Abstract
Considering the diverse clinical presentation and likely polygenic etiology of schizophrenia, this investigation examined the effect of polygenic risk on a well-established intermediate phenotype for schizophrenia. We hypothesized that a measure of cumulative genetic risk based on additive effects of many genetic susceptibility loci for schizophrenia would predict prefrontal cortical inefficiency during working memory, a brain-based biomarker for the disorder. The present study combined imaging, genetic and behavioral data obtained by the Mind Clinical Imaging Consortium study of schizophrenia (n = 255). For each participant, we derived a polygenic risk score (PGRS), which was based on over 600 nominally significant single nucleotide polymorphisms, associated with schizophrenia in a separate discovery sample comprising 3322 schizophrenia patients and 3587 control participants. Increased polygenic risk for schizophrenia was associated with neural inefficiency in the left dorsolateral prefrontal cortex after covarying for the effects of acquisition site, diagnosis, and population stratification. We also provide additional supporting evidence for our original findings using scores based on results from the Psychiatric Genomics Consortium study. Gene ontology analysis of the PGRS highlighted genetic loci involved in brain development and several other processes possibly contributing to disease etiology. Our study permits new insights into the additive effect of hundreds of genetic susceptibility loci on a brain-based intermediate phenotype for schizophrenia. The combined impact of many common genetic variants of small effect are likely to better reveal etiologic mechanisms of the disorder than the study of single common genetic variants.
Key words: schizophrenia, DLPFC, working memory, intermediate phenotype, fMRI, genetic risk score
Introduction
Attempts to identify the underlying genetics of schizophrenia have produced inconsistent results. More often than not either the effects of specific genetic variants on the clinical diagnosis fail to replicate across studies1–3 or have small effects that explain only a minor fraction of the occurrence of the disorder.4
The reasons for limited progress are 2-fold. Firstly, most genetic studies of mental disorders to date are aimed at identifying single or few risk genes for which the selection of genes may be based on ill-conceived pathophysiological models of schizophrenia. Also, a focus on a small number of genes fails to consider the polygenic nature of complex disorders such as schizophrenia.5 Genetic risk for schizophrenia appears to derive from hundreds, if not thousands, of genetic variants with small effects.6–8
Secondly, investigating categorical entities, such as diagnosis, ignores the spectrum of illness, and that psychotic symptoms can be measured at subclinical levels in prodromal patients and in the general population.9 A continuum of abnormality is present for schizophrenia-related traits in healthy controls,10 unaffected relatives of schizophrenia patients,11,12 and across diagnostic boundaries.7,13,14
Researchers have started to focus on genetic factors that may be expressed in continuously distributed traits such as neuropsychological indices15–17 or brain-based intermediate phenotypes.18–22 Studying intermediate phenotypes, which are thought to be closer to the underlying substrate of disease pathophysiology than behavioral measures or disease status, could facilitate the search for susceptibility genes.5 Indeed, 2 meta-analyses indicated that schizophrenia risk variants showed larger effects with brain structure and function indices than cognitive measures.23,24
In the current study, we investigated dorsolateral prefrontal cortex (DLPFC) dysfunction during working memory (WkM) processing, which is a widely acknowledged intermediate phenotype for schizophrenia. Compared with matched healthy controls, patients are characterized by prefrontal neural inefficiency, ie, they need to recruit more neural resources than controls for the same level of task difficulty and may show decreased neural activity (hypofrontality) when task difficulty becomes too great.25–28
Genome-wide association (GWA) studies with large sample sizes have allowed discovery of new risk genes for schizophrenia. Recently, the GWA study approach has been combined with intermediate phenotypes in schizophrenia.29–31 The use of a polygenic risk score (PGRS) to identify genetic associations with intermediate phenotypes represents a similarly promising strategy. A PGRS is based on the additive effects of hundreds or thousands of disease-related gene variants that together may help capture polygenic aspects of the disorder6–8 and is minimally compromised by multiple testing, which often limits GWA studies.
In the present study, we combined PGRS and intermediate phenotype approaches to avoid the limitations of diffuse clinical phenotypes and instead directly characterize neural manifestations of polygenic risk for the disorder. We formed a PGRS based on the results of a large schizophrenia GWA study.7 Next we tested for associations between the PGRS and whole-brain neural activity during a WkM task in a large and independent sample of schizophrenia patients and healthy controls. We hypothesized that polygenic risk would predict DLPFC inefficiency during a WkM task.
