Abstract
During invasion of host cells by Trypanosoma cruzi, the parasite that causes Chagas disease, the elongated, flagellated trypomastigotes remodel into oval amastigotes with no external flagellum. The underlying mechanism of this remodeling and the fate of the flagellum are obscure. We discovered that T. cruzi trypomastigotes discard their flagella via an asymmetric cellular division. The flagellar proteins liberated become among the earliest parasite proteins to enter the MHC-I processing pathway in infected cells. Indeed, paraflagellar rod protein PAR4-specific CD8+ T cells detect infected host cells >20hrs earlier than immunodominant transsialidase-specific T cells. Overexpression of PAR4 in T. cruzi enhanced the subdominant PAR4-specific CD8+ T cell response, resulting in improved control of a challenge infection. These results provide insights into previously unappreciated events in intracellular invasion by T. cruzi and highlight the importance of T cells that recognize infected host cells early in the infectious process, in the control of infections.
INTRODUCTION
Chagas disease, caused by the hemoflagellate protozoan parasite, Trypanosoma cruzi has a large and growing worldwide impact. Globally, it is the leading cause of infectious myocarditis (Feldman and McNamara, 2000), with millions infected in endemic areas of the Americas. Although T. cruzi infection induces robust humoral and cellular immune responses that normally result in the control of the acute infection, the infection is rarely completely resolved. The deficits in the immune response that prevent parasitological cure are of significant interest since it is the chronic persistence of T. cruzi that ultimately leads to the clinical disease decades after the initial infection (Zhang and Tarleton, 1999).
CD8+ T cells are known to be key players in the control of T. cruzi infection (Tarleton et al., 1992; Tarleton et al., 1994), and a significant proportion of the anti-T. cruzi CD8+ T cells are specific for epitopes derived from parasite trans-sialidase family proteins (Martin et al., 2006). Trans-sialidases are encoded by a family of >3000 highly variable genes that differ between isolates and are also capable of intra-isolate recombination. The trans-sialidase specific CD8+ T cell response, despite being surprisingly large numerically, is dispensable for normal control of the infection (Rosenberg et al., 2010). These findings, as well as the enormous potential for epitope variability in the trans-sialidase family have created interest in the identification of other, invariant, non-large gene-family proteins of T. cruzi that could elicit CD8+ T cell responses to epitopes that are shared among all isolates.
Among the multiple factors that govern the strength of CD8+ T cell response to a particular epitope, the access of the protein to the host cell cytoplasm (Garg et al., 1997) and the amount of that protein that reaches the antigen processing machinery (Wherry et al., 2002) are especially critical. In addition to the strength, in frequency, of the T cell response, the ability of T cells to detect and destroy pathogen-infected cells early in the infection cycle, thus restricting parasite replication potential, contributes to success in infection control (Yates et al., 2011). In T. cruzi infection, the movement of parasites from the extracellular space to the cytoplasm of host cells in mammals involves conversion of the infecting flagellated trypomastigotes to the morphologically distinct amastigotes which lack an extended flagellum. We hypothesized that the remodeling required for this conversion would result in the release and subsequent degradation of parasite proteins that gain access to host cell class I MHC presentation pathways. Furthermore, increasing the expression of non-variant proteins released during this remodeling should enhance their immunogenicity and would be expected to augment immune control of T. cruzi.
In this study, we document that T. cruzi sacrifices its flagellum during amastigogenesis in host cells shortly after invasion, making the released flagellar proteins among the earliest parasite proteins available for presentation to CD8+ T cells. The flagellum of trypanosomatids contains a unique network of cytoskeletal filaments referred to as the paraflagellar rod (PFR) that lies alongside the axoneme and is involved in motility, flagellar beat patterns and possibly tissue attachment (Maga and LeBowitz, 1999). The PFR in T. cruzi is composed of four proteins: PARs 1–4, PAR4 being the least studied in terms of its biology and immunogenicity (Luhrs et al., 2003). Here, we show that PAR4 is the target of T. cruzi-specific CD8+ T cell responses and that a T. cruzi strain transgenically overexpressing PAR4 (TcPAR4) induced enhanced PAR4-specific CD8+ T cell responses that provided significantly improved protection from challenge infection. These results validate PAR4 and perhaps other flagellar proteins as potential vaccines for T. cruzi and other intracellular flagellates and support the overexpression of subdominant T cell targets as a strategy for improving live attenuated vaccines.
