Summary
Daughter strand gaps formed upon interruption of replication at DNA lesions in E. coli can be repaired by either translesion DNA synthesis or homologous recombination (HR) repair. Using a plasmid-based assay system that enables a discrimination between the strand transfer and template switching (information copying) modes of HR gap repair, we found that approximately 80% of strand gaps were repaired by physical strand transfer from the donor, whereas approximately 20% appear to be repaired by template switching. HR gap repair operated on both small and bulky lesions, and largely depended on RecA and RecF, but not the RecBCD nuclease. In addition, we found that HR was mildly reduced in cells lacking the RuvABC and RecG proteins involved in resolution of Holliday junctions. These results, obtained for the first time under conditions that detect the two HR gap repair mechanisms, provide in vivo high-resolution molecular evidence for the predominance of the strand transfer mechanism in HR gap repair. A small but significant portion of HR gap repair appears to occur via a template switching mechanism.
Keywords: DNA damage, DNA damage tolerance, DNA repair, DNA replication, translesion DNA synthesis
DNA lesions that have escaped repair may inhibit DNA replication, leading to arrested replication forks and/or the formation of daughter strand gaps (DSG) in DNA 1–4. While there has been considerable research effort in recent years to analyze the resolution of arrested replication forks 5–12, the repair of DSG has received little attention. However, two recent studies had highlighted the significance of DSG. An analysis of DNA replication in UV-irradiated S. cervisiae cells had revealed uncoupling of leading and lagging strand replication at UV lesions, and the formation of gaps on both leading and lagging strands 13. Another study reported, using an in vitro system, that priming occurs on the leading strand thus providing a possible mechanism for daughter strand gap formation at leading-strand lesions, and not only at lagging-strand lesions 14. The study of mechanisms of DSG repair in E. coli dates back nearly four decades, when Howard-Flanders proposed that gap repair occurs by homologous recombination (HR) involving strand transfer from the sister chromatid. His studies and those of others had defined gap repair as a process that depends on RecA and RecF, but not on the RecBCD nuclease 15–18. Although the results of those pioneering studies were consistent with the strand transfer model of gap repair, they did not provide proof for the model due to several reasons: (1) experiments were usually performed on populations of UV-irradiated cells most of which were dead, and the strand exchanges they detected could have been the results of DNA scrambling in dead cells 19 (2) the available techniques were unable to provide proof that exchanged DNA segments were indeed utilized to fill in the gaps (DNA exchanges could have occurred elsewhere), and (3) gap-filling by the alternative mechanism of gap filling via template switching could not been identified by those techniques. Using a two-plasmid assay system in which strand transfer and template switching mechanisms could be discriminated for the first time, we analyzed the mechanism of HR gap repair.
Experimental design
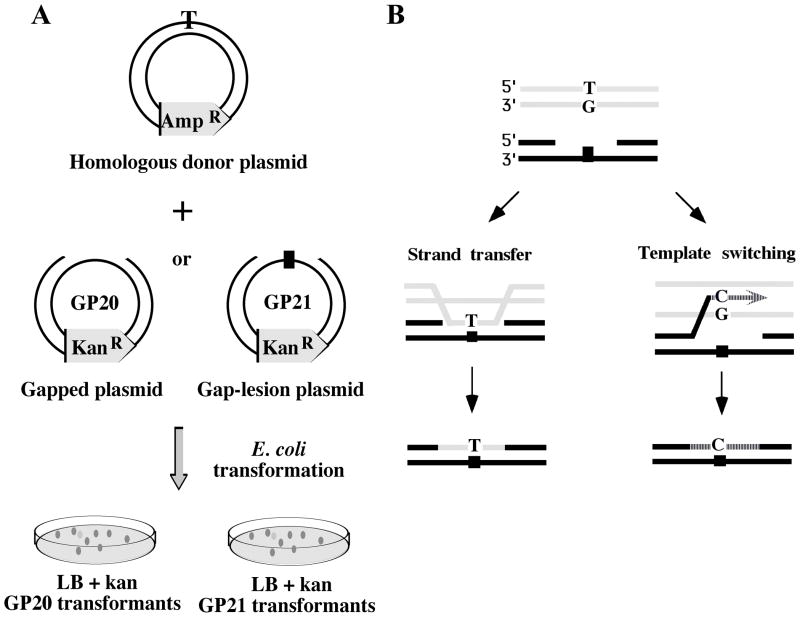