Methods
Participants
Imaging, genetic and behavioral data from 255 participants of the Mind Clinical Imaging Consortium (MCIC) study of schizophrenia from 4 participating sites (the University of New Mexico [UNM], the University of Minnesota [UMN], Massachusetts General Hospital [MGH], and the University of Iowa [UI]) were used to determine genetic polymorphisms in cryo-conserved blood samples and to analyze whole-brain neural activity during a WkM task. All subjects gave written informed consent prior to study enrolment. The human subjects research committees at each of the 4 sites approved the study protocol. Out of a total of 248 participants, who passed genetic quality control procedures (see below), imaging data of 241 participants were available for genetic analysis, resulting in a final dataset of 92 schizophrenia patients and 114 healthy controls after imaging quality control steps (see below). Patients had a Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) diagnosis of schizophrenia (n = 88), schizophreniform disorder (n = 3), or schizoaffective disorder (n = 1), established using a Structured Clinical Interview for DSM disorders (SCID)32 and a review of case files by trained clinicians. In the initial cohort, controls were matched to the patient group for age, gender, and parental education and were excluded if they had a history of a medical or Axis I psychiatric diagnosis. The majority of participants were of Caucasian descent (102 healthy controls and 73 patients). For additional details about the participants and clinical measures, see Ehrlich et al19. For the replication analyses, we used 2 additional datasets from the International Schizophrenia Consortium (ISC) and from the Psychiatric Genomics Consortium (see below).
Case-Control Dataset From the ISC
The ISC served as an independent discovery sample. It consists of 3322 schizophrenia patients and 3587 controls. In this study, we used ISC results based on 739 995 single nucleotide polymorphisms (SNPs) from the Affymetrix Genome-Wide Human SNP 5.0 and 6.0 arrays, which had been tested for association using a case-control design and Cochran-Mantel-Haenszel statistic. Based on 7 different statistical thresholds (P < .01, P < .05, P < .1, P < .2, P < .3, P < .4, and P < .5) in the Cochran-Mantel-Haenszel analyses controlling for site, nominally associated alleles were selected as “score alleles” for the calculation of 7 PGRSs (see below) in the MCIC sample (target sample). For more information on the ISC, see online supplementary http://schizophreniabulletin.oxfordjournals.org/lookup/suppl/doi:10.1093/schbul/sbt174/-/DC1 SM 1.1 or Purcell et al7 and Stone et al33.
Case-Control Dataset From the Psychiatric Genomics Consortium
For the purpose of replication, we used results from another discovery sample, the Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC). Their stage 1 discovery sample consisted of 15 429 subjects (6458 cases and 8971 controls). Over 1.2 million SNPs were tested for disease association using a logistic regression model controlling for the effects of site and population stratification. For more information on the PGC, see Ripke et al8.
Behavioral Task
The Sternberg Item Recognition Paradigm (SIRP) is a WkM task, previously shown to consistently activate the DLPFC in healthy controls and schizophrenia patients.26 The SIRP was administered during six 46-second blocks per run for three 360-second runs. In each block, a memory set, composed of 1 (load 1), 3 (load 3), or 5 (load 5) digits, was presented (2 blocks per load condition). The Encode phase was followed by a presentation of 14 individual digits presented consecutively (the Probe phase), and participants responded to each probe to indicate whether or not the probe digit was in the memory set. For additional details about the paradigm, see online supplementary SM 1.2. The stimuli and responses were presented and collected using E-prime software (E-Prime v1.1; Psychology Software Tools, Inc.). Participants were excluded from further analysis if they completed a block with less than a 75% accuracy rate and/or had more than 6 probes not answered within a block.
Image Acquisition and Preprocessing
Structural magnetic resonance imaging (MRI) data were acquired with either a 1.5T Siemens Sonata (UNM, MGH, and UI) or a 3T Siemens Trio (UMN). Functional MRI (fMRI) data were acquired with either a 1.5T Siemens Sonata (UNM) or a 3T Siemens Trio (UMN, MGH, and UI). Structural data, needed for image registration and DLPFC label generation, were processed with the automated atlas-based FreeSurfer reconstruction software (http://surfer.nmr.mgh.harvard.edu). Functional data were registered to the corresponding structural images using FreeSurfer and analyzed using fMRIB Software Library (FSL) (http://www.fmrib.ox.ac.uk/fsl). We fit a general linear model to the fMRI time course at each voxel in a whole-brain model to estimate the average activation during the 3 loads of the probe condition in all trials. Equal weight was given to all loads. For additional details regarding data acquisition, (pre)processing, and quality assurance, please see online supplementary SM 1.3 and Walton et al34.
Genotyping
Blood samples were obtained from 255 participants and sent to the Harvard Partners Center for Genetics and Genomics for DNA extraction. All DNA extraction and genotyping was done blind to group assignment. Genotyping was performed at the Mind Research Network Neurogenetics Core Lab using the Illumina Human Omni-Quad BeadChip. Quality control steps included the following steps. SNPs on the X or Y chromosome, or those with a genotyping rate of less than 90% or a minor allele frequency of less than 5% were excluded from the analysis. We also removed 7 participants with extreme heterozygosity values (±3 SD) or with a genotyping rate of less than 90%. Using this data set consisting of 749 968 SNPs, additional SNPs were imputed based on the Hapmap3 dataset. Imputation was done using IMPUTE2 with a probability threshold of .95. The imputed data set was then again filtered for a minor allele frequency of 5% and a Hardy-Weinberg equilibrium in controls with a threshold of 10–6. The final data set consisted of 1 073 955 SNPs, and the genotyping rate in remaining individuals was 0.99. Quality control steps were carried out with PLINK, 1.07.35
PGRS Calculation
Using 7 different statistical thresholds (P < .01, P < .05, P < .1, P < .2, P < .3, P < .4, and P < .5),7 we selected all nominally significant SNPs from the discovery sample (ISC study), which were also present in the imputed MCIC dataset, and derived 7 PGRSISC for each MCIC study participant. If a genotype in the score was missing for a particular individual, then the expected value was imputed based on the sample allele frequency. The score was calculated as the sum across SNPs of the number of reference alleles (0, 1, or 2) at that SNP multiplied by the logarithm of the odds ratio (OR) for that SNP. ORs were taken from case-control analysis in the discovery samples as described above.
