Abstract
Long non-coding RNA has been involved in cancer progression, and high HOX transcript antisense intergenic RNA (HOTAIR) is thought to be a poor prognostic indicator in tumorigenesis of multiple types of cancer. Hence, the present study further reveals its prognostic value in tumor malignancy. A systematic review of PubMed and Web of Science was carried out to select literatures relevant to the correlation between HOTAIR expression levels and clinical outcome of various tumors. Overall survival (OS), metastasis-free survival (MFS), recurrence-free survival (RFS), and disease-free survival (DFS) were subsequently analyzed. Data from studies directly reporting a hazard ratio (HR) and the corresponding 95% confidence interval (CI) or a P value as well as survival curves were pooled in the current meta-analysis. A total of 2255 patients from 19 literatures almost published in 2011 or later were included in the analysis. The results suggest that HOTAIR was highly associated with HR for OS of 2.33 (95%CI = 1.77-3.09, P heterogeneity = 0.016). Stratified analyses indicate that elevated levels of HOTAIR appears to be a powerful prognostic biomarker for patients with colorectal cancer (HR = 3.02, 95CI% = 1.84-4.95, P heterogeneity = 0.699) and esophageal squamous cell carcinomas (HR = 2.24, 95CI% = 1.67-3.01, P heterogeneity = 0.711), a similar effect was also observed in analysis method and specimen, except for ethnicity. In addition, Hazard ratios for up-regulation of HOTAIR for MFS, RFS, and DFS were 2.32 (P<0.001), 1.98 (P = 0.369), and 3.29 (P = 0.001), respectively. In summary, the high level of HOTAIR is intimately associated with an adverse OS in numerous cancers, suggesting that HOTAIR may act as a potential biomarker for the development of malignancies.
Introduction
Noncoding RNAs (ncRNA) are initially identified from sequencing and microarray for whole genome and transcriptome, and at least 90% of ncRNAs has been found to be actively transcribed [1], [2]. The transcription of ncRNA revealed its complication in biogenesis than protein-coding RNA, such as extensive antisense, overlapping and non-coding RNA expression [3], [4], [5]. Despite initial argument claimed that ncRNA may be a fake transcriptional noise, increasing evidences suggested that ncRNAs may play a dominant biological role in cell metabolism and survival [6], [7], [8], [9]. Furthermore, the recent studies demonstrated that long non-coding RNAs (lncRNA, 200nt in length) express at tissue-specific patterns and that it is abnormally regulated in a variety of diseases, including cancer [10], [11], [12], [13]. Multiple regulatory bases have been involved in the regulation of lncRNA, such as transcriptional regulation, epigenetic regulation, and posttranscriptional regulation [9], [14]. Moreover, lncRNAs exhibit unique profiles in many kinds of cancers, which represent carcinogenesis and progression regarded as a predictor of patient outcomes [15], [16], [17].
HOTAIR, a prominently focused lncRNA, was initially reported to be implicated in primary breast cancer and breast cancer metastasis, wherein elevated HOTAIR promoted tumor invasiveness and metastasis [18]. HOTAIR overexpression has been shown to be associated with expression of polycomb repressive complex 2 (PRC2), inducing its relating methylation of histone H3 lysine 27 (H3K27) [10], [18]. In addition to breast cancer [18], recent clinical evidences show HOTAIR is also involved in the progression of many other types of cancer, such as hepatocellular carcinomas (HCC) [19], colorectal cancer (CRC) [20], esophageal squamous cell carcinomas (ESCC) [21], suggesting that HOTAIR expression serves as a prognostic factor for tumorigenesis. Although HOTAIR expression is considered to relating to clinical prognosis of multiple cancers, the impact of HOTAIR on the development of cancer still remains elusive. Some studies reported that up-regulation of HOTAIR contributes to tumorigenesis, including bladder cancer [22], cervical cancer [23], colorectal cancer [24], etc., while a few evidences exhibited an adverse effect recognized as a protective factor to against carcinogenesis [25]. It is necessary therefore to clarify the relationship between HOTAIR and cancer. Thus, the present study conducted the first meta-analysis using qualified relevant literatures to achieve a precise evaluation of the association between HOTAIR expression and cancer clinical prognosis.
