Abstract
Mouse Slc39a8 and Slc39a14 genes encode ZIP8 and ZIP14, respectively, which are ubiquitous divalent cation/(HCO3−)2 symporters responsible for uptake of Zn2+, Fe2+ and Mn2+ into cells. Cd2+ and other toxic nonessential metals can displace essential cations, thereby entering vertebrate cells. Whereas Slc39a8 encodes a single protein, Slc39a14 has two exons 4 which, via alternative splicing, give rise to ZIP14A and ZIP14B; why differences exist in cell-type-specific expression of ZIP14A and ZIP14B remains unknown. Inflammatory stimuli have been associated with ZIP8 and ZIP14 up-regulation, but a systematic study of many tissues simultaneously in a laboratory animal following inflammatory cytokine exposure has not yet been reported. Herein we show that C57BL/6J male mice—treated intraperitoneally with lipopolysaccharide (LPS), or the proinflammatory cytokines tumor necrosis factor (TNF) or interleukin-6 (IL6)—exhibited quantatively very different, highly tissue-specific, and markedly time-dependent up- and down-regulation of ZIP8, ZIP14A and ZIP14B mRNA levels in twelve tissues. Magnitude of the inflammatory response was confirmed by measuring the proinflammatory cytokine TNF, IL6 and interleukin-1β (IL1B) mRNA levels in the same tissues of these animals. Our data suggest that most if not all tissues use ZIP8, ZIP14A and/or ZIP14B) for Zn2+ uptake, some tissues under basal conditions and others moreso when inflammatory stressors are present; collectively, this might lead to substantial alterations in plasma Zn2+ levels, due to Zn2+ redistribution not just in liver, but across many vital organs. In the context of cadmium-mediated toxicity, our data suggest that tissues other than liver, kidney and lung should also be considered.
Keywords: ZIP transporter gene family, Slc39 gene family, ZIP8 Zn2+/(HCO3−)2 symporter, ZIP14 Zn2+/(HCO3−)2 symporter, inflammation, tumor necrosis factor, interleukin-6, lipopolisaccharide
Introduction
Characterization of the ZIP8 and ZIP14 transporters has led to greater appreciation of interactions between Zn2+ uptake, intracellular Zn2+ levels, and inflammation. Following discovery of variable response to cadmium toxicity among inbred strains of mice, the Cdm locus was shown to be primarily associated with this phenotypic difference1;2. Subsequently, the mouse Slc39a8 gene was identified3, and its encoded ZIP8 protein found to be a divalent metal/bicarbonate symporter4;5. Direct proof that the Slc39a8 gene was indeed the Cdm locus was confirmed by bacterial artificial chromosome (BAC) transgenesis; a BAC carrying only the Slc39a8 gene plus 87 kb 5′- and 18 kb 3′-flanking regions from a “cadmium-sensitive” mouse—inserted into the genome of a “cadmium-resistant” mouse—reverted the phenotype to cadmium sensitivity6;7.
The SLC39 family comprises 14 genes highly conserved between mouse and human; phylogenetic analysis showed mammalian SLC39A8 was evolutionarily most closely related to SLC39A14, the two genes having diverged from one another less than 420 million years ago7;8. The Slc39a14 gene encodes ZIP14, also a divalent metal/bicarbonate symporter8. Both ZIP85 and ZIP149 transport an electroneutral complex, Zn2+/(HCO3−)2, in which Zn2+ can be displaced by Fe2+ or Mn2+, as well as nonessential metal cations including Cd2+, Hg2+, Pb2+, Pt2+ and U2+9. Others have also demonstrated that ZIP810–13 and ZIP1412–16 function to import endogenous Zn2+, Fe2+ and/or Mn2+ into the cell. In fact, a hypomorphic mouse line—expressing ~10% of wild-type ZIP8 levels in all tissues and exhibiting markedly lowered Zn2+ and Fe2+ tissue levels—suffers severe defects in hematopoiesis and in multiple-organ organogenesis, which are neonatal-lethal17.
From the beginning of mammalian ZIP8 and ZIP14 studies, there has been a close association with inflammatory stimuli. Mammalian ZIP8 was discovered when a cDNA library—prepared from human monocytes stimulated with M. bovis BCG cell wall—was screened to identify novel genes induced by inflammatory danger signals; inflammatory cytokines such as tumor necrosis factor (TNF) were shown to be potent inducers of the ZIP8 transcript18.
Subsequent cytokine-activation studies include: lipopolysaccharide (LPS), TNF and interleukin-6 (IL6) induction of ZIP14 in mouse liver but not small intestine or spleen19; TNF-mediated up-regulation of ZIP8 but not ZIP14 in human lung10; effects of ZIP8 overexpression vs siRNA-mediated ZIP8 knockdown on Zn2+ levels during expression of interferon-γ in human T-cells20; recognition of ZIP14 participation in gluconeogenesis as well as G-protein-coupled receptor-mediated signaling in mouse bone, pituitary gland and liver21; LPS-induced endotoxemia stimulating ZIP14 levels in liver, white adipose tissue and muscle of Slc39a14(−/−) knockout mice22; importance of increased Zn2+ levels during ZIP14- or nitric oxide-mediated resistance to LPS-induced apoptosis in sheep pulmonary artery endothelial cell cultures23; identification of a negative feedback loop in fetal fibroblasts from Slc39a8-hypomorphic mice—involving ZIP8, IκB kinase and NFκB—which directly regulates innate immune function through coordination of Zn2+ metabolism24; demonstration of LPS as a strong inducer of ZIP14 in human macrophages25; and recent demonstration that “the Zn2+-ZIP8-MTF1 axis” represents an essential catabolic regulator during the pathogenesis of osteoarthritis26.
These studies demonstrate the ubiquitous nature of inflammatory cytokine-induced ZIP8 and/or ZIP14 in numerous mammalian tissues and cell types; however, these previous ZIP8 and ZIP14 studies had shown LPS or TNF treatment usually at 32 h, and IL6 treatment at 16 h, and only in a few tissues. Thus, a systematic examination of numerous tissues in the same animal, as a function of time following administration of inflammatory stimuli, has not yet been reported. Herein we show quantitative real-time polymerase chain reaction (qRT-PCR) measurements of ZIP8 and ZIP14 and cytokine mRNA levels in 12 tissues of C57BL/6J mice over a 32-h period after LPS and TNF administration, and over a 16-h period following IL6 treatment.