Statistical Models
We performed whole-brain analyses investigating the relationship between PGRSISC at all 7 statistical thresholds and WkM-induced brain activity for patients and controls using mixed effects models in FSL. For more details, see online supplementary SM 1.3. All models were cluster corrected according to FSL default settings with a z value of 2.3 and Bonferroni corrected with a P value of .007 (.05/7) and controlled for acquisition site and the number of nonmissing genotypes of all SNPs used to calculate the PGRS to control for potential differences in genotyping rate between cases.7 To account for nonrandom sampling of schizophrenia patients, we explicitly modeled the effects of diagnosis in our main model and tested for diagnosis by PGRS interaction effects. To control for population stratification (see below), we included the first 4 principal components (PCs) as covariates.
Since only a risk score at the strictest significance level (PGRSISC(P < .01)) was significantly related to neural activity (see “Results” section), in subsequent models, we (a) used a pruned PGRSISC(P < .01), which included only SNPs that did not show an association with the population structure in our sample (see below), and (b) tested further PGRS(P < .01) variants (PGRSISC(P < .01)-PGC and PGRSPGC(P < .01)) based on a second discovery sample for the purpose of replication (see also below). These models were controlled for the same covariates as in the main models and cluster corrected with a P value of .05. We extracted indices of activation for the DLPFC in percent signal change (%Δ) at the most activated DLPFC location. We then regressed out all relevant covariates and estimated the percent of variance explained by PGRSISC(P < .01). Sample characteristic analyses were carried out with SPSS 17.0.
Replication Analysis
To replicate our original findings, we calculated new scores based on results from the PGC study (see above and Ripke et al8) by applying 2 different strategies:
(1) Following the same procedure as with the ISC discovery sample, we selected nominally associated alleles from the original ISC sample (based on a P value of less than .01) but used PGC ORs and estimated a second risk score version, referred to as PGRSISC(P < .01)-PGC.
(2) We selected nominally associated PGC alleles and their corresponding PGC ORs (based on a P value of less than .01) as “score alleles” for the calculation of a risk score in the MCIC sample (target sample), referred to as PGRSPGC(P < .01).
Population Stratification
To avoid confounding effects due to population stratification, we followed a similar procedure as described in Purcell et al7. First, we applied PC analysis to our genotype data using EIGENSTRAT of the EIGENSOFT 3.0 software package,36,37 extracted 10 PCs, and then included the first 4 components as covariates in our imaging models.7 As an additional measure to control possible effects due to population substructure, we pruned the PGRSISC(P < .01) to include only SNPs that did not show an association with the first 2 PCs and reran our analysis. For additional details, see online supplementary SM 1.4.
Functional Annotation Clustering
To explore the underlying biological processes associated with PGRSISC(P < .01) genes, we used DAVID Bioinformatic Database version 6.7.38 We mapped all 608 SNPs to their corresponding genes using the batch query function in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/batchquery.html) and successfully identified 240 known genes. Seven genes could not be found in the DAVID database. The remaining 233 genes were clustered based on their functional annotations using all 3 gene ontology categories (biological processes, cellular components, and molecular functions). We kept DAVID default settings but applied a high threshold to minimize overlapping categories, as described in Huang et al38.
Results
Sample Characteristics
Demographic variables such as age and handedness did not differ between patients and controls (table 1). There were significantly more female participants in the control group, and patients had a significantly lower WRAT-IIIRT score and lower parental education than controls. There was no effect of acquisition site on gender, WRAT-IIIRT score, and handedness, but sites differed in their participant’s age and parental education (table 1).
Table 1.