The two-plasmid HR gap repair assay system 20 is based on a gap-lesion plasmid (kanR) carrying a site-specific lesion in a ssDNA region, and on a homologous donor plasmid (ampR) (Fig. 1A). The experimental scheme involves one of two protocols: In the first, sequential transformation protocol, cells are transformed with the donor plasmid first, and then used for a secondary transformation with the gapped plasmid. In the second protocol, cells are co-transfected with the gapped and the donor plasmids simultaneously. In both cases selection is made on LB plates containing kanamycin, which is encoded by the gapped plasmid. Only plasmids whose gaps were repaired give rise to kanR colonies. Gap repair can occur either by translesion DNA synthesis (TLS) or by HR, which can be distinguished by the specificity of the nucleotide present in the repaired plasmid at the site corresponding to the lesion. In case of an abasic site lesion, gap repair via TLS leads to incorporation of primarily an A opposite the lesion 21–23. However, because the donor plasmid was engineered to carry a T at the corresponding location, HR gap repair yields a T at the same site.
Fig. 1.
Outline of the gap-filling recombination repair assay.
A. A scheme of the sequential transformation protocol is presented. See text for details. B. A scheme of an assay that discriminates between strand-transfer and template-switching mechanisms of HR gap repair. If a T/G mismatch in the donor DNA is located at the position corresponding to the lesion, then gap filling by strand transfer will place a T opposite the lesion, whereas template switching will place a C at that position.
The adaptation of the two-plasmid system for the discrimination of the two alternative HR gap filling mechanisms, namely strand transfer and template switching, is outlined in Fig. 1B. The key component is a donor plasmid that contains a mismatch at the site corresponding to the abasic site in the gapped plasmid. When a donor containing a TG mismatch is used, HR by strand transfer will place a T across the abasic site, whereas HR via template switching will copy the G and therefore place a C across the abasic site (Fig. 1B; the scheme is meant to illustrate the discrimination principle between strand transfer and template switching, and not to provide a detailed mechanism; see Fig. 3 and its discussion below). In the absence of SOS induction TLS is very low across the abasic site, and expected to lead to the formation of primarily -1 frameshifts in this system 20; 21. The preservation of the mismatch is critical to the success of this approach, meaning that both mismatch repair and replication of the donor plasmid must be disabled. We therefore used a mutS strain, which lacks mismatch repair, but in which HR gap repair was unaffected (data not shown). To disable replication of the donor plasmid, we constructed a synthetic plasmid that had no origin of replication. Such donor plasmid cannot be maintained in E. coli, but upon co-transformation with the gap-lesion plasmid it survives long enough to allow HR gap repair. Under these conditions there is practically one copy of the donor plasmid per cell, which differs from the conditions used in the past, in which a high copy number plasmid was used 20. To examine the effect of the copy number of the donor plasmid on the efficiency of HR we assayed gap repair of the gap-lesion plasmid in the presence of homologous and heterologous donor plasmids, which were maintained at high (50–100), low (10–15) or single copy. The background gap repair, observed with the heterologous donor plasmids or without any donor was very low, and similar in all cases, with an average of 1.7±0.4%. Upon addition of the homologous donor, gap repair increased by 37-, 5-and 3-fold, for plasmids maintained at high, low and singly copies, respectively (Table 1s). The homologous origin-less synthetic plasmid caused a 2.6-fold increase in gap repair, similar to that of the single copy plasmid (Table 1s). DNA sequence analysis verified that whereas the background repair involved -1 frameshifts opposite the abasic site (primarily due to background TLS), the increase in gap repair in the presence of the homologous donors led to insertion of sequences from the donor plasmids (Table 1s), consistent with previous results 20.
