Abstract
Long non-coding RNAs have become the focus of considerable interest over the past few years. Intriguing novel functions have been reported for lincRNAs. Three recent papers identify lincRNAs that work in a more conventional way—encoding protein—in each case a small polypeptide with an interesting biological activity (Magny et al, 2013; Pauli et al, 2014), (Bazzini et al, 2014).
See also: EG Magny et al (September 2013), A Pauli et al (February 2014) and AA Bazzini et al (May 2014)
Conventional protein-coding genes account for only a fraction of the RNA transcribed in animal genomes. Many of us grew up thinking that RNAs came in two flavours: those with protein-coding capacity and non-coding RNAs with structural roles, in the form of ribosomal RNAs, tRNAs, snoRNAs, etc. Interest in other forms of long non-coding RNAs (lincRNAs) has been growing over the past decade, building in part on the fact that many lincRNAs are the precursors for micro-RNA biogenesis. In some cases, the miRNA is the only known product of a primary transcript that can be tens of Kb in length. But there is much more to lincRNAs: functions include X inactivation and other forms of chromatin modification (Gupta et al, 2010; Tian et al, 2010), enhancer-like functions regulating transcription (Orom et al, 2010) and regulation of post-transcriptional gene expression by functioning as micro-RNA sponges (Hansen et al, 2013; Memczak et al, 2013). Recent papers from the Couso, Schier and Giraldez/Rajewsky laboratories now bring us full circle, assigning a protein-coding function to lincRNAs (Magny et al, 2013; Pauli et al, 2014), (Bazzini et al, 2014) (Fig1).
Figure 1. lincRNAs have been thought to act as non-coding molecules.
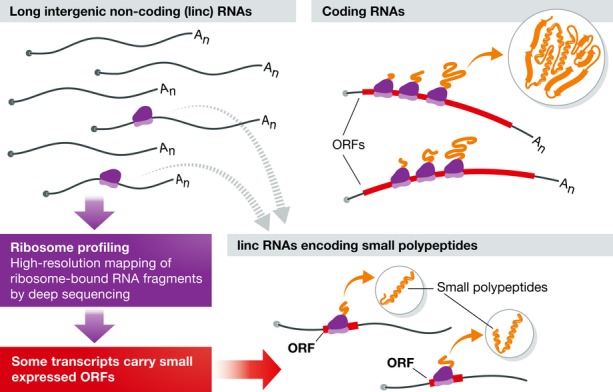
Recent studies identified several lincRNAs that, unexpectedly, encode for small polypeptides.
These stories have a precedent, or two. In 2004, the Drosophila polar granule component gene (pgc) was reported to function as a non-coding RNA that acted in the embryonic primordial germ cells to prevent transcription of the zygotic genome (Martinho et al, 2004). pgc RNA localizes to the nascent germ cells in the embryo to transiently block activation of RNAPolII. A few years later, in 2008, the Nakamura and Ladurner laboratories reported that the functional product of the pgc gene was a peptide that blocks RNAPolII by preventing an activating phosphorylation event mediated by P-Tefb (Hanyu-Nakamura et al, 2008; Timinszky et al, 2008). A second “former lincRNA” produced by the tarsal-less/polished rice/mille-pattes gene turns out to encode small peptides that control epithelial morphogenesis in Drosophila and Tribolium (Savard et al, 2006; Galindo et al, 2007; Kondo et al, 2007). Intriguingly, this peptide promotes N-terminal processing of the transcription factor Shavenbaby, converting it from a repressor to an activator (Kondo et al, 2010).
The recent report from the Couso laboratory built on their previous work on tarsal-less to search for additional Drosophila transcripts that might encode small peptides. They identified a lincRNA that is expressed in muscle and encodes small peptides (Magny et al, 2013). Interestingly, these peptides are related to the vertebrate peptides Sarcolipin and Phospholamban in sequence and predicted structure. Mutants lacking the fly lincRNA, which they name sarcolamban, show a defect in cardiac function. Based on the known role of the human peptides in calcium uptake by the sarcolemmal endoplasmic reticulum, Magny et al were able to assign a molecular function to the fly peptides and showed that the heart problem could be partially corrected by expression of the human peptides in the fly mutant. This example of functional conservation suggests an ancient origin for the regulation of calcium uptake by this family of small peptides.
The Schier and Giraldez/Rajewsky laboratories set out to survey Zebrafish lincRNAs for protein-coding potential. Both groups made use of ribosome profiling, a method that allows high-resolution mapping of ribosome-bound RNA fragments by deep sequencing (Ingolia et al, 2009). The Schier laboratory specifically looked for novel secreted proteins (Pauli et al, 2014). They found 700 predicted open reading frames that had not been annotated previously as protein-coding transcripts. Over 80% were conserved in other vertebrates and many encode polypeptides of considerable length—so it is unclear why they were missed before. Some of these transcripts had been annotated as lincRNAs. 28 of these were predicted to be new secreted proteins, with signal peptides, but lacking transmembrane domains. The new paper focused on the role of one of these loci, now named toddler, which encodes a secreted peptide. Using TALEN technology to produce mutants disrupting the peptide coding sequence, Pauli et al (2014) provide evidence that the peptide functions as a signal to promote cell motility in the early fish embryo. Toddler peptide, also known as ELABELA, acts as an activator of a G protein-coupled receptor called Apelin to promote cell movements required for heart development (Chng et al, 2013; Pauli et al, 2014).
The Giraldez and Rajeswky laboratories used ribosome profiling and developed new computational methods to identify Zebrafish lincRNAs that might encode novel short proteins (Bazzini et al, 2014). The “ORFscore” method performed well with annotated Zebrafish RefSeq transcripts. Applying ORFscore to 2,450 potential non-coding RNAs identified 190 transcripts with the potential to encode small polypeptides (20–100 aa) and further 89 predicted to encode longer peptides. This report introduces a second computational tool, called micPDP, which uses a conservation-based approach to identify novel small peptides. Of 63 conserved short Zebrafish peptides identified by micPDP, 23 were also found by ribosome profiling. Interestingly, a similar analysis of human small ORFs by the two approaches yielded even more limited overlap: seven small ORFs were identified by both methods (out of 173 found by micPDP and 135 by ORFscore).
These studies point to an emerging biology of small peptides. Why have these remained relatively obscure until now? One reason is statistical. Genome annotation has tended to filter out potential short open reading frames because they are simply too numerous. Conservation across genomes can help predict function. But, as geneticists, we know that conservation is not a prerequisite for function. Likewise, there can be reasons other than production of a peptide that might explain ribosome binding to RNA, and indeed phased binding. The methods reported in these studies are tantalizing, but clearly much work is needed to validate the predictions and to explore function. To date, functions have been assigned to only a few of these former lincRNAs, but the versatile tools available for genome manipulation should allow a rapid follow-through. We can expect to be hearing a lot about the small protein world in years to come.
Conflict of interest
The author declares that he has no conflict of interest.
References
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng SC, Ho L, Tian J, Reversade B. ELABELA: A hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27:672–680. doi: 10.1016/j.devcel.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le NF, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF. Toddler: an embryonic signal that promotes cell movement via apelin receptors. Science. 2014:343. doi: 10.1126/science.1248636. , 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Bortfeld M, Ladurner AG. Repression of RNA polymerase II transcription by a Drosophila oligopeptide. PLoS ONE. 2008;3:e2506. doi: 10.1371/journal.pone.0002506. [DOI] [PMC free article] [PubMed] [Google Scholar]