Materials and Methods
Chemicals
LPS, TNF, and IL6 were purchased from Sigma-Aldrich (St. Louis, Missouri). Dilutions were made in 0.09% NaCl (saline) solution and filtered through a 0.22-μm syringe prior to injection of the mice.
Treatment of Animals, Harvesting of Tissues
C57BL/6J male mice between 6 and 8 weeks of age were purchased from The Jackson Laboratory (Bar Harbor, Maine); following acclimation to our mouse colony for 5–7 days, these mice were studied. All mouse experiments were conducted in accordance with the National Institutes of Health standards for the care and use of experimental animals and University Cincinnati Medical Center Institutional Animal Care and Use Committee [protocol #11-09-12-01; approved 5 September 2008 → 4 Sept 2011 → 3 Sept 2014].
Subsequent to intraperitoneal treatment with LPS (5 mg/kg), TNF (5 μg/kg), or IL6 (100 μg/kg), the mice (N=3 per time-point) were killed at 2, 4, 8, 16 or 32 h; untreated mice provided the zero-hour time-point, and all experiments were repeated at least twice. Twelve tissues (collected at the six time-points) included liver, proximal small intestine (PSI), kidney, lung, heart, brain, pancreas, spleen, testis, urinary bladder, skin, and preputial gland duct (PGD). PSI samples represent the first 4 cm beyond the pyloric sphincter. PGD samples were isolated from tissues under the skin of the penis; surrounding fat was carefully separated so that the preputial gland could be harvested without breaking, as well as with virtually no other attached tissue (surrounding skin, cartilage, collagenous tissue or muscle).
Total RNA Preparation
With Tri-Reagent (TR18, Molecular Research Center, Inc.; Cincinnati, OH), total RNA was isolated from the 12 above-mentioned organs or tissues, following the protocol recommended by the vendor.
Reverse Transcription and qRT-PCR
Total RNA (2.5 μg) from each sample was used as template for reverse transcription and primed with oligo(dT), using the (Verso cDNA kit, AB-1453/B, Thermo Scientific; Waltham, MA). Total RNA (2.5 μg) was added to reactions containing 3.8 μM oligo(dT)20 and 0.77 mM dNTP, to a final volume of 13 μL; reactions were incubated at 65°C for 5 min, and then 4°C for 2 min. Next, we added 7 μL of solution containing 14 mM dithiothreitol and 40 units of RNaseOUT Recombinant RNase inhibitor™ (Verso cDNA kit). After incubation at room temperature for 2 min, 1 μL of reverse transcriptase was added to each sample. Reaction tubes were incubated at 42°C for 30 min, followed by 75°C for 10 min (to inactivate reverse transcriptase), and placed immediately on ice. Diethylpyrocarbonate (DEPC)-treated distilled water (80 μL) was added to dilute the cDNA that had been generated, and the resultant mixture was stored at −80°C until use.
We performed qRT-PCR in the Bio-Rad DNA Engine Opticon 2™ (Bio-Rad Laboratories; Hercules, CA), using iQ SYBR Green Supermix™ (170-8882, Bio-Rad Laboratories). The housekeeping gene’s β-actin mRNA (ACTB) was employed as the internal control. Standard curves using serial dilutions of total RNA resulted in excellent linearity (r = 0.99), indicating our qRT-PCR results were valid. All primers used are available upon request.
Statistics
All experiments were confirmed in at least triplicate and repeated twice. Statistical analysis was performed using the sofware SPSS version 13.0 (Microsoft®; Somers, NY). Data are presented as means ± S.E.M. of triplicate samples. P-values <0.05 are regarded as statistically significant.
Results and Discussion
Whereas SLC39A8 and SLC39A14 originated from one ancestral gene in fish, they diverged into two distinct genes (on different chromosomes) after the sea animal/land animal split7;8. Although the gene structures are similar, i.e. both mouse genes having three noncoding alternative exons-1 and eight coding exons, Slc39a14 contains alternative exons 4a and 4b. Thus, different amino acids in exons 4a and 4b were found to cause two distinct proteins having differential expression patterns in various tissues7;8. To date, no antibody has been generated that distinguishes between the two proteins. Km values for Zn2+, Fe2+ and Cd2+ uptake in Xenopus oocytes, nonetheless, were shown to be distinctly very different between ZIP14A and ZIP14B9: 0.36 μM vs 0.78 μM for Zn2+, respectively; 0.19 μM vs 0.64 μM for Fe2+, respectively; and 0.54 μM vs 0.31 μM for Cd2+, respectively. However, the significance of tissue-specific functions of these two ZIP14 distinct proteins remains to be determined. It should be noted that, however, by using different primers, one can quantify ZIP14A and ZIP14B mRNA levels by qRT-PCR in various tissues.