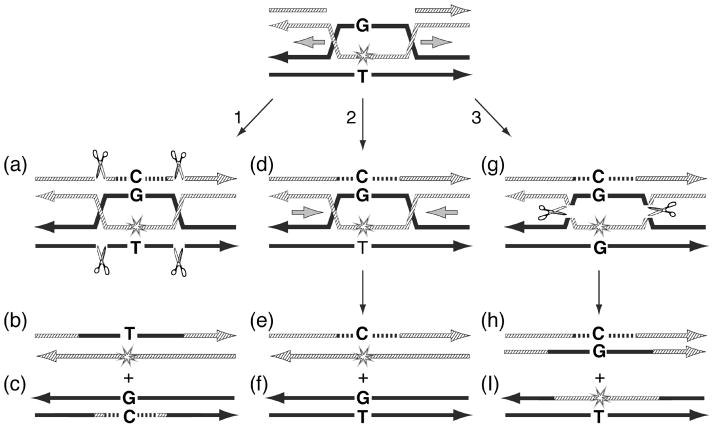
Fig. 3.
Hypothetical HR gap repair mechanisms involving two Holliday Junctions (HJ). HR is shown between a gap-lesion plasmid and a homologous donor containing a T:G mismatch at the site corresponding to the abasic site in the gap-lesion plasmid. (1), resolution of the two HJ in the vertical direction leads to gap filling via a strand transfer mechanism (product b), with a T opposite the site of the abasic site. (2), branch migration of the two HJ towards each other leads to dissolution (perhaps with the help of a topoisomerase), leading to a template switch gap recombination mechanism, which contains a C opposite the site of the abasic site. (3), resolution of the two HJ in the horizontal direction leads to template switching and strand transfer of the lesion-containing strand to the donor plasmid. This is also scored as template switching, because the selection is made for the plasmid that originally carried the gap-lesion. See text for further discussion.
The majority of gaps are repaired via the strand transfer mode of HR, but repair via template switching occurs too
The results of HR gap repair experiments such as described in Fig. 1B are shown in Table 1. Out of the 58 mutants isolated, 36 (62%) contained a T or a C, and could therefore be attributed to HR gap repair. Of these, 89% (32/36) isolates contained a T opposite the site corresponding to the abasic site. In order to rule out the possibility of a mismatch bias during HR, we performed similar experiments with a donor carrying a CA mismatch at the same location. In this case, strand transfer is expected to place a C opposite the abasic site, whereas template switching would put a T at the same location, thus deriving sequencing results that are exactly the opposite of the former donor plasmid. The DNA sequence analysis of 67 isolates, presented in Table 1, shows that 42 carried a C or a T at the relevant site, suggesting that 63% of the gap-lesion plasmids were repaired by HR gap repair, similar to the previous set of experiments. Consistent with the previous donor, a majority of isolates with this donor (32/42; 76%) carried a C opposite the site corresponding to the abasic site. Only 10/42 isolates (24%) had a T at that location. In both cases, all other isolates contained a −1 deletion at the location of the abasic site, consistent with background TLS across the abasic site by pol III holoenzyme 21; 24. Their relative substantial fraction in this type of experiments is due to a lower efficiency of HR caused by the need to use a single-copy and origin-less synthetic donor plasmid, as discussed above. Taken together these results indicate that the majority of HR gap repair events (64/78; 82%) occur via physical transfer of a homologous DNA segment from the donor. However, a substantial fraction of 18% of the gaps appears to be repaired by a template switching mechanism. The clear preference for strand transfer in our system is consistent with the early experiments demonstrating transfer of UV lesions from parental to daughter strands which were explained by a mechanism of HR repair 25; 26, and with studies performed in E. coli 27 with a plasmid carrying a defined site-specific lesion. To our knowledge there is no previous indications for in vivo HR gap repair via template switching.
Table 1.