Materials and Methods
Data sources and searches
The published data searching was performed using a literature review system with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines [26]. The selected literatures were determined via an electronic search of PubMed and Web of Science using these following terms: “HOTAIR”, “cancer or tumor or carcinomas” and “prognosis or outcome”. The last search was updated in July 18, 2014. Citation lists of retrieved articles were searched manually to ensure sensitivity of the search strategy.
Study selection
Studies considered eligible met the following criteria: 1) studied patients with any type of cancers; 2) explored the link between HOTAIR and clinical prognosis; 3) availability of a hazard ratio (HR) and 95% confidence interval (CI) or a P value for overall survival (OS). For a secondary analysis, studies including an HR for metastasis-free survival (MFS), disease-free survival (DFS), or recurrence-free survival (RFS) were also used to further analyze. OS [27], MFS [28], DFS [27], and RFS [29] were described previously; 4) published as a full paper in English. Studies were excluded based on the following criteria: 1) duplicated studies, reviews, letters, unpublished data, and comments; 2) those published in language other than English; 3) lack of key information for further analysis; 4) non-human research.
Data extraction
Two investigators (QWD, HLS) independently evaluated and extracted data from each identified studies based on criteria of inclusion and exclusion. Corresponding authors were contacted to clarify missing or ambiguous data. OS was treated as a dominant outcome of interest, but MFS, RFS and DFS were set as the secondary outcomes. The following information was carefully extracted: name of first author, year of publication, country of origin, ethnicity of the study population, type of specimen, cancer type, number of patients included in analysis, detection method of HOTAIR, cut-off defining high HOTAIR, follow-up period, and HR and corresponding 95% CI for OS, MFS, RFS, or DFS as applicable. Cancer type subgroups were generated for the main outcome if at least two studies on the type of cancer were available; the only one study was pooled in a subgroup termed “Other.” HR was firstly extracted from multivariable analysis where available. Otherwise, HR was extracted from univariate analysis, and calculated from Kaplan-Meier survival curve by HR digitizer software Engauge 4.0 as described previously [30].
Statistical analyses
All extracted data were combined into a meta-analysis using STATA software version 11.0 (STATA Corporation, College Station, TX, USA). Hazard ratios with the corresponding 95% CIs were used to estimate the strength of the link between HOTAIR and clinical prognosis. If HR was not directly reported, a mathematical estimation was conducted by calculating the necessary data on the basis of the previously reported methods [31]. Cochran's Q test and Higgins I-squared statistic were used to estimate the heterogeneity of pooled results. If P <0.05 for Q-test showed significant heterogeneity among studies, the random-effects model (DerSimoian-Laird method) was implemented to calculate the pooled HRs [32]. Otherwise, the fixed-effects model (Mantel-Haenszel method) was used [33]. To further explore the potential source of heterogeneity among studies, meta-regression was performed utilizing variables as cancer type, ethnicity, analysis method, type of specimen. To validate the stability of outcomes in this meta-analysis, sensitivity analysis was performed by sequential omission of each individual study. Publication bias was conducted by Begg's funnel plot and Egger's linear regression test and a P <0.05 was considered representative of statistically significant publication bias.
Results
Characteristics of studies
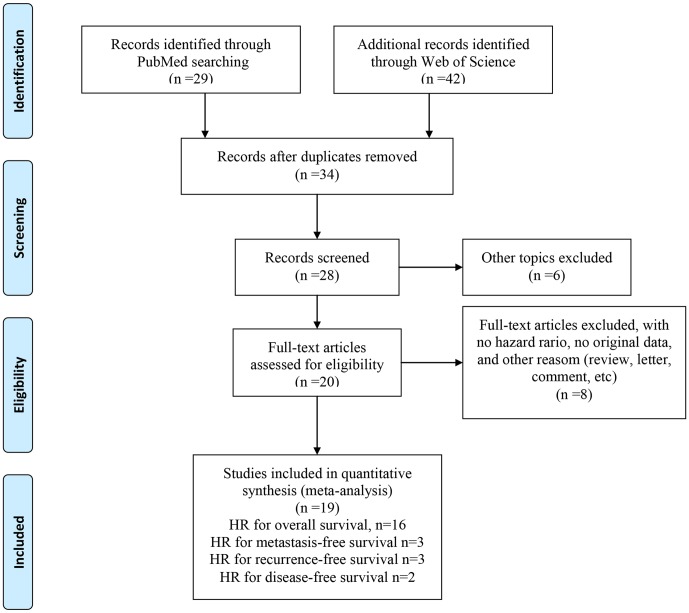
There were 71 papers in the electronic search of PubMed and EMBASE. On the basis of the inclusion criteria, 19 eligible papers were enrolled in this meta-analysis (shown in Figure 1). The main characteristics of included studies are shown in Table 1; all studies were almost published in 2011 or later. There were 16 studies for OS, 3 for MFS, 3 for RFS, and 2 for DFS in the meta-analysis. Participants in 16 studies were Asian and in the other 3 studies were Caucasian. Various cancers were recorded in our study, including BC, HCC, CRC, ESCC, etc. The types of specimen were tissue for twenty-two studies and blood for one study. The cut-off values included in the studies were inconsistent due to different detection methods, even in 6 studies were not reported. Hazard ratios with the corresponding 95% CIs were extracted from univariate analysis and the graphical survival plots in 6 studies, and multivariate analysis in 18 studies.