Effects of LPS, TNF and IL6 on ZIP mRNA Levels
LPS treatment (Figure 1A; Table 1. A; Table S1. A)
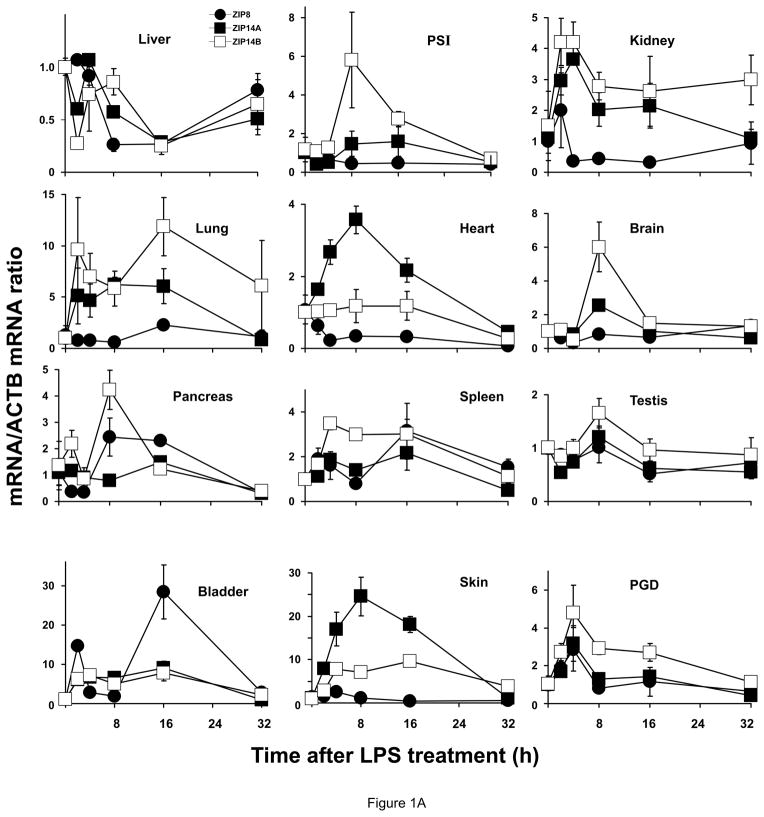
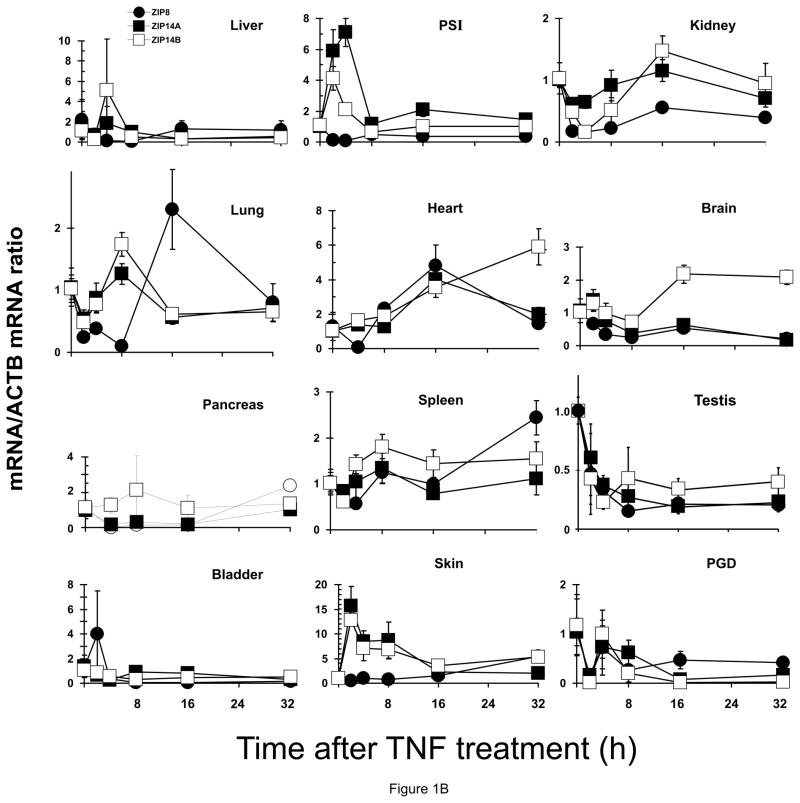
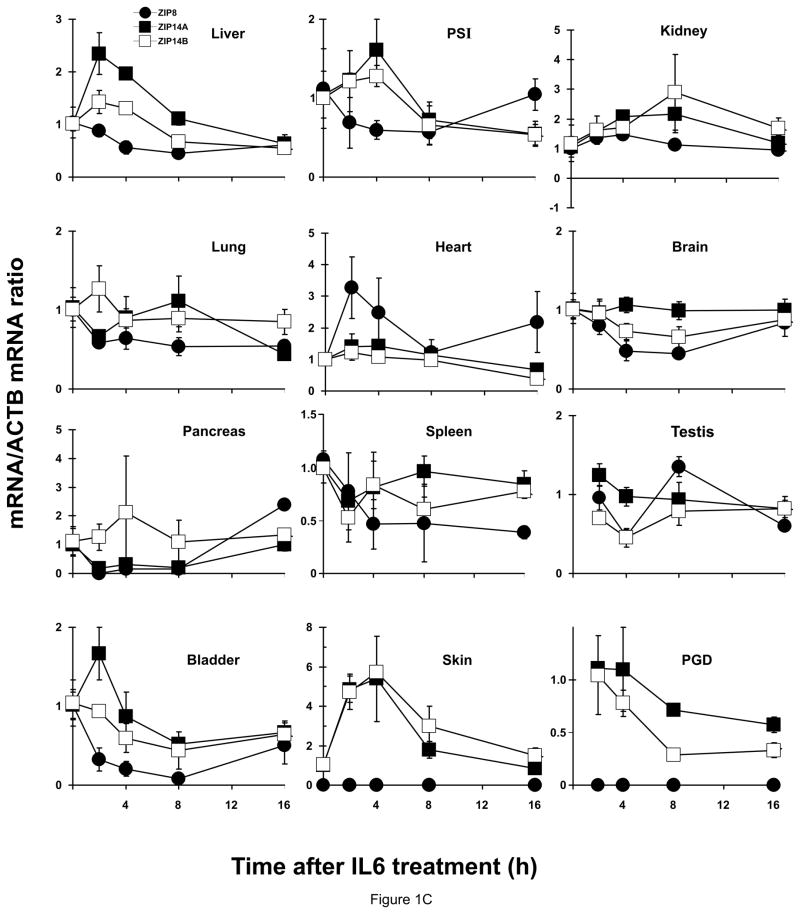
Figure 1.