Sequence analysis of the nucleotide opposite the lesion in gap repair experiments performed with donors carrying mismatches as markers for strand transfer versus template switching homologous recombination mechanisms
| Nucleotide opposite the lesion | Number of isolates
|
|
|---|---|---|
| Type of mismatch in donor plasmid | ||
| T/G1 | C/A2 | |
|
|
||
| T | 32 | 10 |
| C | 4 | 32 |
| A | 0 | 0 |
| G | 0 | 0 |
| −1 | 22 | 25 |
|
| ||
| Total isolates: | 58 | 67 |
| Total HR gap repair events3: | 36 | 42 |
| Strand Transfer HR gap repair events: | 89% | 76% |
The assay was performed essentially as previously described 20. DNA mixtures containing the gap-plasmid GP21 (with a site-specific abasic site; 100 ng), and the homologous origin-less plasmids pOFGP20T/G or pOFGP20C/A (300 ng) were used to co-transform E. coli mutS cells, defective in mismatch repair. Plasmids were isolated from kanR colonies, and subjected to DNA sequence analysis. The table shows the identity of the base located opposite the lesion in individual clones.
The construction of gap-lesion plasmids GP21 (with a synthetic abasic site), GP20 (without a lesion) were previously described 20; 21; 41. The origin-less plasmids were constructed in three steps as follow: (a) Plasmid pSKSL was digested with restriction nucleases BspE1 and BsaH1, and the resulting 3340 bp fragment was ligated to a BspE1-and BsaH1-cleaved 772 bp PCR fragment, which carried BspE1 and BsaH1 restriction sites, and the origin of replication from plasmid pSKSL. This yielded plasmid pOri2 that contained two origin of replication. (b) Plasmid pOri2 was restricted with BanI, deleting a 1021 bp fragment with the original replication origin of the plasmid. The 3092 bp fragment was then self-ligated to form plasmid pOri1. (c) Finally, the ori-less plasmid carrying the mismatch was prepared by a method similar to the preparation of the gapped plasmid, except that a duplex oligonucleotide carrying the T/G or C/A mismatches was used instead of the gapped duplex. Plasmid pOri1 was cleaved with BstX I and Bsa I and the resultant large fragment, which did not contain any origin of replication, was gel-purified, and ligated to the duplex oligonucleotide carrying the mismatch. The ligation product was gel purified again. The presence of mismatches was verified by DNA sequence analysis of the two strands, and the inability to replicate was verified by demonstrating the inability of the plasmid to transform E. coli cells to kanamycin resistance.
In this origin-less donor plasmid the T is located opposite the site corresponding to the lesion.
In this origin-less donor plasmid the C is located opposite the site corresponding to the lesion.
The total HR gap repair events do not include the −1 deletions, which are independent on the presence of a homologous plasmid (Table 1s).
HR gap repair is recA- and recF-dependent, but recB-independent
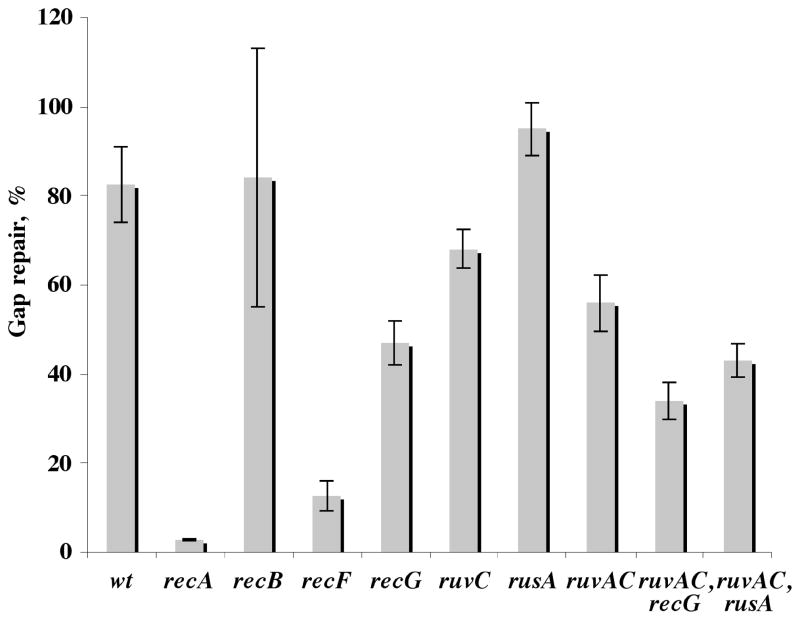