Figure 1. Flow chart for selection of studies for inclusion in this meta-analysis.
Table 1. Main characteristics of included studies.
| First author | Year | Country | Ethnicity | Specimens | Cancer type | Method | Cut-off* | Follow-up (month) | Number | Analysis | Survival |
| Yang [19] | 2011 | China | Asian | Tissue | HCC | RT-qPCR | NR | 18.6 (median) | 60 | Multivariable | RFS |
| Kogo [20] | 2011 | Japan | Asian | Tissue | CRC | RT-qPCR | 0.027 | 36 (mean) | 100 | Multivariable | OS |
| Lu [40] | 2012 | Italy | Caucasian | Tissue | BC | RT-qPCR | NR | 86 (median) | 336 | Multivariable | OS, RFS |
| Li [41] | 2012 | China | Asian | Tissue | LSCC | RT-qPCR | NR | 60 (total) | 72 | Multivariable | OS |
| Nie [42] | 2013 | China | Asian | Tissue | NPC | ISH | 6 | 69 (median) | 160 | Multivariable | OS |
| Nakagawa [43] | 2013 | Japan | Asian | Tissue | NSCLC | RT-qPCR | 2 | 31.4 (median) | 77 | Univariable | DFS |
| Lv [44] | 2013 | China | Asian | Tissue | ESCC | ISH | 6 | 60 (total) | 93 | Univariable | OS |
| Li [45] | 2013 | China | Asian | Tissue | ESCC | RT-qPCR | 125 | 60 (total) | 100 | Multivariable | OS |
| Ge [46] | 2013 | China | Asian | Tissue | ESCC | RT-qPCR | NR | 60 (total) | 137 | Multivariable | OS, MFS |
| Liu [47] | 2013 | China | Asian | Tissue | NSCLC | RT-qPCR | 8.57 | 60 (total) | 42 | Univariable | OS |
| Chen [48] | 2013 | China | Asian | Tissue | ESCC | RT-qPCR | 26.6 | 38 (mean) | 78 | Multivariable | OS |
| Zhang [49] | 2013 | China | Asian | Tissue | GBM | GSEA | NR | 50 (totoal) | 89 | Multivariable | OS |
| Sørensen [50] | 2013 | Denmark | Caucasian | Tissue | BC | Microarray | 0.6 | 217.2 (mean) | 164 | Multivariable | MFS |
| Svoboda [51] | 2014 | Czech | Caucasian | Tissue, Blood | CRC | RT-qPCR | 0.7, 4.4 | 35 (mean) | 73, 84 | Multivariable | OS |
| Qiu [52] | 2014 | China | Asian | Tissue | EOC | RT-qPCR | NR | 50.5 (median) | 64 | Univariable | OS, DFS |
| Huang [53] | 2014 | China | Asian | Tissue | Cervical | RT-qPCR | NR | 42 (mean) | 218 | Multivariable | OS |
| Liu [54] | 2014 | China | Asian | Tissue | GC | RT-qPCR | NR | 50 (totoal) | 78 | Univariable | OS |
| Wu [55] | 2014 | China | Asian | Tissue | CRC | RT-qPCR | 5 | 55.5 (median) | 120 | Multivariable | OS, MFS |
| Yan [29] | 2014 | China | Asian | Tissue | Bladder | RT-qPCR | NR | 60 (total) | 110 | Multivariable | RFS |
*cancerous/noncancerous
HCC, hepatocellular carcinoma; CRC, colorectal cancer; BC, breast cancer; LSCC, laryngeal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell lung cancer; ESCC, esophageal squamous cell carcinoma; GBM, glioblastoma multiforme; EOC, epithelial ovarian cancer; GC, gastric cancer; RT-qPCR, real-time quantitative PCR; ISH, in situ hybridization; GSEA, gene set enrichment analysis; NR, not reported; OS, overall survival; MFS, metastasis-free survival; RFS, recurrence-free survival; DFS, disease-free survival.