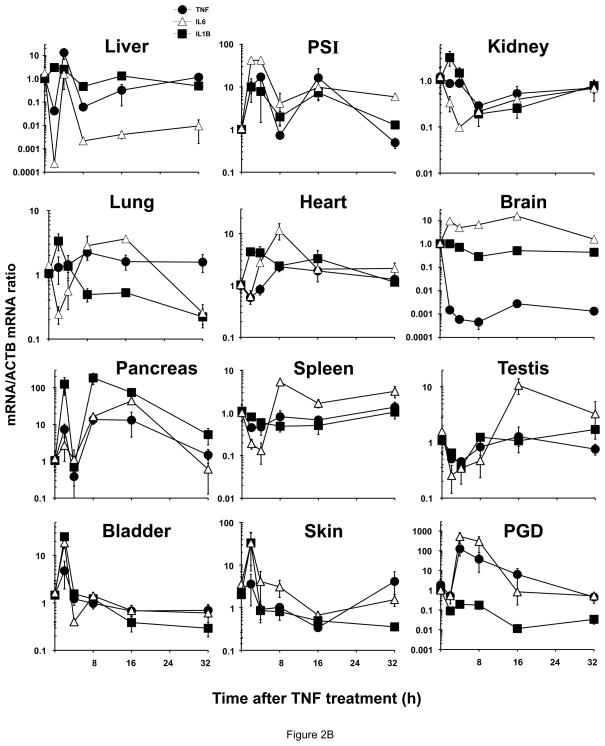
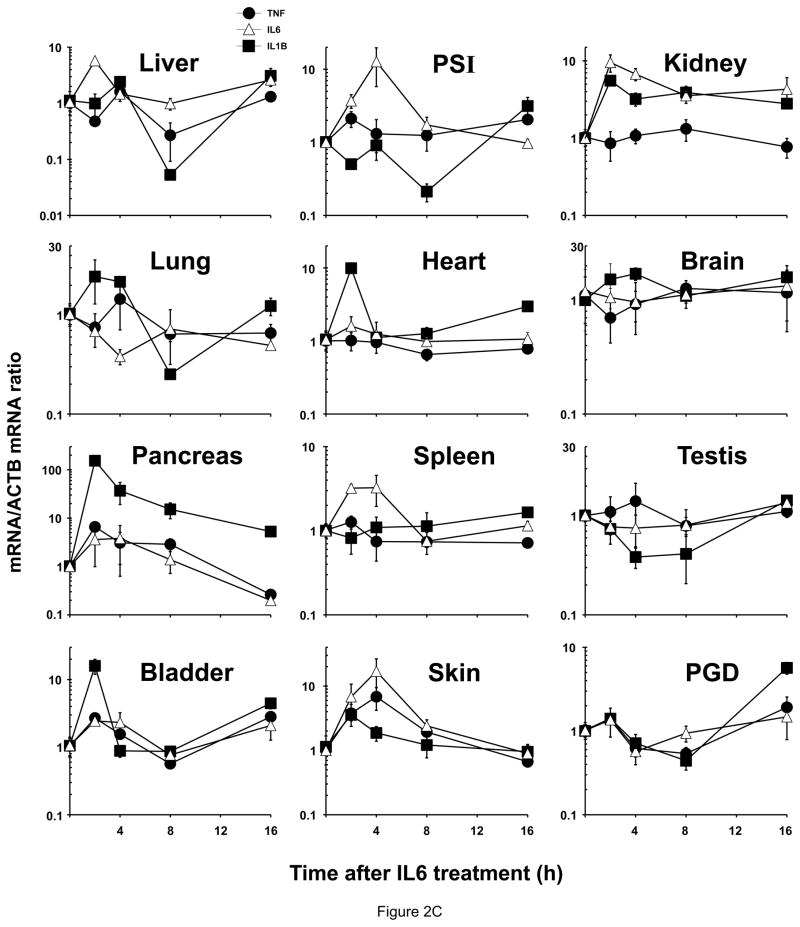
Relative ZIP8, ZIP14A and ZIP14B mRNA levels in mice following treatment with LPS, TNF or IL6. ZIP8 (closed circles, ●), ZIP14A (closed squares, ■) and ZIP14B (open squares, □) mRNA levels were quantified in each of twelve tissues as a function of time after the single dose of LPS or cytokine, as indicated. N=3 per time-point, and all experiments were repeated twice; β-actin (ACTB) mRNA was employed as the control. Results were normalized using the non-treated mRNA/ACTB mRNA ratio. Note the 2- to ~30-fold differences in values on the ordinates. Statistical analysis of all time-points is summarized in Table S1. (A) LPS treatment. (B) TNF treatment. (C) IL6 treatment.
Table 1.
Relative increases in up-regulation of mRNA, as measured by qRT-PCR
Cutoff values were arbitrarily set at: <3 (mRNA/ACTB mRNA ratio) = zero; 3–10 = +1; 10–20 =
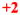 ; 20–50 =
; 20–50 =
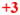 ; 50–200 =
; 50–200 =
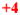 ; and 200–1,000 =
; and 200–1,000 =
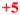 . Red denotes substantial up-regulation. Statistically significant down-regulation =
. Red denotes substantial up-regulation. Statistically significant down-regulation =
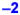 .
.
| A. Data from Figures 1A & 2A (LPS treatment) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hours | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | ||
| Liver | PSI | Kidney | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | −2 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | −2 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | 0 | +1 | 0 | 0 | 0 |
| ZIP14B | 0 | 0 | 0 | −2 | 0 | ZIP14B | 0 | 0 | +1 | 0 | 0 | ZIP14B | +1 | +1 | +1 | +1 | +1 |
| TNF | 0 | 0 | 0 | 0 | 0 | TNF | +5 | +4 | +4 | +3 | +4 | TNF | +1 | +1 | 0 | 0 | 0 |
| IL6 | 0 | 0 | 0 | 0 | 0 | IL6 | +5 | +3 | +1 | +2 | +1 | IL6 | +4 | +2 | 0 | 0 | 0 |
| IL1B | +1 | +1 | +1 | 0 | 0 | IL1B | +5 | +4 | +2 | +1 | +4 | IL1B | +1 | 0 | 0 | 0 | 0 |
| Lung | Heart | Brain | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | −2 | ZIP8 | 0 | −2 | 0 | 0 | 0 |
| ZIP14A | +1 | +1 | +1 | +1 | 0 | ZIP14A | 0 | 0 | +1 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 |
| ZIP14B | +1 | +1 | +1 | +2 | +1 | ZIP14B | 0 | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | +1 | 0 | 0 |
| TNF | +3 | +1 | +1 | +1 | +1 | TNF | +1 | +1 | 0 | 0 | 0 | TNF | +1 | +1 | +1 | +1 | +1 |
| IL6 | +4 | +3 | +1 | +1 | +1 | IL6 | +4 | +4 | +1 | +1 | 0 | IL6 | +2 | +1 | +1 | 0 | 0 |
| IL1B | +2 | +1 | +1 | +1 | +1 | IL1B | +1 | +1 | 0 | 0 | 0 | IL1B | +2 | +2 | +2 | 0 | +1 |
| Pancreas | Spleen | Testis | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | +1 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | −2 | ZIP14A | 0 | 0 | 0 | 0 | −2 |
| ZIP14B | 0 | 0 | +1 | 0 | 0 | ZIP14B | 0 | +1 | +1 | +1 | 0 | ZIP14B | 0 | 0 | 0 | 0 | 0 |
| TNF | 0 | 0 | 0 | 0 | 0 | TNF | +1 | +1 | 0 | 0 | 0 | TNF | +1 | +1 | 0 | 0 | 0 |
| IL6 | +1 | +1 | 0 | 0 | −2 | IL6 | +4 | +3 | +1 | +1 | 0 | IL6 | +3 | +2 | +1 | +1 | 0 |
| IL1B | 0 | 0 | 0 | 0 | −2 | IL1B | +1 | +1 | 0 | 0 | 0 | IL1B | +1 | 0 | 0 | 0 | 0 |
| Bladder | Skin | PGD | |||||||||||||||
| ZIP8 | +2 | 0 | 0 | +3 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | +1 | 0 | 0 | 0 |
| ZIP14A | +1 | +1 | +1 | +1 | 0 | ZIP14A | +1 | +2 | +3 | +2 | 0 | ZIP14A | 0 | +1 | 0 | 0 | 0 |
| ZIP14B | +1 | +1 | +1 | +1 | 0 | ZIP14B | 0 | +1 | +1 | +1 | 0 | ZIP14B | +1 | +1 | +1 | +1 | 0 |
| TNF | +4 | +1 | +1 | +1 | 0 | TNF | +1 | +1 | 0 | 0 | 0 | TNF | +1 | 0 | 0 | 0 | 0 |