The genetic requirement for the HR gap repair reaction was examined by conducting the experiments in a series of mutants and their isogenic parents, using the sequential transformation protocol, a gap-lesion plasmid carrying an abasic site, and a high-copy donor. HR was very effective in repairing the gap-lesion plasmid, leading to a survival of 83%, and was completely recA-dependent (Fig. 2), consistent with our previous results 20. Interestingly, in the absence of RecF, plasmid survival was only 12.7±3.5%, suggesting that the majority of gap repair events are RecF-dependent. The residual gap repair was still via HR, as indicated by DNA sequence analysis that showed a majority of T nucleotide present opposite the site of the lesion (Table 2s). In contrast, in a recB21 mutant, gap repair was as effective as in the wild-type strain (84±29%), and descendents contained the marker of HR (Table 2s). The results of experiments utilizing this mutant suffered from a relatively large standard deviation due to the difficulties of maintaining plasmids in strains defective in the RecBCD nuclease 28; 29. In any case, the fact that gap repair did not require RecB suggests that the process does not require a double-strand break as an intermediate, since the recBCD nuclease is needed for processing of double-strand breaks in HR 30; 31. Our plasmid does not contain a χ site, which is required for the recombination activity of RecBCD 32,33. This is consistent with the notion that gap repair did not proceed via a DSB intermediate, because dsDNA is degraded by RecBCD in the absence of χ and therefore expected to have a big effect on plasmid survival. RecBCD-independent homologous recombination was reported in the past 34; 35.
Fig. 2.
Gap-lesion plasmid repair in recombination-deficient mutants. E. coli cells were transformed with either the heterologous partner plasmid pUC18 (ampR) or the homologous partner plasmid pFGP20/Tamp (ampR). AmpR-resistant colonies were used for a second round of transformation with the gap-lesion plasmid GP21 (kanR). The same colony was used in parallel for transformation with GP20 (gap-plasmid without lesion). All strains where assayed for their survival relative to GP20 transformants. The results presented here are the average of at list three independent experiments each with at list two repetitions. The strains used are identified by genotype and are AB1157 (wt), WBN10 (ΔrecA:: tn10), WBY166 (recB21), WBY220 (ΔrecF349), WBL16 (RecG265::cat), WBL19(RuvC67:: cat) AM821 (ΔrusA::kan), AM547 (ΔruvAC65), TNM1219 (ΔruvAC65, ΔrecG263::kan), AM888 (ΔruvAC65, ΔrusA:: kan).
All strains are isogenic derivatives of E. coli AB1157 (argE3 hisG4 leuB6 proA2 ter1 ara14 galK2 lacY1 mtl1 xyl5 thi1 tsx33 rpsL31 supE44). Strains E. coli WBL16 (recG265::cat), WBL19 (ruvC67::cat), and WBL13 (mutS::Tn10) are derivatives of AB1157 constructed by generalized P1 transudation using as donors the strains N4452 (recG265::cat), N4453 (ruvC67::cat), and MM294 (mutS::Tn10), respectively. Sources of other strains were: AM547 (Δruv(A–C)65), AM821 (ΔrusA::kan), AM888 (Δruv(A–C)65, ΔrusA:: kan), and TNM1219 (Δruv(A–C)65, ΔrecG263::kan), lab collection of R. G. Lloyd; N4453 (RuvC67:: cat), E. coli genetic stock center; MM294mutS (glnV44(AS) λ-rfbC1 0 endA1 spoT1 0 thi-1 hsdR17 creC510 mutS::Tn10), WBN2 (3recA:: Tn10), WBY 100 (Δ (umuDC)595:cat), WBY166 (recB21 ArgA:Tn10), and WBY220 (ΔrecF349 TnaA:Tn10), lab collection of Z. Livneh.
