Abstract
The Hippo pathway plays a critical role in organ size control and tumorigenesis. YAP (and also its homologue TAZ) is a transcription co-activator that mediates the biological functions of the Hippo pathway. YAP activity is inhibited upon phosphorylation by the Lats kinases (reviewed in Yu & Guan, 2013). In a recent study in Cell, Mori et al (2014) uncover a new function of YAP in global microRNA (miRNA) biogenesis by the modulation of the Microprocessor machinery. The results also suggest that this novel YAP function may contribute to cell growth control and tumorigenesis.
See also: M Mori et al (February 2014)
YAP inactivation is involved in cell–cell contact inhibition. When cells are cultured at low density, YAP localizes to the nucleus to regulate gene expression; in contrast, under high cell density, YAP remains inactive in the cytoplasm (Fig1A and B; Zhao et al, 2007). miRNA biogenesis is also affected by cell–cell contact, with increasing levels of mature miRNAs in cells cultured at high density (Hwang et al, 2009). The new paper by Mori et al (2014) nicely connects these so far separate observations to report that YAP acts upstream in the process of miRNA biogenesis and is crucial for miRNA expression in response to cell–cell contact signals. In the absence of cell contact-mediated signals, nuclear YAP interacts with DDX17 (DEAD Box Helicase 17, also known as p72), which sequesters DDX17 away from DROSHA and DGCR8 (two major components of the Microprocessor). DDX17 recognizes a sequence motif in the 3′ flanking sequence of pri-miRNAs, and this facilitates cleavage by Microprocessor. When DDX17 is associated with YAP, the processing of pri-miRNA to pre-miRNA by Microprocessor is reduced, limiting overall miRNA production (Fig1A). Cell contact-induced cytoplasmic retention of YAP prevents DDX17 binding and permits DDX17 interaction with DROSHA and DGCR8 to mediate effective pri-miRNA cleavage and miRNA production (Fig1B).
Figure 1. Hippo pathway regulates miRNA processing.
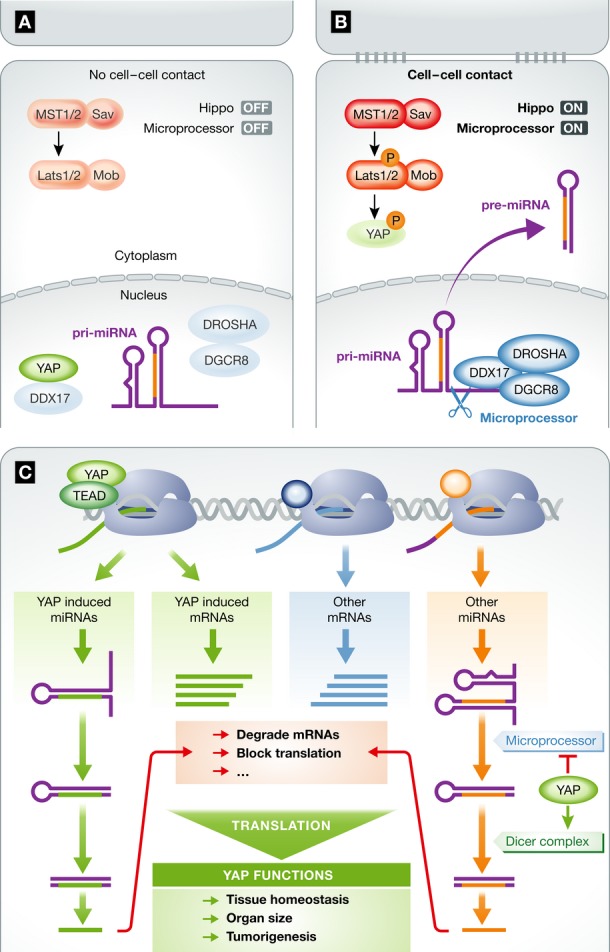
(A) At low cell density, Hippo pathway kinases are inactive, YAP is unphosphorylated and localizes in nucleus. Nuclear YAP sequesters DDX17 and prevents it from forming a complex with DROSHA and DGCR8, thereby resulting inefficient miRNA processing. (B) At high cell density, cell–cell contact activates Hippo pathway kinases, YAP is phosphorylated and retained in cytoplasm. DDX17 is dissociated from YAP and forms an effective Microprocessor with DROSHA and DGCR8, therefore promoting processing of pri-miRNAs to pre-miRNAs. (C) Transcription-dependent and independent roles of YAP in RNA biogenesis. YAP, through TEAD (and maybe other transcription factors), induces expression of many mRNAs and miRNAs. On the other hand, YAP modulates miRNA processing by regulating Microprocessor or Dicer complex. MicroRNAs may regulate stability and translation of diverse mRNAs. Both transcription-dependent and independent mechanisms may contribute to YAP-dependent global gene expression to control organ growth and tumorigenesis.