| IL6 | +5 | +4 | +2 | +1 | 0 | IL6 | +4 | +2 | +1 | 0 | 0 | IL6 | +3 | 0 | 0 | 0 | −2 |
| IL1B | +4 | +2 | +1 | +1 | +1 | IL1B | 0 | 0 | 0 | 0 | 0 | IL1B | +1 | 0 | 0 | 0 | 0 |
| B. Data from Figures 1B & 2B (TNF treatment) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hours | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | ||
| Liver | PSI | Kidney | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | −2 | −2 | −2 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | +1 | +1 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 |
| ZIP14B | 0 | +1 | 0 | 0 | 0 | ZIP14B | +1 | 0 | 0 | 0 | 0 | ZIP14B | 0 | −2 | 0 | 0 | 0 |
| TNF | 0 | +2 | 0 | 0 | 0 | TNF | 0 | +2 | 0 | +2 | 0 | TNF | 0 | 0 | 0 | 0 | 0 |
| IL6 | −2 | 0 | 0 | 0 | 0 | IL6 | +3 | +3 | +1 | 0 | 0 | IL6 | 0 | −2 | 0 | 0 | 0 |
| IL1B | +1 | +1 | 0 | 0 | 0 | IL1B | +1 | +1 | +1 | 0 | 0 | IL1B | +1 | 0 | 0 | 0 | 0 |
| Lung | Heart | Brain | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | +1 | 0 | 0 | ZIP8 | 0 | −2 | −2 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | +1 | 0 | 0 | ZIP14A | 0 | 0 | −2 | 0 | 0 |
| ZIP14B | 0 | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | +1 | +2 | 0 | ZIP14B | 0 | 0 | 0 | 0 | 0 |
| TNF | 0 | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | 0 |
| IL6 | −2 | 0 | +1 | +1 | −2 | IL6 | 0 | 0 | +2 | 0 | 0 | IL6 | +1 | +1 | +1 | +1 | 0 |
| IL1B | +1 | 0 | 0 | 0 | −2 | IL1B | +1 | +1 | 0 | 0 | 0 | IL1B | 0 | 0 | −2 | 0 | 0 |
| Pancreas | Spleen | Testis | |||||||||||||||
| ZIP8 | 0 | +4 | +5 | +4 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 |
| ZIP14B | 0 | +1 | 0 | +3 | 0 | ZIP14B | 0 | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | 0 | 0 | 0 |
| TNF | +1 | 0 | +2 | +2 | 0 | TNF | 0 | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | 0 |
| IL6 | 0 | 0 | +2 | +3 | 0 | IL6 | −2 | −2 | +1 | 0 | 0 | IL6 | 0 | 0 | 0 | +1 | 0 |
| IL1B | +4 | 0 | +4 | +4 | +1 | IL1B | 0 | 0 | 0 | 0 | 0 | IL1B | 0 | 0 | 0 | 0 | 0 |
| Bladder | Skin | PGD | |||||||||||||||
| ZIP8 | +1 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | 0 | ZIP8 | −2 | 0 | 0 | 0 | 0 |
| ZIP14A | 0 | 0 | 0 | 0 | 0 | ZIP14A | +2 | +1 | +1 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | 0 |
| ZIP14B | 0 | 0 | 0 | 0 | 0 | ZIP14B | +2 | +1 | +1 | +1 | +1 | ZIP14B | 0 | 0 | 0 | 0 | 0 |
| TNF | +1 | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | 0 | TNF | 0 | +4 | +3 | +1 | 0 |
| IL6 | +2 | 0 | 0 | 0 | 0 | IL6 | +3 | 0 | 0 | 0 | 0 | IL6 | 0 | +5 | +5 | 0 | 0 |
| IL1B | +2 | 0 | 0 | 0 | 0 | IL1B | +3 | 0 | 0 | 0 | 0 | IL1B | 0 | 0 | 0 | 0 | 0 |
| C. Data from Figures 1C & 2C (IL6 treatment) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hours | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | 2 | 4 | 8 | 16 | 32 | ||
| Liver | PSI | Kidney | |||||||||||||||
| ZIP8 | 0 | 0 | −2 | 0 | ZIP8 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | |||
| ZIP14A | 0 | 0 | 0 | −2 | ZIP14A | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | |||
| ZIP14B | 0 | 0 | 0 | −2 | ZIP14B | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | 0 | 0 | |||
| TNF | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | |||
| IL6 | +1 | 0 | 0 | 0 | IL6 | +1 | +1 | 0 | 0 | IL6 | +1 | +1 | +1 | +1 | |||
| IL1B | 0 | 0 | −2 | 0 | IL1B | 0 | 0 | −2 | 0 | IL1B | +1 | +1 | +1 | +1 | |||
| Lung | Heart | Brain | |||||||||||||||
| ZIP8 | 0 | 0 | −2 | −2 | ZIP8 | +1 | 0 | 0 | 0 | ZIP8 | 0 | −2 | −2 | 0 | |||
| ZIP14A | 0 | 0 | 0 | −2 | ZIP14A | 0 | 0 | 0 | −2 | ZIP14A | 0 | 0 | 0 | 0 | |||
| ZIP14B | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | 0 | −2 | ZIP14B | 0 | 0 | 0 | 0 | |||
| TNF | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | |||
