Abstract
Background:
Twin and multiplex family studies have established significant heritability for schizophrenia (SZ), often summarized as 81%. The Consortium on the Genetics of Schizophrenia (COGS-1) family study was designed to deconstruct the genetic architecture of SZ using neurocognitive and neurophysiological endophenotypes, for which heritability estimates ranged from 18% to 50% (mean = 30%). This study assessed the heritability of SZ in these families to determine whether there is a “heritability gap” between the diagnosis and related endophenotypes.
Methods:
Nuclear families (N = 296) with a SZ proband, an unaffected sibling, and both parents (n = 1366 subjects; mean family size = 4.6) underwent comprehensive endophenotype and clinical characterization. The Family Interview for Genetic Studies was administered to all participants and used to obtain convergent psychiatric symptom information for additional first-degree relatives of interviewed subjects (N = 3304 subjects; mean family size = 11.2). Heritability estimates of psychotic disorders were computed for both nuclear and extended families.
Results:
The heritability of SZ was 31% and 44% for nuclear and extended families. The inclusion of bipolar disorder increased the heritability to 37% for the nuclear families. When major depression was added, heritability estimates dropped to 34% and 20% for nuclear and extended families, respectively.
Conclusions:
Endophenotypes and psychotic disorders exhibit comparable levels of heritability in the COGS-1 family sample. The ascertainment of families with discordant sibpairs to increase endophenotypic contrast may underestimate diagnostic heritability relative to other studies. However, population-based studies also report significantly lower heritability estimates for SZ. Collectively, these findings support the importance of endophenotype-based strategies and the dimensional view of psychosis.
Key words: schizophrenia, psychosis, endophenotypes, cognition, biomarkers, heritability
Introduction
Schizophrenia (SZ) is a severe and persistent psychotic disorder that is characterized by significant cognitive impairments and psychosocial disability.1 Previous meta-analyses of twin studies have indicated a substantial heritability of this disorder ranging from 44% to 87% with a mean of 81%.2 Although a broad range of heritability estimates have been obtained using a variety of sampling strategies (eg, twin concordance rates, parent-offspring correlations, sibling correlations, and multiplex families), it is the estimate of 81% that is perhaps most commonly reported in the literature.3,4 Yet, as some have suggested, twin studies may overestimate the heritability of SZ.5,6 Indeed, recent population-based studies have begun to challenge this figure and suggest that the actual heritability of SZ may be lower.7,8 Other studies of families ascertained through a SZ proband have even found “scant evidence” of heritability of this illness.9 Although the heterogeneous nature of SZ may be in part responsible for the discrepant estimates, variations in ascertainment strategy likely contribute to this wide range of heritability estimates for SZ.5,10,11
The Consortium on the Genetics of Schizophrenia (COGS) explores quantitative neurocognitive and neurophysiological endophenotypes related to SZ as a means for understanding the genetic determinants of SZ. The first phase of COGS (COGS-1) focused on the recruitment of families via SZ probands with at least 1 unaffected sibling, and both parents available for evaluation in order to maximize contrast between and within families. Heritability estimates for 12 SZ-related endophenotypes ranged from 18% to 50% in the COGS-1 families with a mean of 30%.12,13 In parallel, association, linkage, and copy number variation (CNV) studies have begun to elucidate the genomic substrates of these endophenotypes and of SZ itself in this cohort.12,14,15
Considering the often cited heritability estimate of SZ of 81%, there appears to be a significant “heritability gap” between illness and endophenotype heritability. This study, therefore, aimed to examine the heritability of SZ (alone and in combination with related serious psychiatric illnesses) in the COGS-1 families for a comparison with established heritability estimates of the endophenotypes in the same families. Given the COGS-1 ascertainment criteria, we hypothesized that SZ would be significantly heritable, albeit at lower levels than previously derived from twin- and multiplex family-based designs. We also hypothesized that expanding the diagnostic category in relatives to include bipolar disorder (BPD) would increase the illness heritability observed in accordance with the emerging dimensional view of psychotic disorders. In contrast, with the diagnostic group in relatives further expanded to include major depressive disorder (MDD), we anticipated a reduction in the observed heritability through a broadening of the diagnostic boundary beyond psychosis. Lastly, as study inclusion criteria required the enrollment of intact families, we hypothesized that COGS-1 patients would exhibit significantly lower levels of clinical symptoms and have a higher functional status than a cohort of patients enrolled at the same COGS laboratories as part of subsequent ongoing COGS-2 case-control study that does not require family member participation.