Overall survival
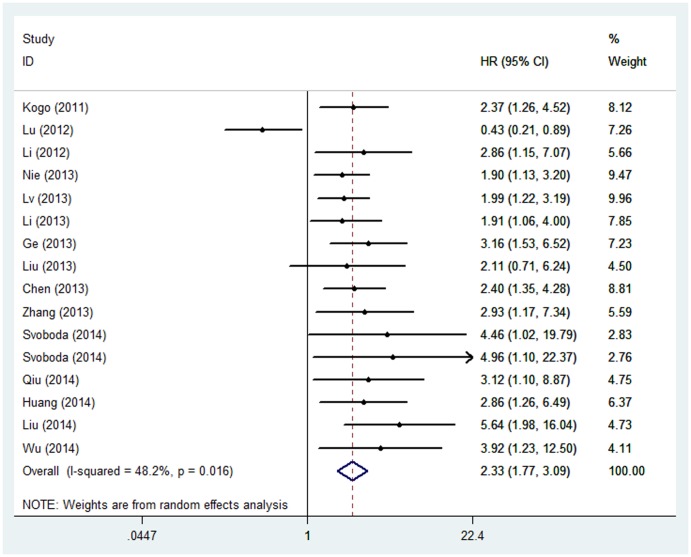
The main results of this meta-analysis are shown in Table 2. 16 studies comprising 1844 patients reported HR for OS. It is suggested that elevated HOTAIR predicted a poor outcome for OS (HR = 2.33, 95%CI = 1.77-3.09, P H = 0.016; Figure 2). Stratified analyses by cancer type indicated that the prognostic effect of HOTAIR was highest in CRC (HR = 3.02, 95%CI = 1.84-4.95, P H = 0.699), followed by ESCC (HR = 2.24, 95%CI = 1.67-3.01, P H = 0.016). HR for the subgroup of other cancers was 2.18 (95%CI = 1.25-3.78, P H = 0.001).
Table 2. The main results of pooled analyses.
| Survival | Variables | NO. of studies | NO. of patients | P value | Regression model | |||
| P H | P Z | P E | Random | Fixed | ||||
| OS | All | 16 | 1844 | 0.016 | 0.000 | 0.110 | 2.34 (1.77–3.09) | 2.20 (1.82–2.66) |
| Cancer type | ||||||||
| CRC | 4 | 377 | 0.699 | 0.000 | — | 3.02 (1.84–4.95) | 3.02 (1.84–4.95) | |
| ESCC | 4 | 408 | 0.711 | 0.000 | — | 2.24 (1.67–3.01) | 2.24 (1.67–3.01) | |
| Other | 8 | 1059 | 0.001 | 0.006 | — | 2.18 (1.25–3.78) | 1.95 (1.46–2.60) | |
| Ethnicity | ||||||||
| Asian | 13 | 1351 | 0.894 | 0.000 | — | 2.43 (1.99–2.97) | 2.43 (1.99–2.97) | |
| Caucasian | 3 | 493 | 0.001 | 0.482 | — | 1.92 (0.31–11.91) | 0.92 (0.51–1.67) | |
| Analysis method | ||||||||
| Univariable | 4 | 277 | 0.329 | 0.000 | — | 2.53 (1.64–3.89) | 2.13 (1.71–2.65) | |
| Multivarible | 12 | 1567 | 0.009 | 0.000 | — | 2.26 (1.59–3.20) | 2.44 (1.67–3.55) | |
| Specimen | ||||||||
| Tissue | 15 | 1760 | 0.015 | 0.000 | — | 2.29 (1.72–3.04) | 2.17 (1.79–2.63) | |
| Blood | 1 | 84 | — | 0.037 | — | 4.96 (1.10–22.37) | 4.96 (1.10–22.37) | |
| MFS | All | 3 | 421 | 0.080 | 0.000 | — | 2.81 (1.44–5.57) | 2.32 (1.62–3.33) |
| RFS | All | 3 | 506 | 0.000 | 0.369 | — | 1.98 (0.44–8.85) | 1.96 (1.37–2.81) |
| DFS | All | 2 | 141 | 0.969 | 0.001 | — | 3.29 (1.61–6.70) | 3.29 (1.61–6.70) |
P H, P value of heterogeneity test; P Z, P value of Z test; P E, P value of Egger's test; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; OS, overall survival; MFS, metastasis-free survival; RFS, recurrence-free survival; DFS, disease-free survival.