HR gap repair is partially dependent on proteins involved in the resolution of Holliday junction (HJ) intermediates
Analysis of HR gap repair was examined also in ruvA–C, recG and rusA mutants, which are involved in the resolution of Holliday junction (HJ) intermediates 7; 9; 36. As can be seen in Fig. 2, deleting the rusA resolvase gene had no effect on HR gap repair. Inactivation of the recG helicase gene, the ruvC resolvase gene, or the entire ruv operon had each a small effect, of up to 1.8-fold decrease, on HR gap repair. In the absence of both the ruv genes and recG HR gap repair decreased by 2.4-fold, down to 34±4%. Thus, HR gap repair involves the proteins of HJ resolution RuvA–C and RecG, but in their absence there seem to be alternative pathways that enable the recombination reaction. Indeed, DNA sequence analysis of plasmids isolated from these mutant cells showed that the majority contained the maker of HR (T nucleotide opposite the site of the abasic site), indicating an HR reaction (Table 2s). The involvement of RuvABC in HR repair is indicative of nucleolytic resolution of HJ, as expected for a strand exchange mechanism, whereas RecG is usually associated with an alternative pathway, not involving endonucleolytic resolution. The mild phenotypes in the absence of RuvC may be explained by the action of an alternative resolvase, with a contribution from RecG, acting perhaps as an alternative to RuvAB. In this context it should be mentioned that topoisomerase III was recently shown to act in a recombination pathway alternative to RuvABC 37. Alternatively, new proteins, such as YqgF, which has a predicted similarity to RuvC, might be involved in the process 38. The purified YqgF protein, however, shows no resolvase activity on synthetic HJ structures in vitro (R. G. Lloyd, G. J. Sharples and A. A. Mahdi, unpublished work). In addition, backup may be provided also by hitherto undiscovered resolvase activities of other known helicases, nucleases or topoisomerases 39; 40.
HR efficiently repairs a gap opposite a benzo[a]pyrene-guanine adduct
In order to examine the generality of the HR gap repair reaction with regards to the type of the DNA lesion, we used a gapped plasmid carrying a site-directed benzo[a]pyrene-guanine (BP-G) adduct. As can be seen in Table 2, the presence of a homologous donor plasmid increased the survival of the BP-G-containing gapped plasmid from a background level of 0.8±0.3% up to 69±7%, comparable to the effect of HR on the repair of a gapped abasic site DNA lesion 20 (Fig. 2). When a donor plasmid with a short heterology of 12 nucleotides at the site of the lesion was used for the repair of a BP-G gapped-plasmid the survival of the acceptor decreased to 27±3%, indicating the importance of sequence homology for HR on one hand, and the ability of RecA to overcome both DNA damages and heterologies during HR on the other hand, consistent with in vitro studies 7. It should be noted that the HR gap repair reaction is effective also for the very bulky DNA lesion BP-G. This is a significant finding since this lesion was bypassed very poorly by TLS in the same assay system (~1.5%), even when the SOS response was induced 41. It suggests that HR gap repair operates on a wide range of DNA lesions, including those that are poorly bypassed by TLS.
Table 2.
Gaps opposite the bulky benzo[a]pyrene-guanine adduct are efficiently repaired by homologous recombination
| Donor plasmid | Gap repair, % |
|---|---|
| None | 0.8±0.3 |
| pFGPB-cm | 68.5±7.1 |
| pFGP20/T-amp | 27.2±3.3 |
E. coli WBY100 cell where transformed with either pFGPBP-cm or pFGP20/T-amp, and selected on LB-cm or LB-amp plates, respectively. Cells carrying the donor plasmid were then, re-transformed with the gap-lesion plasmid GP-BPG1 carrying a benzo[a]pyrene-G adduct. BP-G1 Tansformants where assayed for their ability to survive on kanamycin plats relative to GP20 transformants. The efficiency of gap repair was calculated by dividing the number of colonies obtained with the gap-lesion plasmid, by the number of colonies obtained with GP20. Typically plates with 50–200 colonies were counted, except for the background plates, where colony counts were lower. The results are the averages of 3 independent experiments.