Methods
Subjects
COGS-1 families were ascertained at 7 sites through the identification of probands who met the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) criteria for SZ via the administration of the Diagnostic Interview for Genetic Studies (DIGS).16 Each family consisted of a proband with SZ, at least 1 unaffected sibling, and both parents, with blood samples for DNA extraction required for all subjects and endophenotype testing required for the proband and unaffected sibling. Additional affected and unaffected siblings were included whenever possible. Families missing 1 or both parents were accepted if 2 or more additional siblings were available, regardless of affected status. This ascertainment strategy was designed to provide greater potential for phenotypic contrasts between and within families. The ascertainment and screening methods, inclusion/exclusion criteria, and continuous clinical quality assurance procedures are detailed elsewhere.17 The final COGS-1 data set, included in this article, consisted of 296 predominantly nuclear families comprising 1366 subjects with an average family size of 4.6 members (range 4–14). The majority of the families (62%) consisted of a single sibling pair discordant for SZ, with sibships of 3 accounting for 26% of families and larger sibships accounting for 12%. A total of 1286 subjects had DNA available for genotyping, and of these, 1004 subjects were assessed for the 12 quantitative neurocognitive and neurophysiological endophenotypes.12 Detailed descriptions of the assessment procedure for the endophenotypes are provided elsewhere.13,17–22
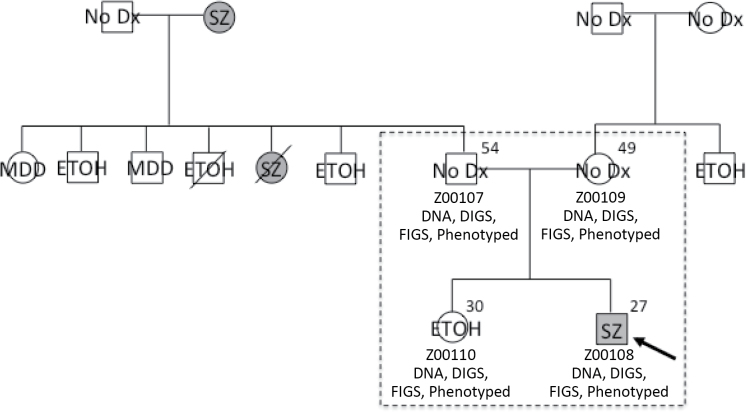
The Family Interview for Genetic Studies (FIGS)23 was administered to all participants in order to obtain convergent psychiatric diagnoses for first-degree relatives of interviewed participants. Although 1297 subjects were directly assessed via the DIGS, psychiatric diagnoses were obtained from the FIGS for an additional 1952 family members who did not participate in endophenotype testing. Using best-estimate consensus diagnostic information, we were able to extend the nuclear families to comprise 3304 subjects with a mean family size of 11.2 and diagnoses available for 3249 subjects. Table 1 illustrates the differences between the nuclear families used for our previous endophenotype studies and the extended families that we were able to construct using FIGS information provided by participating subjects. FIGS information was only used when information was obtained from reliable first-degree informants in accordance with the original purpose of this instrument.23 For example, the mother of the proband could provide diagnostic information for her parents and siblings, none of whom were directly interviewed for this study. Figure 1 shows an example of the best-estimate consensus summary information obtained and how FIGS information was used to extend the nuclear families.
Table 1.
Sample Description
| Nuclear Familiesa | Extended Pedigreesb | |
|---|---|---|
| Family information | ||
| Number of families | 296 | 296 |
| Average size | 4.6 | 11.2 |
| Average generations | 2.0 | 2.7 |
| Relative pairs | ||
| Sibling | 714 | 4113 |
| Half-sibling | 17 | 40 |
| Parent-child | 1530 | 4644 |
| Grandparent-grandchild | 18 | 2368 |
| Avuncular | 8 | 3294 |
| Cousin | 0 | 235 |
| Subjects | ||
| Total subjects | 1366 | 3304 |
| Males/females | 736/630 | 1508/1795 |
| SZ | 321 | 391 |
| BPD | 25 | 60 |
| MDD | 201 | 296 |
| Other disorder | 2 | 53 |
| Never mentally ill | 748 | 2449 |
| Unknown diagnosis | 69 | 49 |
| Relative frequency | ||
| SZ | 24.8% | 12.0% |
| SZ + BPD | 26.7% | 13.9% |
| SZ + BPD + MDD | 42.2% | 23.0% |
Note: BPD, bipolar disorder; MDD, major depressive disorder; SZ, schizophrenia.
aNuclear families include 1297 subjects directly diagnosed via the Diagnostic Interview for Genetic Studies, as well as 1337 subjects that provided Family Interview for Genetic Studies information for all included family members and additional nonparticipating subjects.
bExtended pedigrees include additional subjects indirectly diagnosed via the Family Interview for Genetic Studies information obtained from directly assessed first-degree relatives.
Fig. 1.