Italic indicates HR with 95% CI used to analyses.
The bold represents statistically significant results.
Figure 2. Forest plots of studies evaluating hazard ratios (HRs) of HOTAIR for overall survival.
The effect of elevated HOTAIR on OS among different races is shown in Table 2. The hazard ratios were 2.43 (95%CI = 1.99-2.97, P H = 0.894) for Asian, and 1.92 (95%CI = 0.31-11.91, P H = 0.001) for Caucasian. When different analysis methods were considered, HOTAIR was a strong prognostic marker both by univariate analysis (HR = 2.13, 95%CI = 1.71-2.65, P H = 0.329) and by multivariate analysis (HR = 2.26, 95%CI = 1.59-3.20, P H = 0.009). Performing subgroup analyses stratified by specimen, increased HOTAIR was closely associated with poor prognosis both in tissue (HR = 2.29, 95%CI = 1.72-3.04, P H = 0.015) and in blood (HR = 4.96, 95%CI = 1.10-22.37).
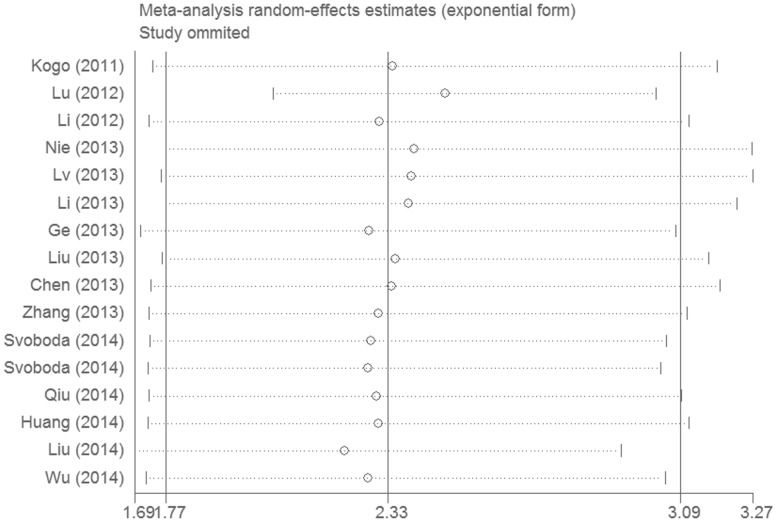
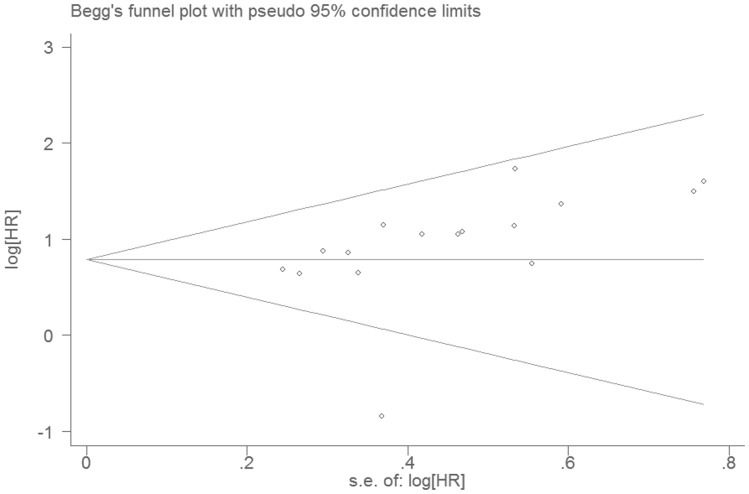
Sensitivity analysis is presented in Figure 3. The result pattern was not significantly impacted by removing single study each time. Begg's funnel plot and the Egger's linear regression test were conducted to evaluate publication bias. The shape of the funnel plot showed no significant asymmetry in Figure 4. Subsequently, Egger's test also suggested no evidence of publication bias (P = 0.110).