| IL6 | 0 | 0 | 0 | 0 | IL6 | 0 | 0 | 0 | 0 | IL6 | 0 | 0 | 0 | 0 | |||
| IL1B | 0 | 0 | −2 | 0 | IL1B | +1 | 0 | 0 | +1 | IL1B | 0 | 0 | 0 | 0 | |||
| Pancreas | Spleen | Testis | |||||||||||||||
| ZIP8 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | |||
| ZIP14A | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | |||
| ZIP14B | 0 | 0 | 0 | 0 | ZIP14B | 0 | 0 | 0 | 0 | ZIP14B | 0 | −2 | 0 | 0 | |||
| TNF | +1 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | |||
| IL6 | 0 | 0 | 0 | 0 | IL6 | 0 | 0 | 0 | 0 | IL6 | 0 | 0 | 0 | 0 | |||
| IL1B | +4 | +3 | +2 | +1 | IL1B | 0 | 0 | 0 | 0 | IL1B | 0 | 0 | 0 | 0 | |||
| Bladder | Skin | PGD | |||||||||||||||
| ZIP8 | 0 | 0 | −2 | 0 | ZIP8 | 0 | 0 | 0 | 0 | ZIP8 | 0 | 0 | 0 | 0 | |||
| ZIP14A | 0 | 0 | 0 | 0 | ZIP14A | +1 | +1 | 0 | 0 | ZIP14A | 0 | 0 | 0 | 0 | |||
| ZIP14B | 0 | 0 | 0 | 0 | ZIP14B | +1 | +1 | 0 | 0 | ZIP14B | 0 | 0 | 0 | 0 | |||
| TNF | 0 | 0 | 0 | 0 | TNF | +1 | +1 | 0 | 0 | TNF | 0 | 0 | 0 | 0 | |||
| IL6 | 0 | 0 | 0 | 0 | IL6 | +1 | +2 | 0 | 0 | IL6 | 0 | 0 | 0 | 0 | |||
| IL1B | +2 | 0 | 0 | +1 | IL1B | +1 | 0 | 0 | 0 | IL1B | 0 | 0 | 0 | +1 | |||
ZIP8 mRNA induction by LPS was highest in bladder at 2 & 16 h; ZIP14A mRNA highest in skin at 8 h but was elevated during the first 16 h; and ZIP14B mRNA elevated highest in lung at 16 h but slightly elevated at all time-points. All induced ZIP mRNA levels peaked between 2 and 16 h, and usually were headed toward baseline by 32 h; exceptions included ZIP14B in lung and kidney, which remained substantially elevated at 32 h. Other than bladder, LPS-induced ZIP8 was negligible in the other 11 tissues; actually, ZIP8 was significantly down-regulated in liver, heart and brain. In liver both ZIP14A and ZIP14B mRNA levels were also down-regulated by LPS treatment, significantly at the 16-h time-point.
TNF treatment (Figure 1B, Table 1. B; Table S1. B)
ZIP8 mRNA levels were extremely high in pancreas at 4, 8 & 16 h; and both ZIP14A mRNA and ZIP14B mRNA highest in skin at all early time-points, and ZIP14B in pancreas at 16 h. TNF-induced ZIP mRNAs were negligible over the 32-h period in: for ZIP8 in liver, PSI, lung, spleen, testis and skin; for ZIP14A in liver, kidney, lung, pancreas, spleen, testis, bladder and PGD; for ZIP14B in lung, brain, spleen, testis, bladder and PGD. ZIP8 mRNA levels were significantly down-regulated at early time-points in kidney, brain and PGD. ZIP14A was significantly down-regulated by TNF in brain at 8 h, and ZIP14B in kidney at 4 h.
IL6 treatment (Figure 1C; Table 1. C; Table S1. C)
There was no significant up-regulation by IL6 of ZIP8, ZIP14A or ZIP14B mRNA in any of the 12 tissues over the 16-h experiment. Significant down-regulation was observed for ZIP8 in liver, lung, brain and urinary bladder at 8 and/or 16 h; for ZIP14A in liver, lung and heart at 16 h; and for ZIP14B in liver and heart at 16 h and testis at 4 h.
Effects of LPS, TNF and IL6 on Cytokine mRNA Levels
LPS treatment (Figure 2A; Table 1. A; Table S1. A)
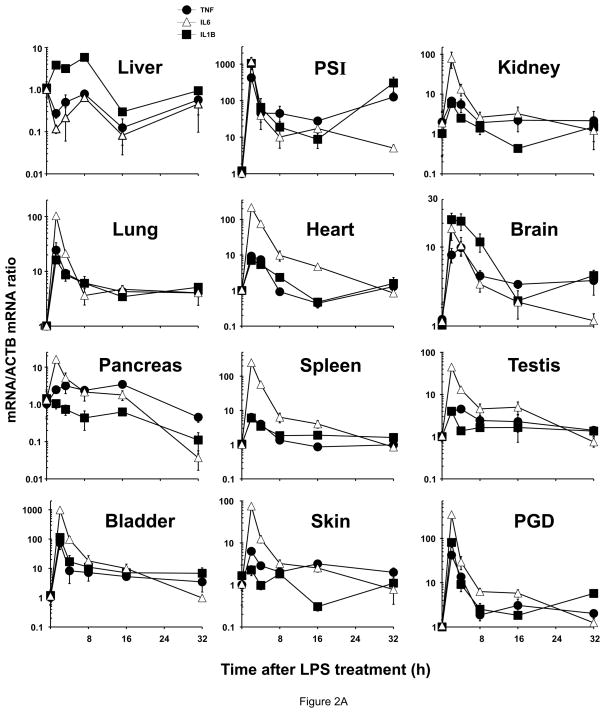
Figure 2.
Relative cytokine mRNA levels in mice following treatment with LPS, TNF or IL6. The same mice shown in Fig. 1 were used to address the correlation between ZIP mRNA and cytokine mRNA levels. TNF mRNA (closed circles, ●), IL6 mRNA (open triangles, △) and IL1B mRNA (closed squares, ■). Results again were normalized using the non-treated mRNA/ACTB mRNA ratio. Note the 2- to ~1,000-fold differences in values on the ordinates; whereas Fig. 1 data were plotted on a linear scale, the data in Fig. 2 are plotted on a logarithmic scale. Statistical analysis is included in Table S1. (A) LPS treatment. (B) TNF treatment. (C) IL6 treatment.