Example of a nuclear family from the COGS-1 database that was extended using FIGS information. A dashed box indicates members of the nuclear family, all of whom directly participated in the study. The age of each participating subject is indicated in the upper righthand corner and subject IDs are given, along with an indication of whether DNA was collected and whether DIGS, FIGS, or endophenotypes are available for the subject. DIGS-based diagnoses are indicated for each participating member, and FIGS-based diagnoses are indicated for additional first-degree relatives of a participating member with additional diagnostic information below as relevant. Squares indicate males and circles indicate females, and diagonal lines indicate the subject is deceased. COGS-1, Consortium on the Genetics of Schizophrenia; DIGS, Diagnostic Interview for Genetic Studies; ETOH, alcohol dependence; FIGS, Family Interview for Genetic Studies; MDD, major depressive disorder; no Dx, never mentally ill; SZ, schizophrenia.
To determine whether the COGS-1 requirement of participation of at least 3 additional family members, including at least 1 unaffected sib, resulted in a study ascertainment bias that favored a less-severe form of SZ, COGS-1 SZ probands were compared to a sample of 1189 SZ patients who were recruited and evaluated as part of the subsequent and ongoing 3000 person COGS-2 case-control study, which does not require the participation of family members. In this context, clinical symptoms for both COGS-1 and COGS-2 SZ patients were assessed using the Scale for the Assessment of Negative Symptoms24 and the Scale for the Assessment of Positive Symptoms.25 A modified Global Assessment of Functioning Scale26 was used for assessing participants’ overall level of functional status across psychological, social, and occupational domains via an anchored measure in accordance with our previously published methods.26–28
Statistical Analyses
Pedstats was used to determine the number of informative relative pairs for each endophenotype.29 Heritability analyses of the diagnostic categories were conducted via SOLAR v.4.3.1,30 consistent with our previous methods for estimating the heritability of the quantitative endophenotypes.12,13 Although variance component methods were used for the endophenotypes, SOLAR uses a liability threshold model to accommodate discrete traits, implemented in separate maximization routines that are generally identical to those used for quantitative trait maximization. The null hypothesis of no heritability (h 2 = 0) is tested by comparing a full model, which assumes that some fraction of the phenotypic variation is explained by genetic factors, to a reduced model, which assumes that no variation is explained by genes, using likelihood ratio tests. The bias introduced by our discordant sibpair ascertainment strategy necessitated a correction to better reflect the true mean of the disorder in the analysis. We, therefore, fixed the mean of each diagnostic category to more accurately reflect the population prevalence using the corresponding critical z values. We estimated the prevalence of SZ and BPD each as 1% and MDD as 16.2%.31 We used z values of 2.33 for the SZ model, 2.05 for the SZ + BPD model, and 0.91 for the SZ + BPD + MDD model. Comparisons of severity of clinical symptoms, functional status, and endophenotype deficits were performed in SPSS version 20 using ANOVA adjusted for age, sex, and site of ascertainment as necessary.12
Results
The heritability of SZ was determined to be 31% for the nuclear and 44% for the extended families (table 2). When the clinical diagnostic phenotype was broadened to include a history of a serious psychotic mental disorder (ie, SZ or BPD), heritability estimates rose to 37% for the nuclear families and remained at 44% for the extended families. In contrast, when MDD was added as a nonpsychotic, clinically impaired group, heritability estimates for the nuclear and extended families dropped to 34% and 20%, respectively (table 2).
Table 2.
Heritability (h 2) Estimates for Each Diagnostic Category and Family Set, Indicating the Numbers of Unaffected, Discordant, and Affected Relative Pairs for Each Model
| Family Set | Diagnoses | Informative Relative Pairsa | ||||
|---|---|---|---|---|---|---|
| Sibling | Parent-Offspring | Otherb | h 2 ± SE | P value | ||
| Nuclear | SZ | 219/466/17 | 747/561/8 | 16/21/1 | 0.31±0.02 | <.0001 |
| SZ + BPD | 206/464/32 | 712/580/24 | 16/21/1 | 0.37±0.32 | <.0001 | |
| SZ + BPD + MDD | 121/422/159 | 461/669/186 | 13/12/13 | 0.34±0.03 | <.0001 | |
| Extended | SZ | 3141/876/42 | 3713/759/16 | 3648/2017/90 | 0.44±0.04 | <.0001 |
| SZ + BPD | 3032/953/74 | 3571/873/44 | 3411/2205/139 | 0.44±0.17 | <.0001 | |
| SZ + BPD + MDD | 2523/1261/275 | 2925/1310/253 | 2624/2779/352 | 0.20±0.04 | <.0001 | |
Note: Abbreviations are explained in the first footnote to table 1.
aInformative relative pairs are listed as unaffected/discordant/affected for each diagnostic and relationship category.
bOther includes half-sibling, grandparent-grandchild, avuncular, and cousin pairs.