RESULTS
T. cruzi sacrifices its flagellum during amastigogenesis
Though one of the most conspicuous changes to T. cruzi during intracellular amastigogenesis is the conversion of the extensively flagellated trypomastigote to an amastigote with only a remnant of a flagellum, little is known concerning the underlying mechanism by which this happens or the fate of the flagellum during this process. To investigate these questions, we first visualized the flagellum of T. cruzi in the trypomastigote stage by expression of a tagged version of PAR4 in T. cruzi (PAR4-tdTomato/PAR4-HA) or using anti-PAR4 antibody. PAR4 localized only to the flagellar compartment of the trypomastigotes (Figure1A) unless the carboxy- terminal flagellar localization signal was truncated (tr.PAR4) (Figures S1 and S2), consistent with the distribution of the PAR4 orthologue in T. brucei (Bastin et al., 1999). Upon invasion of host cells, the flagellum appears disassociated from the developing amastigote, but retains a connection to both a basal body and DNA (presumably kinetoplast DNA) (Figure 1B–C), suggesting a duplication of these 2 organelles prior to release of the flagellum. Further analysis of this transformation both inside host cells (Figure S3) and axenically (Figure 2) demonstrate that during amastigognesis, trypomastigotes release their flagella via an asymmetrical division resulting in two daughter cells, one with a barely noticeable flagellum devoid of the PFR (Bastin et al., 1996), that matches the classical description of a newly formed amastigote (Andrews et al., 1987) and a second daughter with a smaller cell body and an extensive flagellum.
Figure 1. T. cruzi sacrifices its flagellum during intracellular amastigogenesis.
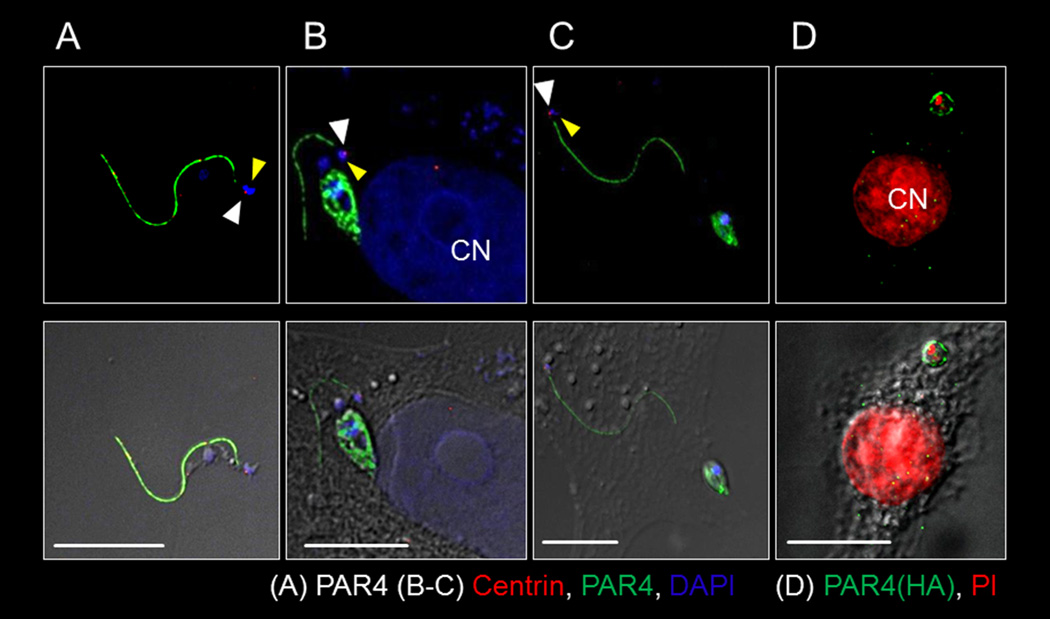
(A–C) Fluorescent (top panels) and DIC overlay (bottom panels) showing T. cruzi flagellum in the extracellular trypomastigote stage (A) and separated from the newly formed amastigote (6–9 hrs post infection) in the host cell cytoplasm (B,C). The white and yellow arrows respectively indicate the basal body and DNA associated with the flagellum. (D) An intact flagellum is not evident in an infected host cell with single amastigote 9–12 hrs post-infection but flagellar origin PAR4 (HA-tagged) ‘particles’ appear in the host cell cytoplasm. Anti-PAR4 (a–c) or PAR4-HA used to track T. cruzi PAR4. 4',6-diamidino-2-phenylindole (DAPI) (A–C) or Propidium Iodide (PI) (D) stained DNA and anti-centrin marked the basal body (A–C). CN indicates host cell nucleus. Scale bar = 10µ. See also figures S1 and S2.
Figure 2. Asymmetric division during T. cruzi amastigogenesis yields 1K1N and 1K0N daughter cells.