TNF, IL6 and IL1B mRNA levels were all highest in PSI and urinary bladder at 2 h, remaining significantly elevated throughout the 32-h experiment in PSI, and through the first 16 h in bladder for TNF and IL6 mRNA. TNF mRNA was also significantly up-regulated in lung at 2 h. IL6 mRNA was also was elevated at early time-points in kidney, lung, heart, brain, spleen, testis, skin and PGD. LPS-mediated stimulation of IL1B mRNA was also significantly increased in lung and brain persistently through the 32-h experiment. Significant down-regulation was seen only for IL6 mRNA in pancreas and PGD at 32 h, and for IL1B mRNA in pancreas at 32 h.
TNF treatment (Figure 2B; Table 1. B; Table S1. B)
TNF-induced TNF mRNA levels were greatest in PGD at 4 & 8 h, next highest in pancreas at 8 & 16 h, and smaller increases in PSI at 4 & 16 h, and in liver at 4 h. TNF-mediated induction of IL6 mRNA was significantly highest in PGD at 4 & 8 h, in PSI at 2 & 4 h, and in pancreas at 8 & 16 h. Significant IL6 mRNA induction was also seen in skin and bladder at 2 h, and in heart at 8 h. IL1B mRNA induction after TNF was highest in pancreas throughout the 32 h experiment, and in skin and bladder at the 2-h time-point. Significant down-regulation by TNF was found only: for IL6 mRNA in liver at 2 h, kidney at 4 h, in lung at 2 & 32 h, and for spleen at 2 & 4 h; for IL1B mRNA in lung at 32 h and brain at the 8-h time-point.
IL6 treatment (Figure 2C; Table 1. c; Table S1. C)
TNF mRNA levels following IL6 administration were not significantly up-regulated in any of the 12 tissues—except for small increases in pancreas and skin at early time-points. IL6-induced IL6 mRNA was significantly increased only in skin at 4 h. IL6-stimulated IL1B mRNA levels were highest in pancreas at 2 & 4 h, which persisted throughout the 16-h experiment. IL1B mRNA induction by IL6 was also significantly increased in bladder at 2. Down-regulation by IL6 treatment was found in liver, PSI and lung—in each case only at the 8-h time-point.
Brain-Specific Differences
Interestingly, in brain, we observed that LPS and TNF but not IL6 enhanced ZIP14B mRNA selectively (Fig. 1), and similarly, LPS and TNF but not IL6 altered the proinflammatory cytokine profile (Fig. 2). It is well known that the ligand and its corresponding receptor include: LPS and toll receptor-4 (TLR4); TNF and TNF receptor; IL6 and IL6 receptor. Of what relevance might this be in brain?
Fever reflects a multiphasic response (elevation and decline of the body core temperature) that is regulated by central thermoregulatory mechanisms localized to the preoptic area of hypothalamus. Several proinflammatory cytokines including TNF and IL6 act as endogenous pyrogens, whereas other cytokines can act as antipyretic agents; these findings confirm the link between the immune and central nervous systems27. Cytokine levels in serum and cerebral spinal fluid are associated with fever. In the brain, presence of specific cytokine receptors on various cell types—and the effects of pharmacological application of cytokines and of their neutralizing antibodies—are known to alter the fever response via eicosanoid and other second-messenger lipid mediators28;29; these findings are supported by cytokine- and cytokine receptor- transgenic mouse models27.
Overall, data in the present study imply that tissues have different levels of responsiveness to ligands probably as a result of the receptors to which they specifically bind; moreover, ZIP14B is likely to be transcriptionally regulated in a manner differently from ZIP14A, perhaps at the splliceosome level, as had been previously suggested by observed differences in tissue-specific regulation of ZIP14B vs ZIP14A7;8. Further investigations—using chromatin immunoprecipitation and electrophoretic mobility shift assays and reporter genes, which has been previously done for ZIP824—will be required to tease out regulatory differences between ZIP14B and ZIP14A. It is exciting to speculate that ZIP14B vs ZIP14A expression might be triggered by tissue- and cell type-specific cytokines and lipid mediators.
Conclusions
LPS-, TNF- and IL6-mediated induction of ZIP8, ZIP14A and ZIP14B mRNA levels, as well as stimulation of the inflammatory cytokine TNF, IL6 and IL1B mRNA levels, were found to be extremely variable depending on the inflammatory stressor given, quantitatively very different, highly tissue-specific, and also markedly time-dependent. It is also worth noting that expression of ZIP14A vs ZIP14B mRNA concentrations varied widely among the different treatments, tissues and time-points—providing further evidence that these two splice variants from the same gene must be associated with distinct functions in each cell type7;8.
In summary, these data suggest that all these tissues likely use ZIP8 and ZIP14 transporters for Zn2+ uptake—some tissues without cytokine treatment, others moreso when inflammatory danger (cytokine treatment) is present. Collectively, inflammatory-stimulated alterations in ZIP8-, ZIP14A-and ZIP14B-mediated transporter expression levels would probably lead to substantial decreases in plasma Zn2+ concentrations, due to Zn2+ redistribution not just in liver but across most vital organs. An extension of the present study would be to precisely measure intracellular Zn2+ levels at all time-points following LPS, TNF or IL6 treatment and in all twelve tissues. It seems intuitive that all major organs would require Zn2+ uptake, to varying degrees, in response to conditions of stress such as cytokine exposure. In the context of Cd2+-mediated (or other toxic environmental-metal-mediated) toxicity, this opens the door for studying pathogenesis across a broad range of tissues, beyond simply kidney or lung.
Supplementary Material
Acknowledgments
We thank our colleagues, especially Lei He, for valuable discussions and careful reading of this manuscript. We are grateful to Dr. Marian Miller for excellent help with graphics. We thank Cynthia M. Williams for technical help.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded, in part, by NIH R01 HL118268 (DLK), R01 ES010416 (DWN), T32 ES016646 (MG-P), and P30 ES06096 (DWN).
Footnotes
Declaration of Conflicting Interests
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary files located online at http://ijt.sagepub.com/supplemental.