Anticipating that the discordant sibpair design may have selected for families with fewer affected individuals and perhaps more sporadic vs familial cases of SZ, we examined the effects of positive vs negative family history. An assessment of the extended families revealed 62 (21%) of COGS-1 families exhibited a positive family history of SZ, defined as a minimum of 2 family members with SZ. When family history was broadened to include SZ or BPD, 97 families (33%) were defined as having a positive family history. As one would expect, restricting the analysis to the subset of families with multiple affected members significantly increased the heritability estimates for SZ and SZ + BPD to approximately 80%, consistent with estimates observed in previous twin and multiplex family studies. Another 96 families (32%) had no evidence of family history for any psychiatric illness, and the SZ proband was the only affected family member. These probands may thus represent sporadic cases of SZ. The remaining 103 families (35%) had a family history of MDD in addition to the SZ proband, and some portion of probands from these families likely represent sporadic cases of SZ as well. The COGS-1 families thus seem to reflect a SZ sample that includes combinations of both familial and sporadic cases of SZ.
We further assessed whether the recruitment of intact families may have biased the sample to favor the recruitment of SZ probands with a milder form of illness. A comparison of all COGS-1 probands, regardless of family history, with SZ patients collected as singleton cases in COGS-2 revealed no differences in symptom severity or global functioning (see table 3), providing further evidence that COGS-1 SZ probands are symptomatically representative of a population-based sample that includes both familial and sporadic cases of SZ.
Table 3.
Comparison of Demographic and Clinical Characteristics of SZ Patients Recruited as Part of COGS-1 Family Study vs the COGS-2 Case-Control Design
| Comparison of COGS-1 vs COGS-2 Schizophrenia Patients | Effect Size | ||||||
|---|---|---|---|---|---|---|---|
| COGS-1: 3 Additional Family Members Required | COGS-2: No Additional Family Members Required | ||||||
| N | Mean | SD | N | Mean | SD | (d) | |
| Age* | 295 | 34.39 | 11.02 | 1189 | 46.42 | 11.06 | 1.00 |
| Education* | 294 | 13.73 | 2.09 | 1189 | 12.55 | 2.10 | 0.55 |
| Age of onset | 290 | 22.56 | 5.52 | 1174 | 21.89 | 7.07 | 0.10 |
| GAF | 279 | 45.56 | 13.43 | 1172 | 43.99 | 8.53 | 0.01 |
| SANS total | 289 | 10.17 | 5.91 | 1187 | 10.91 | 5.64 | 0.13 |
| SAPS total | 289 | 6.56 | 4.15 | 1183 | 6.69 | 4.05 | 0.16 |
Note: COGS, Consortium on the Genetics of Schizophrenia; GAF, Global Assessment of Functioning; SANS, Scale for the Assessment of Negative Symptoms; SAPS, Scale for the Assessment of Positive Symptoms.
No significant differences in global clinical symptoms or functional characteristics were detected. The COGS-1 family study, however, enrolled SZ patients who were significantly younger and had higher levels of education.
*P < .001.
Conclusions
Heritability of SZ in the COGS-1 family sample was estimated at 31% and 44%, derived from nuclear and extended families, respectively. At first blush, this appears to be discordant with the commonly cited figure of SZ heritability as 81%, derived primarily from twin studies,2 although more recent population-based studies have yielded lower heritability estimates for SZ.7,8 Specifically, the a priori inclusion of BPD in combination with SZ as a broad psychosis phenotype caused a modest increase in the heritability from 31% to 37% in the nuclear families. This finding supports the concept of psychosis as a dimensional phenotype32 and may reflect the shared genetic architecture among SZ and BPD, which was recently estimated to be 68% from common variation.33 Further broadening of the clinical phenotype via the addition of nonpsychotic MDD for a more general assessment of severe mental illness, however, significantly deflated the heritability estimates in both the nuclear and extended families relative to the SZ + BPD diagnostic category. This is perhaps not surprising, because the shared genetic etiology between MDD and SZ or BPD appears to be much lower (47% and 43%, respectively).33
Many researchers now utilize quantitative endophenotypes to deconstruct the genetic architecture of SZ.34 Endophenotypes are laboratory-based neurocognitive, neurophysiological, imaging, or metabolic measures that show deficits in SZ patients and their clinically unaffected relatives.3,34–36 Endophenotypes need not necessarily be more heritable than the clinical disorders. Their utility lies in the presumably “cleaner” and measurable signals they produce as a result of being more closely tied to the underlying neurobiological processes, compared to the conglomeration of traits and symptoms embedded in the diagnosis. As objective and reliable quantitative traits, endophenotypes also have more power to detect genetic associations than categorical clinical diagnoses. Many endophenotypes are even amenable to translational animal model studies for revealing neurobiological substrates associated with diseases.37–40 In the COGS-1 sample, the heritability estimates for SZ are equivalent to the mean heritability estimate for the 12 endophenotypes of 30% (range: 18%–50%) with 7 of the COGS-1 endophenotypes exhibiting higher heritability estimates than the diagnosis of SZ in the nuclear families.