Basic Demographics According to Acquisition Site
| Site | Sample | Gender (Female) |
Age (y) |
Cognitive Function (WRAT-IIIRT) |
Parental Education | Handedness | PGRSISC(P < .01) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [0–12] | [10−4] | [10−3] | ||||||||||||
| n | n | % | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| UI | HC | 52 | 25 | 48.1 | 30.24a | 10.46 | 50.08 | 4.07 | 14.67a | 2.64 | 0.69 | 2.57 | 8.14 | 1.40 |
| SCZ | 22 | 3 | 13.6 | 31.81a | 8.91 | 48.38 | 5.04 | 15.55a | 3.43 | 0.82 | 2.81 | 3.64 | 1.53 | |
| MGH | HC | 23 | 10 | 43.5 | 40.04a | 9.59 | 51.96 | 3.98 | 14.70a | 3.27 | 1.04 | 2.93 | 4.45 | 1.82 |
| SCZ | 25 | 7 | 28.0 | 37.92a | 9.81 | 45.09 | 8.49 | 11.04a | 6.52 | 0.61 | 1.92 | 9.31 | 1.51 | |
| UMN | HC | 17 | 7 | 41.2 | 31.12a | 11.30 | 50.94 | 4.09 | 16.00a | 2.55 | 0.47 | 0.80 | 4.02 | 1.75 |
| SCZ | 27 | 8 | 29.6 | 31.63a | 10.63 | 46.22 | 5.43 | 15.22a | 2.83 | 1.78 | 3.59 | 5.51 | 1.50 | |
| UNM | HC | 22 | 4 | 18.2 | 30.81 | 12.90 | 51.50 | 3.79 | 16.09 | 5.19 | 1.05 | 2.42 | 3.17 | 1.74 |
| SCZ | 18 | 5 | 27.8 | 35.83 | 14.09 | 45.53 | 7.05 | 12.50 | 5.63 | 1.39 | 3.13 | 8.52 | 1.24 | |
| Total | HC | 114 | 46b | 40.4 | 32.49 | 11.44 | 50.86b | 4.02 | 15.15b | 3.40 | 0.80 | 2.43 | 5.82 | 1.60 |
| SCZ | 92 | 23b | 25.0 | 34.23 | 11.01 | 46.31b | 6.60 | 13.63b | 5.09 | 1.17 | 2.94 | 6.68 | 1.46 | |
Note: WRAT-IIIRT, reading subtest of the Wide Range Achievement Test-III; handedness, Annett Handedness Scale; MGH, Massachusetts General Hospital; UI, University of Iowa; UMN, University of Minnesota; UNM, University of New Mexico; SCZ, patients with schizophrenia; HC, healthy controls; PGRS, polygenic risk score; ISC, International Schizophrenia Consortium. A series of ANOVA and logistic regression analyses were performed to detect significant differences of gender, age, WRAT-IIIRT score, parental education, handedness and PGRSISC( P < .01) between acquisition sites and diagnostic groups. PGRSs according to the other 7 statistical thresholds are not displayed, since they were not related to neural activity, see “Results” section.
aSignificantly different between acquisition sites on the basis of a linear regression (P < .05) with subsequent Bonferroni post hoc tests (P < .05).
bSignificantly different between SCZ and HC on the basis of a linear or logistic regression (P < .05).
WkM-Related Neural Activity
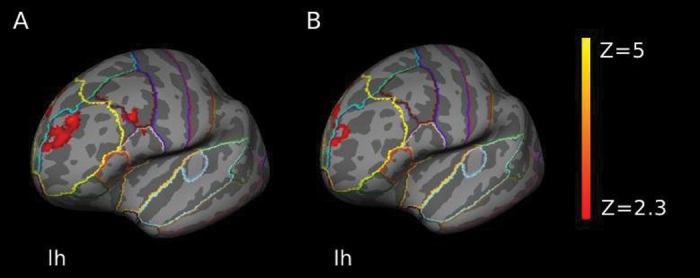
The SIRP task reliably activated WkM-associated brain regions including the DLPFC, striatal, and parietal regions as described previously.34,39 A positive association between PGRSISC(P < .01) and neural activity was evident in an area including the left DLPFC and left ventrolateral prefrontal cortex (VLPFC) (z-max [−6 38 48] = 3.75; P = 6.91 × 10−6, cluster corrected; figure 1A and online supplementary figure 2) in a model covarying for the effects of acquisition site, diagnosis, population stratification, and number of nonmissing genotypes per individual. No effect was found for any of the other PGRSISC thresholds in a whole-brain model including the same covariates. PGRSISC(P < .01) accounted for 4.3% of the total variance (adjusted R 2; R 2 change(1, 204) = 0.048, P = .002) at the most activated DLPFC location (x, y, z: −6, 38, 48), after regressing out all other covariates. Furthermore, this effect was also independent of gender or parental education and remained stable after excluding 4 patients with a diagnosis of schizoaffective disorder or schizophreniform disorder (see online supplementary SM 2.1). There was no significant diagnosis by PGRSISC(P < .01) interaction effect.
Fig. 1.
(A) Functional map illustrating increased neural activity with increasing polygenic risk for schizophrenia (PGRSISC( P < .01)) in the left DLPFC. Results were additionally controlled for acquisition site, diagnosis, population structure, and the number of nonmissing genotypes per individual. (B) Results for a PGRSISC( P < .01), which was pruned for the possibly confounding effect of population stratification. This model was additionally controlled for acquisition site, diagnosis, and the number of nonmissing genotypes per individual. Both models were cluster corrected. The z values are represented according to the color code. PGRS, polygenic risk score; ISC, International Schizophrenia Consortium; DLPFC, dorsolateral prefrontal cortex; lh, left hemisphere.