Figure 3. Sensitivity analysis of effect of individual studies on the pooled HRs for HOTAIR and overall survival of patients.
Figure 4. Funnel plot of HR for overall survival for high HOTAIR (vertical axis) and the standard error (SE) for HR (horizontal axis).
Each study represented by one circle. The horizontal line represented the pooled effect estimate.
Metastasis-free survival, recurrence-free survival and disease-free survival
Three studies comprising 421 patients reported HRs for MFS. Overall, HOTAIR greater than the cut-off was associated with an HR for MFS of 2.32 (95%CI = 1.62-3.33, P H = 0.080). Three studies comprising 506 patients showed HRs for RFS. HOTAIR was not linked with poor RFS. Two studies comprising 141 patients reported HRs for DFS. Up-regulation of HOTAIR predicted a poor clinical outcome for DFS (HR = 3.29, 95%CI = 1.61-6.70, P H = 0.969).
Discussion
As a novel molecular basis, the study of lncRNA has focused on the impact of lncRNA on cancer pathogenesis and prognosis, providing a new insight into cancer therapeutic strategy [34], [35]. Despite substantial progress of lncRNAs in cancer nosogenesis and prognosis, the prognostic effect of lncRNAs is still confused. To explore the prognostic impact of lncRNAs in cancer, this system review and meta-analysis was performed to investigate the impact of HOTAIR on tumor prognosis for achieving more consistent and precise conclusion.
Here we undertook meta-analysis of 18 literatures comprising 2255 patients with tumors to assess the prognostic effect of HOTAIR. We found that an accordant effect of an elevated HOTAIR on OS (HR = 2.33) with the similar hazard ratios among various cancer type subgroups and across analytical methods, or specimens, except for ethnicity subgroups. HOTAIR has been shown to contribute to the progression of many types of cancer and is nowadays considered as a hallmark of cancer [36]. The strong impact on OS was highest in CRC, which is further supported by evidences that HOTAIR plays a critical role in the carcinogenesis of CRC as a result of promoted multipotent cell differentiation [20]. In particular, the elevated levels of HOTAIR are highly associated with worse OS in Asian, but not in Caucasian, suggesting that interaction between genetic and environmental factors may contribute to cancer development. In addition, clinicopathological features and drug treatment status could modify the effect of HOTAIR [25]. However, a study reported that HOTAIR induces repressive chromatin status by promoting the formation of H3K27, suggesting that HOTAIR may function as a tumor suppressor gene by inhibiting proliferation of cancer stem cells [18]. The present study further verify that the levels of HOTAIR is highly associated with cancer development, although the prognostic effect of HOTAIR on MFS and DFS is still remained for the future studies. Importantly, the results are consistent in the tested types of cancer regardless of different data resources and analysis methods.
The mechanisms underlying the association between the high levels of HOTAIR and poor outcome of cancers are still unclear. This study may predict the following potential mechanisms involved in the prognostic impact of HOTAIR on carcinogenesis. HOTAIR regulates the chromatin methylation state by inducing genome-wide retargeting of PRC2 and promotes metastasis of breast cancer by silencing multiple metastasis-suppressing genes [18]. Consequently, PRC2 has been reported to be involved in stem cell pluripotency and progression ofEZH2, SUZ12, and EED [36], [37]. Consistently, HOTAIR knockdown not only suppressed cell invasion, but also decreased cell proliferation, altered cell cycle progression, and induced apoptosis [38].
Physicians prefer to use prognostic data when speaking to patients. As HOTAIR offers independent prognostic information, we may incorporate HOTAIR in a simple score to provide an appropriate therapeutic strategy. In recent years, a few clinical studies with cancer patients showed that reducing levels of HOTAIR is closely associated with a good response to treatment [39]. Furthermore, HOTAIR has been verified to be an independent prognostic indicator in CRC patients [24]. It was suggested that changing blood HOTAIR levels might be useful for tailoring of therapy for cancer patients.