Heritability reflects the genetic contributions to a disorder. In the narrow sense, heritability refers to additive genetic factors, which are typically common variants of relatively small effect that can be detected by association methods.12,14,41–45 These common variants are passed vertically through multiple generations of family members putatively leading to familial forms of SZ, but they also contribute to sporadic cases of the disorder. A large population-based study recently suggested that additive genetic effects account for 64% of SZ risk,8 and another study suggested that common single nucleotide polymorphisms explain approximately 23% of the total risk.33 In the broad sense, heritability refers to all genetic sources of risk, irrespective of allele frequency or effect size, and also includes rare copy number variants (CNVs) or single nucleotide variants (SNVs) of large effect, nonadditive genetic effects, and gene-environment interactions. Although highly penetrant, variants arising de novo appear to play a significant role in SZ risk,46–51 they do not segregate within families and are thus not reflected in heritability estimates. They may, however, explain some of the variation attributed to environment and contribute to some sporadic cases of SZ. It is also possible that some de novo mutations occur in “hotspots,” or vulnerable regions, that are sources of frequent mutation, which may mimic the relatively low-risk profile of common variation and serve as a mechanism whereby risk variation subject to negative selective pressure can be replenished.52 Thus, SZ liability likely involves a combination of segregating variants (common single nucleotide polymorphisms, rare SNVs, and CNVs) and de novo variation (highly penetrant CNVs and damaging coding SNVs) in the context of environmental factors.15,52 This model is consistent with familial vs sporadic cases, evolution, and observed phenotypic heterogeneity, because each case would presumably display a unique risk profile.15
The COGS-1 sample allowed for a direct comparison of the heritability estimates for SZ and endophenotypes. The COGS-1 ascertainment strategy of recruiting intact families with siblings discordant for SZ in an extensive study could bias the sample in a number of ways.17 First, this discordant sibpair design, which was chosen to enhance the study of quantitative endophenotypes associated with an illness, may lead to lower heritability estimates of the illness itself, as we have found. Second, families exhibiting evidence of bilineal transmission were excluded, which may have similarly created a downward bias on the heritability of SZ in our sample. Thus, our selection strategy may have inadvertently excluded more densely affected families for which unaffected siblings might be rare or for which affected members potentially show greater impairment and would therefore be less willing to participate in this study. Such families may also be less likely to be intact due to the dysfunction caused by multiple cases of illness. In contrast to our expectations, however, when COGS-1 probands were compared with COGS-2 singleton SZ cases, there were no apparent differences in clinical symptom severity or functional status, suggesting that the COGS-1 probands are representative of broader population-based samples, at least based on these more global characteristics. One might also speculate that this COGS-1 ascertainment strategy would yield a preponderance of sporadic rather than familial cases of SZ. Of the SZ probands, 33% had at least 1 other family member with a psychotic illness and likely represent familial cases; 32% of families exhibited no family history of any serious mental illness and were likely sporadic. Many of the remaining probands likely also represented sporadic cases with no family history of psychosis. In fact, sporadic cases may be the norm for SZ. Recently, Yang et al53 showed that for a disease such as SZ, 83%–90% of cases are predicted to appear sporadic with a lack of affected first-, second-, or third-degree relatives, which is consistent with both common polygenic and highly penetrant de novo mutation models. The prevalence of sporadic vs familial SZ was recently examined in the Swedish population study of more than 9 million people; only 3.8% of families had more than 1 affected member.8 Thus, the COGS-1 sample does not appear to contain a preponderance of sporadic SZ, but rather reflects a mixture of both sporadic and familial forms of illness. This characterization of families will serve as a valuable platform for future molecular and endophenotypic studies of these samples.