Plasmid pFGPBP-cm is a descendent of the gap-lesion plasmid GP-BPG1-cm obtained by introducing the latter into E. coli and selecting for cmR colonies. It has a G at the position corresponding to the lesion in GP-BPG1–cm. FGP20/T-amp was previously described 20. It contains a short stretch of 12 nucleotides that is heterologous to the cognate sequence surrounding the lesion in the gap-lesion plasmid GP-BPG1, and contains a T at the position corresponding to the lesion in plasmid GP-BPG1-cm.
Mechanisms of HR gap repair
The results presented in this study indicate that strand transfer predominates over template switching, however the detailed mechanisms of these two alternative pathways are not fully understood yet. For example, an alternative to the HR scheme described in Fig. 1B, is a mechanism involving two HJ (Fig. 3). In this scheme, like in Fig. 1B, the donor plasmid contains a G:T mismatch at the site corresponding to the lesion in the gap-lesion plasmid, such that strand transfer and strand switching can be discriminated. Vertical resolution of the two HJ (Fig. 3 pathway 1 structure a) results in a strand transfer from the donor, leading to a T opposite the site of the lesion (Fig. 3, structure b). The two HJ can resolve also by dissolution, with the help of a topoisomerase 38, as shown in pathway 2 (Fig. 3). This pathway involves a template switch and DNA synthesis, which after migration of the two HJ towards each other (dissolution; structure d in Fig. 3) will lead to the formation of a strand switch repair product, with a C opposite the site of the lesion (structure e in Fig. 3). Pathway 3 shows the products expected from horizontal resolution of the two HJ (structures g-i in Fig. 3). In this case, which also involves strand switching (structure g in Fig. 3), the lesion region is transferred to the donor plasmid. However, since the selection is made to the plasmid that originally contained the gap-lesion, the result will be an isolate with the marker of strand switching, namely a C opposite the site of the lesion (structure h in Fig. 3). Vertical resolution of one HJ and horizontal resolution of the other was expected to lead to the fusion of the donor and acceptor plasmids. However, such fused plasmids were not observed in experiments conduced with the gap-lesion plasmid and donor plasmid containing a mismatch. Further studies are needed to elucidate the details of such putative mechanisms during HR gap repair.
Conclusions
Multiple mechanisms were proposed for the tolerance of DNA damages depending in part on whether the encounter of replication complexes with lesions leads to arrested replication forks, or the formation of DSG 5–12. The two new studies mentioned in the Introduction section 13; 14 highlight again the importance of DNA gap formation following DNA damage and the need to address the mechanisms of their formation and repair. Our model assay system clearly shows that HR gap repair does effectively occur in E. coli, even across bulky lesions such as BP-G, and that it is RecA-and RecF- dependent as was previously proposed by indirect evidence for chromosomal recombination repair of daughter strand gaps. It also suggests the existence of an alternative resolution pathway, which does not involve RuvABC and RecG. To our knowledge this is the first quantitative assessment and direct evidence in vivo for both the strand transfer and the template switching mechanisms of HR gap repair. The challenge is now to develop new strategies, with similar resolution, that could be used to study these mechanisms in the chromosome.
Supplementary Material
Acknowledgments
This research was supported by grants from The US-Israel Binational Science Foundation (No. 1999141 to Z. Livneh & R. Kolodner), from FAMRI, the Flight Attendants Medical Research Institute, Florida (to Z. Livneh), the UK Medical Research Council (to R. G. Lloyd), and the National Institutes of Health, USA (grant CA 099194 to Nicholas Geacintov).