Some limitations in this study should be acknowledged. Firstly, only summarized data rather than individual patient data were used. Secondly, a part of studies, especially in subgroups analyses, was lightly relative. Thirdly, because some cut-off values were not reported and the criteria of calculating cut-off value were inconsistent, stratified analysis by cut-off values was not conducted to suggest whether cut-off values were the origin of heterogeneity. Fourthly, we only included studies reporting HR or survival curves, and consequently some publications reporting on the prognostic value of HOTAIR were excluded. For example, only odds ratios were reported, so the selection bias might be appeared. Finally, most of the included studies do not explicitly control for such concurrent conditions, and these may confound the measurement of HOTAIR.
In summary, up-regulation of HOTAIR is associated with adverse survival in many types of cancer, and HOTAIR may serve as an effective prognostic biomarker for diagnosis of cancer. Therefore, clinical checking the levels of HOTAIR expression may provide a promising approach to identify patients who would require more intimately care for personally tailored medical inspection to monitor cancer prevention and treatment.
Supporting Information
Flow chart.
(TIF)
Forest plots.
(TIF)
Sensitivity analysis.
(TIF)
Funnel plot.
(TIF)
Main characteristics.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was supported by grants from The National Nature Science Foundation of China (no. 81172141, 81200401), Nanjing Science and Technology Committee Project (no. 201108025), Nanjing Medical Technology Development Project (no. ZKX11025), Nanjing Health Young Talent Project, Jiangsu Provincial Key Medical Talents to S.K.W., and Nanjing Medical Science and Technique Development Foundation to Y.Q.P. (no. QRX11255) and B.S.H. (no. QRX11254). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Costa FF (2010) Non-coding RNAs: Meet thy masters. Bioessays 32: 599–608. [DOI] [PubMed] [Google Scholar]
- 3. Kapranov P, Willingham AT, Gingeras TR (2007) Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423. [DOI] [PubMed] [Google Scholar]
- 4. Frith MC, Pheasant M, Mattick JS (2005) The amazing complexity of the human transcriptome. Eur J Hum Genet 13: 894–897. [DOI] [PubMed] [Google Scholar]
- 5. Guttman M, Amit I, Garber M, French C, Lin MF, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Zhang J, Zheng H, Li J, Liu D, et al. (2004) Mouse transcriptome: neutral evolution of 'non-coding' complementary DNAs. Nature 431: 1 p following 757; discussion following 757. [PubMed]
- 7. Struhl K (2007) Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105. [DOI] [PubMed] [Google Scholar]
- 8. Ebisuya M, Yamamoto T, Nakajima M, Nishida E (2008) Ripples from neighbouring transcription. Nat Cell Biol 10: 1106–1113. [DOI] [PubMed] [Google Scholar]
- 9. Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159. [DOI] [PubMed] [Google Scholar]
- 10. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, et al. (2010) Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, et al. (2008) Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 17: 642–655. [DOI] [PubMed] [Google Scholar]
- 13. Maruyama R, Shipitsin M, Choudhury S, Wu Z, Protopopov A, et al. (2012) Altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci U S A 109: 2820–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J, Zhang A, Wang Y, Liu N, You Y, et al. (2012) New insights into the roles of ncRNA in the STAT3 pathway. Future Oncol 8: 723–730. [DOI] [PubMed] [Google Scholar]
- 15. Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361. [DOI] [PubMed] [Google Scholar]
- 17. Deng Q, He B, Gao T, Pan Y, Sun H, et al. (2014) Up-Regulation of 91H Promotes Tumor Metastasis and Predicts Poor Prognosis for Patients with Colorectal Cancer. PLoS ONE 9: e103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, et al. (2011) Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 18: 1243–1250. [DOI] [PubMed] [Google Scholar]
- 20. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, et al. (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71: 6320–6326. [DOI] [PubMed] [Google Scholar]
- 21. Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, et al. (2013) Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog 52: 908–915. [DOI] [PubMed] [Google Scholar]
- 22.Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, et al. (2014) Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. [DOI] [PubMed]
- 23. Huang L, Liao LM, Liu AW, Wu JB, Cheng XL, et al. (2014) Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet [DOI] [PubMed] [Google Scholar]
- 24. Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, et al. (2014) HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 35: 1510–1515. [DOI] [PubMed] [Google Scholar]
- 25. Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, et al. (2012) Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat 136: 875–883. [DOI] [PubMed] [Google Scholar]