For over a century,54–56 family studies of psychiatric disorders have demonstrated the substantial contribution of genetic factors to risk of illness,57–61 forming the basis for contemporary molecular approaches. The first family study of SZ—or rather dementia praecox—was conducted by Rudin54 in Kraepelin’s clinic from 1907–1911, reported in 1916,57 see also.62–64 An important strength of the current study is the method of directly interviewing as many relatives as possible (n = 1297) using the DIGS and FIGS. This resource-intensive approach allowed us to later obtain convergent psychiatric information for first-degree relatives of directly interviewed individuals and substantially expand the total sample (N = 3304) for heritability analyses of SZ. Although reliance on informants can result in an underreporting of psychiatric disorders in families with some inherent diagnostic uncertainty,58 the FIGS was developed to minimize these possibilities through the use of structured assessment procedures anchored with specific diagnostic criteria.23 Rather than a decrease in heritability that would be expected with either an underreporting of psychiatric illness in families or a loss of diagnostic fidelity, FIGS information actually increased the heritability estimates in our sample, particularly for models using a broad psychosis phenotype (SZ + BPD).
The COGS, Bipolar and Schizophrenia Network on Intermediate Phenotypes, and other studies attempt to address the imprecision in our current diagnostic symptom through advancing the use of reliable, laboratory-based neurocognitive and neurophysiological measures for use as endophenotypes in genomic studies and biomarkers in clinical outcome studies.65–67 The current analyses are only one step in a larger strategy of examining endophenotype structure. Even if there were to be an illness vs endophenotype “heritability gap,” the endophenotypes are still extremely informative regarding the pathophysiology and genetics of this illness. These results support the continued use of endophenotypes to illuminate the mechanisms contributing to SZ and related psychoses. We anticipate that the ongoing COGS-2 case-control study will shed further light on the genomic and neurobiological substrates of SZ.
Acknowledgments
The authors thank all of the COGS-1 participants and support staff that made this study possible, including the following key personnel: University of California San Diego (R01-MH065571; MH042228, MH079777, MH087889): Joyce Sprock, Katrin Meyer-Gomes, Barbara Haugeland, Kari Tweedale, Sheldrick Holmes, Marlena Pela, Maria Bongiovanni; Harvard University (RO1-MH065562; MH43518; Commonwealth Research Center of the Massachusetts Department of Mental Health): Stephen J. Glatt, Lynda Tucker, Monica Landi, Erica Lee, and Frances Schopick; Mount Sinai School of Medicine (RO1-MH065554): Rui Ferreira, Robert Fieo, Christopher Smith, and Rebecca West; University of California Los Angeles (RO1-MH65707): Mark McGee, William Horan and Mark Sergi; University of Colorado (RO1-MH65588): Jamey Ellis, Jeff Hollis, Vicki Pender, Bernadette Sullivan, Bettye Clement, Christopher Cason, and Alexis Ritvo; University of Pennsylvania (RO1-MH65578): Alexandra Duncan Ramos, Jarrod Gutman, CarlaAnn Henry, Paul Hughett, Jennifer Jackson, Adrienne Mishkin, J. Dan Ragland, Leslie Ramsey, David Rice, Jan Richard, Devon Seward, Felipe Silva, and Robert Witalec; University of Washington (R01-MH65558): Kate B. Alvey, Andrew C. David, Sean P. Meichle, and Denise O. Pritzl. The authors also thank Dr John Blangero for his expert advice and guidance with respect to the heritability analyses. Drs Greenwood, Braff, Calkins, Radant, Seidman, Siever, Silverman, Stone, Sugarb DW. Tsuang, and MT. Tsuang report no financial relationships with commercial interests. Dr Light reports having been a consultant to Neuroverse, Envivo, and Astellas. Dr Green reports having been a consultant to Abbott Laboratories (AbbVie), Biogen, and Roche; he is a member of the scientific board for Mnemosyne, and he has received research funds from Amgen. Drs Gur and Turetsky have received unrelated research support for investigator-initiated grants from Pfizer and AstraZeneca. Dr Nuechterlein has received unrelated research support from Ortho-McNeil Janssen Scientific Affairs and has consulted to Wyeth/Pfizer. Dr Swerdlow has been a paid consultant for Neurocine Inc.