References
- 1.Taylor BA, Heiniger HJ, Meier H. Genetic analysis of resistance to cadmium-induced testicular damage in mice. Proc Soc Exp Biol Med. 1973;143:629–633. doi: 10.3181/00379727-143-37380. [DOI] [PubMed] [Google Scholar]
- 2.Dalton TP, Miller ML, Wu X, et al. Refining the mouse chromosomal location of Cdm, the major gene associated with susceptibility to cadmium-induced testicular necrosis. Pharmacogenetics. 2000;10:141–151. doi: 10.1097/00008571-200003000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Dalton TP, He L, Wang B, et al. Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc Natl Acad Sci U S A. 2005;102:3401–3406. doi: 10.1073/pnas.0406085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Girijashanker K, Dalton TP, et al. ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol Pharmacol. 2006;70:171–180. doi: 10.1124/mol.106.024521. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z, Li H, Soleimani M, et al. Cd2+ versus Zn2+ uptake by the ZIP8 HCO3−-dependent symporter: kinetics, electrogenicity and trafficking. Biochem Biophys Res Commun. 2008;365:814–820. doi: 10.1016/j.bbrc.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Schneider SN, Dragin N, et al. Enhanced cadmium-induced testicular necrosis and renal proximal tubule damage caused by gene-dose increase in a Slc39a8-transgenic mouse line. Am J Physiol Cell Physiol. 2007;292:C1523–C1535. doi: 10.1152/ajpcell.00409.2006. [DOI] [PubMed] [Google Scholar]
- 7.He L, Wang B, Hay EB, Nebert DW. Discovery of ZIP transporters that participate in cadmium damage to testis and kidney. Toxicol Appl Pharmacol. 2009;238:250–257. doi: 10.1016/j.taap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girijashanker K, He L, Soleimani M, et al. Slc39a14 gene encodes ZIP14, a metal/bicarbonate symporter: similarities to the ZIP8 transporter. Mol Pharmacol. 2008;73:1413–1423. doi: 10.1124/mol.107.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nebert DW, Galvez-Peralta M, Hay EB, et al. ZIP14 and ZIP8 zinc/bicarbonate symporters in Xenopus oocytes: characterization of metal uptake and inhibition. Metallomics. 2012;4:1218–1225. doi: 10.1039/c2mt20177a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besecker B, Bao S, Bohacova B, Papp A, Sadee W, Knoell DL. The human zinc transporter SLC39A8 (ZIP8) is critical in zinc-mediated cytoprotection in lung epithelia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1127–L1136. doi: 10.1152/ajplung.00057.2008. [DOI] [PubMed] [Google Scholar]
- 11.Wang CY, Jenkitkasemwong S, Duarte S, et al. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J Biol Chem. 2012;287:34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkitkasemwong S, Wang CY, Mackenzie B, Knutson MD. Physiologic implications of metal-ion transport by ZIP14 and ZIP8. Biometals. 2012;25:643–655. doi: 10.1007/s10534-012-9526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujishiro H, Yano Y, Takada Y, Tanihara M, Himeno S. Roles of ZIP8, ZIP14, and DMT1 in transport of cadmium and manganese in mouse kidney proximal tubule cells. Metallomics. 2012;4:700–708. doi: 10.1039/c2mt20024d. [DOI] [PubMed] [Google Scholar]
- 14.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. ZIP14 (SLC39A14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci U S A. 2006;103:13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao N, Gao J, Enns CA, Knutson MD. ZRT/IRT-like protein 14 (ZIP14) promotes the cellular assimilation of iron from transferrin. J Biol Chem. 2010;285:32141–32150. doi: 10.1074/jbc.M110.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinilla-Tenas JJ, Sparkman BK, Shawki A, et al. ZIP14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am J Physiol Cell Physiol. 2011;301:C862–C871. doi: 10.1152/ajpcell.00479.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gálvez-Peralta M, He L, Jorge-Nebert LF, et al. ZIP8 zinc transporter: indispensable role for both multiple-organ organogenesis and hematopoiesis in utero. PLoS One. 2012;7:e36055. doi: 10.1371/journal.pone.0036055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begum NA, Kobayashi M, Moriwaki Y, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80:630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 19.Liuzzi JP, Lichten LA, Rivera S, et al. Interleukin-6 regulates the zinc transporter ZIP14 in liver and contributes to hypozincemia of the acute-phase response. Proc Natl Acad Sci U S A. 2005;102:6843–6848. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aydemir TB, Liuzzi JP, McClellan S, Cousins RJ. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-γ expression in activated human T-cells. J Leukoc Biol. 2009;86:337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hojyo S, Fukada T, Shimoda S, et al. The zinc transporter SLC39A14/ZIP14 controls G-protein coupled receptor-mediated signaling required for systemic growth. PLoS One. 2011;6:e18059. doi: 10.1371/journal.pone.0018059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beker Aydemir T, Chang SM, Guthrie GJ, et al. Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia) PLoS One. 2012;7:e48679. doi: 10.1371/journal.pone.0048679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thambiayya K, Wasserloos K, Kagan VE, Stoyanovsky D, Pitt BR. A critical role for increased labile zinc in reducing sensitivity of cultured sheep pulmonary artery endothelial cells to LPS-induced apoptosis. Am J Physiol Lung Cell Mol Physiol. 2012;302:L1287–L1295. doi: 10.1152/ajplung.00385.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu MJ, Bao S, Galvez-Peralta M, et al. ZIP8 regulates host defense through zinc-mediated inhibition of NF-κB. Cell Rep. 2013;3:386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayadi A, Nguyen AT, Bard FA, Bard-Chapeau EA. ZIP14 expression induced by lipopolysaccharides in macrophages attenuates inflammatory response. Inflamm Res. 2013;62:133–143. doi: 10.1007/s00011-012-0559-y. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Jeon J, Shin M, et al. Regulation of the catabolic cascade in osteoarthritis by ‘the zinc-ZIP8-MTF1 axis’. Cell. 2014;156:730–743. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Conti B, Tabarean I, Andrei C, Bartfai T. Cytokines and fever. Front Biosci. 2004;9:1433–1449. doi: 10.2741/1341. [DOI] [PubMed] [Google Scholar]
- 28.Romanovsky AA, Almeida MC, Aronoff DM, et al. Fever and hypothermia in systemic inflammation: recent discoveries and revisions. Front Biosci. 2005;10:2193–2216. doi: 10.2741/1690. [DOI] [PubMed] [Google Scholar]
- 29.Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.