- 26. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, et al. (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6: e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Izzo JG, Malhotra U, Wu TT, Ensor J, Luthra R, et al. (2006) Association of activated transcription factor nuclear factor kappab with chemoradiation resistance and poor outcome in esophageal carcinoma. J Clin Oncol 24: 748–754. [DOI] [PubMed] [Google Scholar]
- 28. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, et al. (2014) Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 32: 395–402. [DOI] [PubMed] [Google Scholar]
- 29. Yan T-H, Lu S-W, Huang Y-Q, Que G-B, Chen J-H, et al. (2014) Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumor Biology [DOI] [PubMed] [Google Scholar]
- 30. Wang W, Chen Y, Deng J, Zhou J, Zhou Y, et al. (2014) The prognostic value of CD133 expression in non-small cell lung cancer: a meta-analysis. Tumour Biol [DOI] [PubMed] [Google Scholar]
- 31. Parmar MK, Torri V, Stewart L (1998) Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 32. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 33. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 34. Sun M, Jin FY, Xia R, Kong R, Li JH, et al. (2014) Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 14: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, et al. (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640. [DOI] [PubMed] [Google Scholar]
- 36. Woo CJ, Kingston RE (2007) HOTAIR lifts noncoding RNAs to new levels. Cell 129: 1257–1259. [DOI] [PubMed] [Google Scholar]
- 37. Richly H, Aloia L, Di Croce L (2011) Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis 2: e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, et al. (2013) HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Milhem MM, Knutson T, Yang S, Zhu D, Wang X, et al. (2011) Correlation of MTDH/AEG-1 and HOTAIR Expression with Metastasis and Response to Treatment in Sarcoma Patients. J Cancer Sci Ther S5. [PMC free article] [PubMed] [Google Scholar]
- 40. Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, et al. (2012) Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Research and Treatment 136: 875–883. [DOI] [PubMed] [Google Scholar]
- 41. Li D, Feng J, Wu T, Wang Y, Sun Y, et al. (2013) Long Intergenic Noncoding RNA HOTAIR Is Overexpressed and Regulates PTEN Methylation in Laryngeal Squamous Cell Carcinoma. The American Journal of Pathology 182: 64–70. [DOI] [PubMed] [Google Scholar]
- 42. Nie Y, Liu X, Qu S, Song E, Zou H, et al. (2013) Long non-coding RNAHOTAIRis an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Science 104: 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, et al. (2013) Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochemical and Biophysical Research Communications 436: 319–324. [DOI] [PubMed] [Google Scholar]
- 44. Lv XB, Lian GY, Wang HR, Song E, Yao H, et al. (2013) Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE 8: e63516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Wu Z, Mei Q, Guo M, Fu X, et al. (2013) Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. British Journal of Cancer 109: 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ge X-S, Ma H-J, Zheng X-H, Ruan H-L, Liao X-Y, et al. (2013) HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Science 104: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, et al. (2013) The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 13: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen F-J, Sun M, Li S-Q, Wu Q-Q, Ji L, et al. (2013) Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Molecular Carcinogenesis 52: 908–915. [DOI] [PubMed] [Google Scholar]
- 49. Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, et al. (2013) HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-Oncology 15: 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sørensen KP, Thomassen M, Tan Q, Bak M, Cold S, et al. (2013) Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Research and Treatment 142: 529–536. [DOI] [PubMed] [Google Scholar]
- 51. Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, et al. (2014) HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 35: 1510–1515. [DOI] [PubMed] [Google Scholar]
- 52. Qiu J-j, Lin Y-y, Ye L-c, Ding J-x, Feng W-w, et al. (2014) Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecologic Oncology 134: 121–128. [DOI] [PubMed] [Google Scholar]
- 53. Huang L, Liao L-M, Liu A-W, Wu J-B, Cheng X-L, et al. (2014) Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Archives of Gynecology and Obstetrics [DOI] [PubMed] [Google Scholar]
- 54. Liu X-h, Sun M, Nie F-q, Ge Y-b, Zhang E-b, et al. (2014) Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Molecular Cancer 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu Z-H, Wang X-L, Tang H-M, Jiang T, Chen J, et al. (2014) Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncology Reports [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart.
(TIF)
Forest plots.
(TIF)
Sensitivity analysis.
(TIF)
Funnel plot.
(TIF)
Main characteristics.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.