References
- 1. Green MF, Kern RS, Braff DL, Mintz J. Neurocognitive deficits and functional outcome in schizophrenia: are we measuring the “right stuff”? Schizophr Bull. 2000;26:119–136 [DOI] [PubMed] [Google Scholar]
- 2. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192 [DOI] [PubMed] [Google Scholar]
- 3. Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645 [DOI] [PubMed] [Google Scholar]
- 4. Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Torrey EF. Are we overestimating the genetic contribution to schizophrenia? Schizophr Bull. 1992;18:159–170 [DOI] [PubMed] [Google Scholar]
- 6. Lazzeroni LC, Ray A. A generalized Defries-Fulker regression framework for the analysis of twin data. Behav Genet. 2013;43:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wray NR, Gottesman II. Using summary data from the Danish national registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front Genet. 2012;3:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lichtenstein P, Yip BH, Björk C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coryell W, Zimmerman M. The heritability of schizophrenia and schizoaffective disorder. A family study. Arch Gen Psychiatry. 1988;45:323–327 [DOI] [PubMed] [Google Scholar]
- 10. Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker E, Downey G, Caspi A. Twin studies of psychopathology: why do the concordance rates vary? Schizophr Res. 1991;5:211–221 [DOI] [PubMed] [Google Scholar]
- 12. Greenwood TA, Swerdlow NR, Gur RE, et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2013;170:521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenwood TA, Braff DL, Light GA, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the Consortium on the Genetics of Schizophrenia. Arch Gen Psychiatry. 2007;64:1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PLoS One. 2012;7:e29630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gulsuner S, Walsh T, Watts AC, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nurnberger JI, Jr, Blehar MC, Kaufmann CA, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–59; discussion 863 [DOI] [PubMed] [Google Scholar]
- 17. Calkins ME, Dobie DJ, Cadenhead KS, et al. The Consortium on the Genetics of Endophenotypes in Schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33:33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calkins ME, Ray A, Gur RC, et al. Sex differences in familiality effects on neurocognitive performance in schizophrenia. Biol Psychiatry. 2013;73:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radant AD, Dobie DJ, Calkins ME, et al. Successful multi-site measurement of antisaccade performance deficits in schizophrenia. Schizophr Res. 2007;89:320–329 [DOI] [PubMed] [Google Scholar]
- 20. Swerdlow NR, Sprock J, Light GA, et al. Multi-site studies of acoustic startle and prepulse inhibition in humans: initial experience and methodological considerations based on studies by the Consortium on the Genetics of Schizophrenia. Schizophr Res. 2007;92:237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turetsky BI, Greenwood TA, Olincy A, et al. Abnormal auditory N100 amplitude: a heritable endophenotype in first-degree relatives of schizophrenia probands. Biol Psychiatry. 2008;64:1051–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turetsky BI, Calkins ME, Light GA, Olincy A, Radant AD, Swerdlow NR. Neurophysiological endophenotypes of schizophrenia: the viability of selected candidate measures. Schizophr Bull. 2007;33:69–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maxwell E. Manual for the FIGS. Bethesda, MD: Clinical Neurogenetics Branch, Intramural Research Program NIMH; 1992 [Google Scholar]
- 24. Andreasen NC. Scale for the Assessment of Negative Symptoms (SANS). Iowa City, IA: University of Iowa; 1984 [Google Scholar]
- 25. Andreasen NC. Scale for the Assessment of Positive Symptoms (SAPS). Iowa City, IA: University of Iowa; 1984 [Google Scholar]
- 26. Hall RC. Global assessment of functioning. A modified scale. Psychosomatics. 1995;36:267–275 [DOI] [PubMed] [Google Scholar]
- 27. McGlashan TH, Zipursky RB, Perkins D, et al. Randomized, double-blind trial of olanzapine versus placebo in patients prodromally symptomatic for psychosis. Am J Psychiatry. 2006;163:790–799 [DOI] [PubMed] [Google Scholar]
- 28. McGlashan TH, Zipursky RB, Perkins D, et al. The PRIME North America randomized double-blind clinical trial of olanzapine versus placebo in patients at risk of being prodromally symptomatic for psychosis. I. Study rationale and design. Schizophr Res. 2003;61:7–18 [DOI] [PubMed] [Google Scholar]
- 29. Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447 [DOI] [PubMed] [Google Scholar]
- 30. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA. 2003;289:3095–3105 [DOI] [PubMed] [Google Scholar]
- 32. Keshavan MS, Morris DW, Sweeney JA, et al. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale. Schizophr Res. 2011;133:250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee SH, Ripke S, Neale BM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2007;33:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Braff DL, Light GA. The use of neurophysiological endophenotypes to understand the genetic basis of schizophrenia. Dialogues Clin Neurosci. 2005;7:125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gottesman II, Shields J. Genetic theorizing and schizophrenia. Br J Psychiatry. 1973;122:15–30 [DOI] [PubMed] [Google Scholar]