Abbreviations
- HR
homologous recombination
- DSG
daughter strand gap
- DSB
double-strand break
- BP-G
benzo[a]pyrene-guanine adduct
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 3.Meneghini R. Gaps in DNA synthesized by ultraviolet light-irradiated WI38 cells. Biochim Biophys Acta. 1976;425:419–427. doi: 10.1016/0005-2787(76)90006-x. [DOI] [PubMed] [Google Scholar]
- 4.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3, and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 5.Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. Replication of damaged DNA and the molecular mechanism of ultraviolet light mutagenesis. CRC Crit Rev Biochem Mol Biol. 1993;28:465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- 6.Kreuzer KN. Interplay between DNA replication and recombination in prokaryotes. Annu Rev Microbiol. 2005;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- 7.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington DC: 2006. [Google Scholar]
- 8.Friedberg EC. Suffering in silence: The tolerance of DNA damage. Nature Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 9.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nature Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 10.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 11.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 13.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cel. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 15.Smith KC, Meun DHC. Repair of radiation-induced damage in Escherichia coli: I. Effect of rec mutations on post-replication repair of damage due to ultraviolet light. J Mol Biol. 1970;51:459–472. doi: 10.1016/0022-2836(70)90001-x. [DOI] [PubMed] [Google Scholar]
- 16.Sedgwick SG. Genetic and kinetic evidence for different types of postreplication repair in Escherichia coli B. J Bacteriol. 1975;117:1077–1081. doi: 10.1128/jb.123.1.154-161.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang TV, Smith KC. Mechanisms of recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol. 1983;156:1093–1098. doi: 10.1128/jb.156.3.1093-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesan AK, Seawell PC. The effect of lexA and recF mutations on post-replication repair and DNA synthesis in Escherichia coli K-12. Mol Gen Genet. 1975;141:189–205. doi: 10.1007/BF00341799. [DOI] [PubMed] [Google Scholar]
- 19.Courcelle J, Ganesan AK, Hanawalt PC. Therefore, what are recombination proteins there for? BioEssays. 2001;23:463–470. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- 20.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 21.Reuven NB, Tomer G, Livneh Z. The mutagenesis proteins UmuD’ and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 22.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA and SSB, and specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 23.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 24.Tomer G, Reuven NB, Livneh Z. The b subunit sliding DNA clamp is responsible for unassisted mutagenic translesion replication by DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1998;95:14106–14111. doi: 10.1073/pnas.95.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganesan AK. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K12. J Mol Biol. 1974;87:103–119. doi: 10.1016/0022-2836(74)90563-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang TV, Smith KC. RecF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J Bacteriol. 1984;158:727–729. doi: 10.1128/jb.158.2.727-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandya GA, Yang IY, Grollman AP, Moriya M. Escherichia coli responses to a single DNA adduct. J Bacteriol. 2000;182:6598–6604. doi: 10.1128/jb.182.23.6598-6604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen A, Clark AJ. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986;167:327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biek DP, Cohen SN. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith GR. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 31.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 32.Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: Location and orientation-dependence of the cutting. Cell. 1985;41:153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- 33.Dixon DA, Kowalczykowski SC. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 34.James AA, Morrison PT, Kolodner R. Genetic recombination of bacterial plasmid DNA. Analysis of the effect of recombination-deficient mutations on plasmid recombination. J Mol Biol. 1982;160:411–430. doi: 10.1016/0022-2836(82)90305-9. [DOI] [PubMed] [Google Scholar]
- 35.Corre J, Cornet F, Patte J, Louran JM. Unraveling a region-specific hyper-recombination phenomenon: Genetic control and modalities of terminal recombination in Escherichia coli. Genetics. 1997;147:979–989. doi: 10.1093/genetics/147.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eggleston AK, West SC. Exchanging partners: recombination in E. coli. Trends Genet. 1996;12:20–26. doi: 10.1016/0168-9525(96)81384-9. [DOI] [PubMed] [Google Scholar]
- 37.Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, DiGate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 38.Aravind L, Makarova KS, Koonin EV. Holliday junction resolvases and related nucleases: identificatin of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Karow JK, Hickson ID. Genetic recombination: Helicases and topoisomerases link up. Curr Biol. 1999;9:R518–R520. doi: 10.1016/s0960-9822(99)80325-x. [DOI] [PubMed] [Google Scholar]
- 41.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells. The Role of DNA polymerase k. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.