Characteristics of the PGRSISC(P < .01)
PGRSISC(P < .01) did not differ by sex, diagnostic group, or acquisition site (table 1) and did not correlate with age, SIRP performance, WRAT-IIIRT score, handedness, or parental education (online supplementary t able 1). Furthermore, there were no significant correlations between PGRSISC(P < .01) and cumulative or current antipsychotic drug dose as well as positive or negative symptoms in the patient group (online supplementary table 1). For linkage disequilibrium patterns between PGRSISC(P < .01) SNPs, please see online supplementary SM 2.3.
Additional Analyses
In a subsequent whole-brain model including a pruned PGRSISC(P < .01), which was derived solely from SNPs unaffected by population stratification (see online supplementary SM 1.4) and with the same covariates as in the main model, the effect of polygenic risk on left DLPFC activity remained significant (z-max [−12 48 30] = 3.55; P = .00584, cluster corrected; figure 1B). There was a second significant cluster in the left frontal medial cortex including the anterior cingulate gyrus with the effect of PGRS pointing in the same direction as in the DLPFC cluster (z-max [−4 46 -18] = 4.01; P = .00972, cluster corrected). As in the first model, there was no significant diagnosis by PGRS interaction effect.
We conducted additional analyses to address the question whether the observed PGRSISC(P < .01) effect may indeed be caused by a small number of SNPs with large effect sizes. If that would be the case, the weighted PGRS for these few SNPs should explain most of the variance of the original PGRSISC(P < .01). We calculated risk scores based on 5, 10, 50, 100, and 500 SNPs of the same dataset, each time iterating through 10 000 random combinations of SNPs. We then estimated the amount of variance of the total PGRSISC(P < .01), which was explained by these PGRS subscores. Scores based on 5 SNPs could only explain 0%–12% of total PGRSISC(P < .01) variance, followed by 0%–13%, 0%–24%, 3%–36%, and 70%–88% for 10, 50, 100, and 500 SNPs (online supplementary figure 4). The low, but steadily increasing amount of explained variance supports the claim that the observed effect is due to the additive impact of a large number of SNPs.
Replication Based on a Second Discovery Sample (PGC)
We sought replication of our original findings using scores based on results from the PGC study (see “Methods” section and Ripke et al8). PGRSISC(P < .01)-PGC correlated significantly with WkM-elicited neural activity in an area including the right DLPFC and the right caudate (z-max [16 12 48] = 3.72; P = .0448, cluster corrected; online supplementary figure 5A). Results were cluster corrected and controlled for the same covariates as in the original model. We also found a positive association between PGRSPGC(P < .01) and neural activity in the right and left DLPFC and in the anterior cingulate cortex (uncorrected results; online supplementary figure 5B) controlling for the same covariates.
Functional Annotations of PGRS Genes
In order to understand which genes the PGRSISC(P < .01) is composed of and what their underlying functional annotations are, we used the functional annotation cluster tool in DAVID to explore the associated gene ontology. The top 5 clusters associated with all mapped genes were related to axonogenesis and neuron projection development, ion binding, cell motility and migration, channel activity, and guanosine triphosphatase regulator activity (table 2). For a full list of gene names, see online supplementary table 2.
Table 2.
Gene Ontology Analysis
| Cluster | Functional Annotation | Enrichment Score | Number of Associated Genes |
|---|---|---|---|
| 1 | Axonogenesis and neuron projection development | 4.58 | 21 |
| 2 | Ion binding | 2.98 | 73 |
| 3 | Cell motility and migration | 2.78 | 12 |
| 4 | Channel activity | 2.32 | 13 |
| 5 | GTPase regulator activity | 2.23 | 13 |
Note: GTP, guanosine triphosphatase; The top 5 clusters associated with PGRSISC( P < .01) genes are listed. Clustering of genes was based on all 3 gene ontology categories (biological processes, cellular components, and molecular functions) using the functional annotation cluster tool in DAVID.38
Discussion
The results of our study suggest an association between increased polygenic risk for schizophrenia and WkM-related neural inefficiency in the left DLPFC. This effect was not attributable to population stratification and was supported by results from additional analyses based on another large GWA study. Gene ontology analysis of the PGRS highlighted loci involved in brain development and synaptic transmission, processes which have been implicated in the etiology of schizophrenia.