YAP is a transcription co-activator, which modulates gene expression by interacting with transcription factors such as TEAD1-4 (Fig1C). It is believed that YAP exerts its biological functions by regulating the transcription of genes involved in cell proliferation and cell death (Yu & Guan, 2013). YAP can directly induce expression of some miRNAs such as miR-29 (Tumaneng et al, 2012). The novel findings by Mori et al now suggest an even broader, regulatory role of YAP by functioning as a master regulator for miRNA processing. Additionally to its described effect in regulating mRNA and miRNA transcription, YAP appears to regulate miRNA processing in a transcription-independent manner. Indeed, one could infer that that YAP-regulated miRNAs may form a regulatory loop by modulating the stability or translation efficiency of mRNAs induced by YAP and other transcription factors (Fig1C). It's, however, noteworthy that some findings in Mori et al (2014) are not consistent with a recent report (Chaulk et al, 2014) suggesting some mature miRNAs being rather negatively correlated with cell density, and YAP induces the activity of DICER complex to enhance the processing of pre-miRNAs to mature miRNAs (Fig1C). Though further studies would be needed to clarify these differences, one simple explanation is that YAP may induce processing of some while inhibit other pre-miRNAs.
A global repression of miRNA expression is frequently observed in many different types of cancers (Lu et al, 2005); however, it's not clear what contributes to the miRNA biogenesis defects in cancers. Mori et al (2014) link YAP activation and miRNA repression in cancers, which is consistent with an oncogenic role of YAP. Using genetic tools, Mori et al (2014) demonstrate that miRNA production is decreased in cells with high YAP activity and also in YAP-driven tumors, and the miRNA repression results in the activation of oncoproteins such as MYC, suggesting an important role of miRNA biogenesis in YAP-induced tumorigenesis.
Further, the down-regulation of miRNA processing can play a causative role in tumorigenesis. As one example, depletion of Microprocessor components or the DICER complex leads to cellular transformation (Kumar et al, 2007). It is therefore reasonable to speculate that YAP induces tumorigenesis via global miRNA suppression. Previously, it has been shown that the YAP-TEAD binding is critical for YAP-induced cell transformation, and a YAP-TEAD binding-defective YAP mutant (S94A) failed to transform NIH3T3 cells (Zhao et al, 2009). In addition, a dominant-negative TEAD completely abrogates the tumor-promoting effect of YAP in mice liver (Liu-Chittenden et al, 2012). These data suggest an essential role of YAP-TEAD interaction and its transcriptional output on tumorigenesis. In contrast, Mori et al (2014) suggest additional, transcription-independent mechanisms for YAP, based on a YAP mutant (S94A) that effectively represses miRNA expression. Thus, it would be critical to assess the importance of YAP-DDX17 interaction and the transcription-independent miRNA biogenesis in the biology controlled by the Hippo pathway. For instance, does DDX17 function as a bona fide tumor suppressor? Is DDX17 involved in organ size control downstream of the Hippo pathway? Answers to these questions will be critical to corroborate the broader significance of these new and really intriguing findings.
The rather unexpected discoveries by Mori et al (2014) and Chaulk et al (2014) indicate a crucial role of YAP not only in the expression of mRNAs but also as a regulator in microRNA processing. There is no doubt that these results widen our understanding of the Hippo pathway. The exact function of the multitude of YAP target genes for its respective biological effects is far from being fully understood. One may speculate that both the genes directly induced by YAP and those from its new involvement in miRNA processing collectively contribute to the physiological output of YAP and the Hippo pathway.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Chaulk SG, Lattanzi VJ, Hiemer SE, Fahlman RP, Varelas X. The hippo pathway effectors TAZ/YAP regulate dicer expression and microRNA biogenesis through Let-7. J Biol Chem. 2014;289:1886–1891. doi: 10.1074/jbc.C113.529362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci U S A. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates Microprocessor and links cell density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]


