 5.Tibbitt MW, Han BW, Kloxin AM, Anseth KS. J. Biomed. Mater. Res. Part A. 2012;100A:1647. doi: 10.1002/jbm.a.34107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anseth KS, Metters AT, Bryant SJ, Martens PJ, Elisseeff JH, Bowman CN, Control J. Release. 2002;78:199. doi: 10.1016/s0168-3659(01)00500-4. [DOI] [PubMed] [Google Scholar]; Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv. Mater. 2006;18:1345. [Google Scholar]
- 7.Ishii D, Ying TH, Mahara A, Murakami S, Yamaoka T, Lee W.-k., Iwata T. Biomacromolecules. 2009;10:237. doi: 10.1021/bm8009363. [DOI] [PubMed] [Google Scholar]
- 8.Zustiak SP, Leach JB. Biotechnol. Bioeng. 2011;108:197. doi: 10.1002/bit.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard CD, O'Shea TM, Siegwart DJ, Calo E, Anderson DG, Reynolds FM, Thomas JA, Slotkin JR, Woodard EJ, Langer R. Biomaterials. 2011;32:587. doi: 10.1016/j.biomaterials.2010.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin AD, Kiick KL. Polym. Chem. 2013;4:133. doi: 10.1039/C2PY20576A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]; Chung BG, Lee K-H, Khademhosseini A, Lee S-H. Lab Chip. 2012;12:45. doi: 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- 12.Merkel TJ, Herlihy KP, Nunes J, Orgel RM, Rolland JP, DeSimone JM. Langmuir. 2009;26:13086. doi: 10.1021/la903890h. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. Journal of the American Chemical Society. 2005;127:10096. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]; Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 13.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv. Mater. 2009;21:5005. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCall JD, Anseth KS. Biomacromolecules. 2012;13:2410. doi: 10.1021/bm300671s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aimetti AA, Machen AJ, Anseth KS. Biomaterials. 2009;30:6048. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Mariner PD, Nahreini JN, Anseth KS, Control J. Release. 2012;162:612. doi: 10.1016/j.jconrel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih H, Lin C-C. Biomacromolecules. 2012;13:2003. doi: 10.1021/bm300752j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rydholm AE, Reddy SK, Anseth KS, Bowman CN. Biomacromolecules. 2006;7:2827. doi: 10.1021/bm0603793. [DOI] [PubMed] [Google Scholar]
- 19.Salinas CN, Anseth KS. Macromolecules. 2008;41:6019. [Google Scholar]
- 20.Valdebenito A, Espinoza P, Lissi EA, Encinas MV. Polymer. 2010;51:2503. [Google Scholar]
- 21.Peppas NA, Bures P, Leobandung W, Ichikawa H. Eur. J. Pharm. Biopharm. 2000;50:27. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]