Results lend support to an approach that takes into account polygenic etiology in schizophrenia. Polygenic approaches have been used to understand genetic contributions to psychopathology such as bipolar disorder,14,40,41 as well as neurodegenerative42 and neurodevelopmental disorders.43 For schizophrenia, a polygenic risk model has been supported by 3 large GWA studies, all of which showed that polygenic risk load differed between large samples of patients and controls.6–8 Investigating quantitative markers, one study confirmed the effect of a PGRS on continuously distributed clinical measures of psychosis in a sample of schizophrenia patients and healthy controls,44 and we have previously analyzed the relationship between neural activity during a WkM task and a risk score, which combined the additive effects of 41 candidate SNPs for schizophrenia.34 The PGRS used in the current study includes a much larger number of SNPs, which were at least nominally significant in a recent GWA study, and is thus based on hundreds of susceptibility loci for schizophrenia. The fact that we found a significant brain-based effect only at the strictest ISC threshold of P <.01 supports the idea that DLPFC dysfunction represents a more circumscribed phenotype than clinical phenotypes or diagnostic categories. Investigating an intermediate phenotype bears the additional advantage of addressing the problem of symptom heterogeneity within a given psychiatric diagnosis and the occurrence of (attenuated) risk markers in healthy controls.10 Since PGRS was associated with brain function, but not with task performance or with diagnosis, we assume that our cumulative genetic risk measure represents unique genetic aspects of dysfunctional neuronal responses related to schizophrenia that are not easy to capture at the level of behavior or symptoms. Thus, we were not only able to use a well-defined continuous measure to describe brain-based deficits in patients, but characterize subtle abnormalities in healthy controls as well. The fact that (a) the genes in this analysis have been shown to be nominally associated with schizophrenia in a large GWA study,7 (b) their cumulative impact correlated with a well-replicated intermediate phenotype for schizophrenia, and (c) we found supporting evidence for our main findings based on results from another large schizophrenia GWA study8 indicates a robust relationship between the proposed PGRS and schizophrenia.
Functional annotation clustering of PGRS genes revealed major biological pathways associated with the investigated risk genes. Impaired axonogenesis and neuron projection development as well as aberrant cell motility and migration point toward aberrant neurodevelopmental processes. These processes have been repeatedly linked to schizophrenia. Subtle alterations in early brain development may ultimately lead to a variety of psychiatric symptoms as well as cognitive deficits, eg, reduced WkM.45–48 For instance, a study by Gay et al49 found that especially patients with increased neurological soft signs (ie, observable defects in motor coordination, motor integration, and sensory integration) display reduced sulcation, an early indicator of later abnormal functional development,50 in the left DLPFC. Consistent with the results from our gene ontology analysis, numerous studies reported an altered cortical architecture of the DLPFC in schizophrenia patients, including altered dendritic spine density in DLPFC layer 3 pyramidal cells51 and reduced size and density of other large neurons in the same layer.52 In line with that the expression of genes involved in synaptic transmission processes, myelin sheaths formation, and neurotrophic signaling (affecting the development and survival of axons) seems to be altered in the DLPFC of schizophrenia patients,51–55 which could also translate into altered WkM processing.56–60
The findings of our study have to be considered in the light of the following limitations. First, our target sample was only of moderate size to investigate the effect of risk variants on brain function. However, the 2 GWA study samples, which were the basis for the risk gene discovery, were well powered, and additional analyses provided supporting evidence for our results. Second, we cannot distinguish between the potential effects of antipsychotic treatment vs those of the underlying disease process on brain function. However, we did not find a correlation between PGRS and measures of antipsychotic medication, and brain dysfunction has been shown to occur in neuroleptic-naive patients61 as well as in high-risk individuals,27,62 suggesting that the reported association is likely to be medication independent. Third, our approach of deriving a risk score (as implemented in PLINK)35 is one among many possibilities. Previous studies have explored a range of different risk scores including unweighted risk scores.63,64 Although a valid approach, in our study, we were not able to detect an effect in a submodel investigating the effect of an unweighted PGRS (post hoc analysis, data not shown), suggesting that additional fine-tuning of risk allele effects may be important. Fourth, we did not investigate how and to what degree rare de novo variants, gene-gene interactions, or environmental risk factors aggravate the observed effect, independent of disease status. General effects of common risk variants on brain-based phenotypes are a well-replicated finding,15,19,65 and it has long been assumed that rare variants, gene-gene interactions, and environmental risk factors may influence disease manifestation. To disentangle these complicated relationships, future studies should investigate gene-environment interactions.
We combined the effects of several hundred genetic risk variants for schizophrenia into a single risk score, and we were able to show that this score predicted DLPFC inefficiency during a WkM task, a common intermediate phenotype for schizophrenia. The finding supports a growing number of reports, which demonstrate a polygenic etiology of schizophrenia and related phenotypes. Identifying neural correlates of cumulative genetic risk could help to understand dysfunctions of underlying brain-based networks and define system neuroscience models of schizophrenia.
Supplementary Material
Supplementary material is available at http://schizoph reniabulletin.oxfordjournals.org.
Funding
This work was supported by the National Institutes of Health (NIH/NCRR P41RR14075 and R01EB005846 to V.D.C.), Department of Energy (DE-FG02-99ER62764), Mind Research Network, Morphometry Bioinformatics Research Network (1U24, RR021382A), Function BIRN (U24RR021992-01, NIH.NCRR MO1 RR025758-01, NIMH 1RC1MH089257 to V.D.C.), the Deutsche Forschungsgemeinschaft (research fellowship to S.E.), and the National Alliance for Research on Schizophrenia and Depression Young Investigator Award (to S.E.).