- 37. Swerdlow NR, Light GA, Trim RS, Breier MR, Hines SR, Powell SB. Forebrain gene expression predicts deficits in sensorimotor gating after isolation rearing in male rats. Behav Brain Res. 2013;257:118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swerdlow NR, Powell SB, Breier MR, Hines SR, Light GA. Coupling of gene expression in medial prefrontal cortex and nucleus accumbens after neonatal ventral hippocampal lesions accompanies deficits in sensorimotor gating and auditory processing in rats. Neuropharmacology. 2013;75:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swerdlow NR, Light GA, Breier MR, et al. Sensory and sensorimotor gating deficits after neonatal ventral hippocampal lesions in rats. Dev Neurosci. 2012;34:240–249 [DOI] [PubMed] [Google Scholar]
- 40. Swerdlow NR, Weber M, Qu Y, Light GA, Braff DL. Realistic expectations of prepulse inhibition in translational models for schizophrenia research. Psychopharmacology. 2008;199:331–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055 [DOI] [PubMed] [Google Scholar]
- 43. Consortium SPG-WASG. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rees E, Kirov G, O’Donovan MC, Owen MJ. De novo mutation in schizophrenia. Schizophr Bull. 2012;38:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rees E, Moskvina V, Owen MJ, O’Donovan MC, Kirov G. De novo rates and selection of schizophrenia-associated copy number variants. Biol Psychiatry. 2011;70:1109–1114 [DOI] [PubMed] [Google Scholar]
- 48. Consortium TIS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465 [DOI] [PubMed] [Google Scholar]
- 52. Owen MJ, Craddock N, O’Donovan MC. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch Gen Psychiatry. 2010;67:667–673 [DOI] [PubMed] [Google Scholar]
- 53. Yang J, Visscher PM, Wray NR. Sporadic cases are the norm for complex disease. Eur J Hum Genet. 2010;18:1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rudin E.Zur Vererbung und Neuentstehung der Dementia Praecox. [On the Inheritance and Fresh Occurrence of Dementia Praecox.] . Berlin, Germany: Springer-Verlag; 1916. [Google Scholar]
- 55. Kendler KS, Zerbin-Rüdin E. Abstract and review of “Zur Erbpathologie der Schizophrenie” (Contribution to the genetics of schizophrenia). 1916. Am J Med Genet. 1996;67:343–346 [DOI] [PubMed] [Google Scholar]
- 56. Kendler KS, Zerbin-Rüdin E. Abstract and review of “Studien Uber Vererbung und Entstehung Geistiger Störungen. I. Zur Vererbung und Neuentstehung der Dementia praecox.” (Studies on the inheritance and origin of mental illness: I. To the problem of the inheritance and primary origin of dementia praecox). 1916. Am J Med Genet. 1996;67:338–342 [DOI] [PubMed] [Google Scholar]
- 57. Gottesman II, Shields J. A critical review of recent adoption, twin, and family studies of schizophrenia: behavioral genetics perspectives. Schizophr Bull. 1976;2:360–401 [DOI] [PubMed] [Google Scholar]
- 58. Andreasen NC, Endicott J, Spitzer RL, Winokur G. The family history method using diagnostic criteria. Reliability and validity. Arch Gen Psychiatry. 1977;34:1229–1235 [DOI] [PubMed] [Google Scholar]
- 59. Tsuang MT, Winokur G. The Iowa 500: field work in a 35-year follow-up of depression, mania, and schizophrenia. Can Psychiatr Assoc J. 1975;20:359–365 [DOI] [PubMed] [Google Scholar]
- 60. Winokur G, Morrison J, Clancy J, Crowe R. The Iowa 500. II. A blind family history comparison of mania, depression, and schizophrenia. Arch Gen Psychiatry. 1972;27:462–464 [DOI] [PubMed] [Google Scholar]
- 61. Kendler KS, McGuire M, Gruenberg AM, O’Hare A, Spellman M, Walsh D. The Roscommon Family Study. I. Methods, diagnosis of probands, and risk of schizophrenia in relatives. Arch Gen Psychiatry. 1993;50:527–540 [DOI] [PubMed] [Google Scholar]
- 62. Weber MM. Ernst Rüdin, 1874-1952: a German psychiatrist and geneticist. Am J Med Genet. 1996;67:323–331 [DOI] [PubMed] [Google Scholar]
- 63. Gershon ES. Ernst Rüdin, a Nazi psychiatrist and geneticist. Am J Med Genet. 1997;74:457–458; author reply 461 [DOI] [PubMed] [Google Scholar]
- 64. Gejman PV. Ernst Rüdin and Nazi euthanasia: another stain on his career. Am J Med Genet. 1997;74:455–456; author reply 461 [DOI] [PubMed] [Google Scholar]
- 65. Light GA, Näätänen R. Mismatch negativity is a breakthrough biomarker for understanding and treating psychotic disorders. Proc Natl Acad Sci USA. 2013;110:15175–15176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Light GA, Swerdlow NR, Rissling AJ, et al. Characterization of neurophysiologic and neurocognitive biomarkers for use in genomic and clinical outcome studies of schizophrenia. PLoS One. 2012;7:e39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perez VB, Swerdlow NR, Braff DL, Näätänen R, Light GA. Using biomarkers to inform diagnosis, guide treatments and track response to interventions in psychotic illnesses. Biomark Med. 2014;8:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]