Supplementary Material
Acknowledgments
V.R. has received lecture fees from Eli Lilly, Janssen-Cilag, Medice, and Novartis and was a member of advisory boards of Eli Lilly and Novartis. All other authors declare no biomedical financial interests or other potential conflict of interests.
References
- 1. Betcheva ET, Mushiroda T, Takahashi A, et al. Case-control association study of 59 candidate genes reveals the DRD2 SNP rs6277 (C957T) as the only susceptibility factor for schizophrenia in the Bulgarian population. J Hum Genet. 2009;54:98–107 [DOI] [PubMed] [Google Scholar]
- 2. Munafò MR, Bowes L, Clark TG, Flint J. Lack of association of the COMT (Val158/108 Met) gene and schizophrenia: a meta-analysis of case-control studies. Mol Psychiatry. 2005;10:765–770 [DOI] [PubMed] [Google Scholar]
- 3. Kishi T, Fukuo Y, Okochi T, et al. Serotonin 6 receptor gene and schizophrenia: case-control study and meta-analysis. Hum Psychopharmacol. 2012;27:63–69 [DOI] [PubMed] [Google Scholar]
- 4. O’Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12 [DOI] [PubMed] [Google Scholar]
- 5. Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci U S A. 1967;58:199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ikeda M, Aleksic B, Kinoshita Y, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478 [DOI] [PubMed] [Google Scholar]
- 7. Purcell SM, Wray NR, Stone JL, et al. ; International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ripke S, Sanders AR, Kendler KS, et al. et al. ; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Linscott RJ, van Os J. Systematic reviews of categorical versus continuum models in psychosis: evidence for discontinuous subpopulations underlying a psychometric continuum. Implications for DSM-V, DSM-VI, and DSM-VII. Annu Rev Clin Psychol. 2010;6:391–419 [DOI] [PubMed] [Google Scholar]
- 10. Tan HY, Callicott JH, Weinberger DR. Intermediate phenotypes in schizophrenia genetics redux: is it a no brainer? Mol Psychiatry. 2008;13:233–238 [DOI] [PubMed] [Google Scholar]
- 11. Docherty AR, Sponheim SR. Anhedonia as a phenotype for the Val158Met COMT polymorphism in relatives of patients with schizophrenia. J Abnorm Psychol. 2008;117:788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silberschmidt AL, Sponheim SR. Personality in relation to genetic liability for schizophrenia and bipolar disorder: differential associations with the COMT Val 108/158 Met polymorphism. Schizophr Res. 2008;100:316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamshere ML, O’Donovan MC, Jones IR, et al. Polygenic dissection of the bipolar phenotype. Br J Psychiatry. 2011;198:284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Need AC, Attix DK, McEvoy JM, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zilles D, Meyer J, Schneider-Axmann T, et al. Genetic polymorphisms of 5-HTT and DAT but not COMT differentially affect verbal and visuospatial working memory functioning. Eur Arch Psychiatry Clin Neurosci. 2012;262:667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donohoe G, Rose E, Frodl T, et al. ZNF804A risk allele is associated with relatively intact gray matter volume in patients with schizophrenia. Neuroimage. 2011;54:2132–2137 [DOI] [PubMed] [Google Scholar]
- 19. Ehrlich S, Morrow EM, Roffman JL, et al. The COMT Val108/158Met polymorphism and medial temporal lobe volumetry in patients with schizophrenia and healthy adults. Neuroimage. 2010;53:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rasetti R, Weinberger DR. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev. 2011;21:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827 [DOI] [PubMed] [Google Scholar]
- 22. Voineskos AN, Lett TA, Lerch JP, et al. Neurexin-1 and frontal lobe white matter: an overlapping intermediate phenotype for schizophrenia and autism spectrum disorders. PLoS One. 2011;6:e20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mier D, Kirsch P, Meyer-Lindenberg A. Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol Psychiatry. 2010;15:918–927 [DOI] [PubMed] [Google Scholar]
- 24. Rose EJ, Donohoe G. Brain vs behavior: an effect size comparison of neuroimaging and cognitive studies of genetic risk for schizophrenia. Schizophr Bull. 2013;39:518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003;160:2209–2215 [DOI] [PubMed] [Google Scholar]
- 26. Manoach DS, Press DZ, Thangaraj V, et al. Schizophrenic subjects activate dorsolateral prefrontal cortex during a working memory task, as measured by fMRI. Biol Psychiatry. 1999;45:1128–1137 [DOI] [PubMed] [Google Scholar]
- 27. Karlsgodt KH, Glahn DC, van Erp TG, et al. The relationship between performance and fMRI signal during working memory in patients with schizophrenia, unaffected co-twins, and control subjects. Schizophr Res. 2007;89:191–197 [DOI] [PubMed] [Google Scholar]
- 28. Schlösser RG, Koch K, Wagner G, et al. Inefficient executive cognitive control in schizophrenia is preceded by altered functional activation during information encoding: an fMRI study. Neuropsychologia. 2008;46:336–347 [DOI] [PubMed] [Google Scholar]
- 29. Bakken TE, Bloss CS, Roddey JC, et al. Association of genetic variants on 15q12 with cortical thickness and cognition in schizophrenia. Arch Gen Psychiatry. 2011;68:781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stein JL, Medland SE, Vasquez AA, et al. Alzheimer’s Disease Neuroimaging Initiative; EPIGEN Consortium; IMAGEN Consortium; Saguenay Youth Study Group; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium; Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potkin SG, Turner JA, Guffanti G, et al. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn Neuropsychiatry. 2009;14:391–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. First M, Spitzer A, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Nonpatient Edition. New York: New York State Psychiatric Institute; 2002 [Google Scholar]
- 33. Stone JL, O’Donovan MC, Gurling H, et al. ; International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walton E, Turner J, Gollub RL, et al. Cumulative genetic risk and prefrontal activity in patients with schizophrenia. Schizophr Bull. 2013;39:703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 38. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 39. Ehrlich S, Yendiki A, Greve DN, et al. Striatal function in relation to negative symptoms in schizophrenia. Psychol Med. 2012;42:267–282 [DOI] [PubMed] [Google Scholar]
- 40. Middeldorp CM, de Moor MH, McGrath LM, et al. The genetic association between personality and major depression or bipolar disorder. A polygenic score analysis using genome-wide association data. Transl Psychiatry. 2011;1:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whalley HC, Papmeyer M, Sprooten E, et al. The influence of polygenic risk for bipolar disorder on neural activation assessed using fMRI. Transl Psychiatry. 2012;2:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA; Alzheimer’s Disease Neuroimaging Initiative. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jung JY, Kohane IS, Wall DP. Identification of autoimmune gene signatures in autism. Transl Psychiatry. 2011;1:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Derks EM, Vorstman JA, Ripke S, Kahn RS, Ophoff RA; Schizophrenia Psychiatric Genomic Consortium. Investigation of the genetic association between quantitative measures of psychosis and schizophrenia: a polygenic risk score analysis. PLoS One. 2012;7:e37852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35:528–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193 [DOI] [PubMed] [Google Scholar]
- 47. Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449 [DOI] [PubMed] [Google Scholar]
- 48. van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645 [DOI] [PubMed] [Google Scholar]
- 49. Gay O, Plaze M, Oppenheim C, et al. Cortex morphology in first-episode psychosis patients with neurological soft signs. Schizophr Bull. 2013;39:820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dubois J, Benders M, Borradori-Tolsa C, et al. Primary cortical folding in the human newborn: an early marker of later functional development. Brain. 2008;131:2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73 [DOI] [PubMed] [Google Scholar]
- 52. Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224 [DOI] [PubMed] [Google Scholar]
- 53. Hakak Y, Walker JR, Li C, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kozlovsky N, Shanon-Weickert C, Tomaskovic-Crook E, Kleinman JE, Belmaker RH, Agam G. Reduced GSK-3beta mRNA levels in postmortem dorsolateral prefrontal cortex of schizophrenic patients. J Neural Transm. 2004;111:1583–1592 [DOI] [PubMed] [Google Scholar]
- 55. Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67 [DOI] [PubMed] [Google Scholar]
- 56. Eastwood SL. The synaptic pathology of schizophrenia: is aberrant neurodevelopment and plasticity to blame? In: Smythies J, ed. International Review of Neurobiology. Vol. 59 Amsterdam: Academic Press; 2004:47–72 [DOI] [PubMed] [Google Scholar]
- 57. McCullumsmith RE, Clinton SM, Meador-Woodruff JH. Schizophrenia as a disorder of neuroplasticity. In: Smythies J, ed. International Review of Neurobiology. Vol. 59 Amsterdam: Academic Press; 2004:19–45 [DOI] [PubMed] [Google Scholar]
- 58. Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486 [DOI] [PubMed] [Google Scholar]
- 59. Stephan KE, Baldeweg T, Friston KJ. Synaptic plasticity and dysconnection in schizophrenia. Biol Psychiatry. 2006;59:929–939 [DOI] [PubMed] [Google Scholar]
- 60. Yin D-M, Chen Y-J, Sathyamurthy A, Xiong W-C, Mei L. Synaptic dysfunction in schizophrenia. In: Kreutz MR, Sala C, eds. Synaptic Plasticity. Vol. 970 Vienna, Austria: Springer Vienna; 2012:493–516 [DOI] [PubMed] [Google Scholar]
- 61. van Veelen NM, Vink M, Ramsey NF, Kahn RS. Left dorsolateral prefrontal cortex dysfunction in medication-naive schizophrenia. Schizophr Res. 2010;123:22–29 [DOI] [PubMed] [Google Scholar]
- 62. Callicott JH, Egan MF, Mattay VS, et al. Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry. 2003;160:709–719 [DOI] [PubMed] [Google Scholar]
- 63. Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morrison AC, Bare LA, Chambless LE, et al. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35 [DOI] [PubMed] [Google Scholar]
- 65. Ho BC, Milev P, O’Leary DS, Librant A, Andreasen NC, Wassink TH. Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Arch Gen Psychiatry. 2006;63:731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.