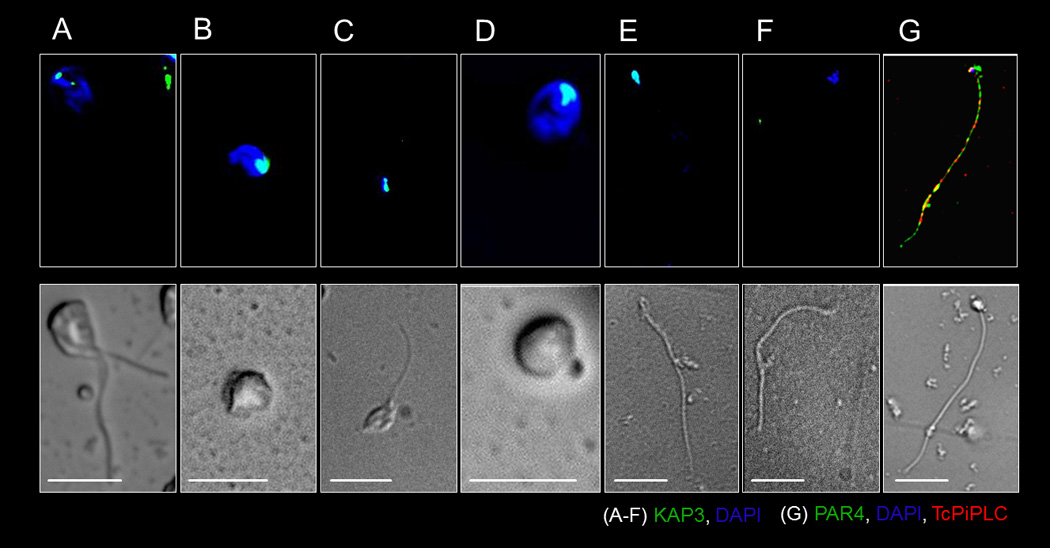
Fluorescent (top panels) and DIC imaging (bottom panels) showing the major stages in T. cruzi amastigogenesis induced by axenic conversion. In 2–4 hrs T. cruzi trypomastigotes converts to biflagellated intermediate stage (A) that in 4–9 hrs gives rise to morphologically dissimilar 1K1N daughter cells with intact kinetoplast and nucleus (B) and 1K0N daughter cells with kinetoplast but and no distinct nucleus (C). Over the next 9–16 hrs, the nucleus and kinetoplast of the 1K1N daughter remains intact (D) but the kinetoplast and cell body of the 1K0N daughter progressively loses its integrity (E-F). The 1K0N daughter is enclosed by a plasma membrane (6hrs post induction of amastigogenesis, G). T. cruzi DNA (A–G), kinetoplast (A–F), PAR4 (G) or plasma membrane (G) were marked using DAPI, anti-KAP3, -PAR4 or -TcPiPLC respectively. Scale bar = 5µ. See also figures S3, S4 and movie S1.
During this process of asymmetric division, T. cruzi trypomastigotes first transform into biflagellated intermediates with two kinetoplasts and two flagella (Figure 2A, S4I), but only a single nucleus, before cytokinesis into the two morphologically distinct daughter cells. The nucleated daughter cell (1K1N) has a functional kinetoplast identifiable by kinetoplastassociated protein and mitotracker staining (Figures 2B and S4A) and a short flagellum (Figures 1D–E, S4A). The nucleus inherited by the first generation 1K1N daughter amastigote from the parent trypomastigote has not undergone replication during this process, as indicated by the absence of BrdU incorporation during this early host cell invasion period (Figure S4E). These fully competent daughter amastigotes presumably progress further in their intra-cellular development over 4–5 days with discernible morphological changes and replication (with nuclear BRdU incorporation; Figures 2D, S3 and S4C, F).
The 1K0N daughter cell possesses an intact kinetoplast (Figures 2C, S4B), a flagellum with associated basal body (Figure 1B–C) and is initially surrounded by a plasma membrane, as indicated by detection of the plasma membrane PI-PLC (Figure 2G). However the only discernable DNA in the 1K0N daughter is associated with the kinetoplast (Figure 2C). In axenic culture, these 1K0N daughter cells showed a clear loss of integrity over time, with a reduction in cell volume (Figure 2E), loss of kinetoplast (Figures 2E–F, S4D), and flagellar motility. The progressive conversion of the population of trypomastigotes into biglagellated intermediates and then 1K1N and 1K0N daughter cells can be monitored axenically (Figure S4G–H) and within host cells (Figure S4I) and is nearly complete by 8 hrs. Evidence for the eventual disintegration of the flagellum in the infected host cell includes the failure to detect intact 1K0N daughters after 9–12 hrs post-infection and the observation of minute HA- tagged (PAR4) ‘particles’ in the cytosol of TcPAR4 infected macrophages (Figure 1D).
Thus, upon host cell infection, T. cruzi trypomastigotes undergo cytokinesis without nuclear division as a means of discharging the large but now unnecessary flagellum into the host cell cytoplasm where it then is degraded.
T. cruzi flagellar component PAR4 is recognized by CD8+ T cells
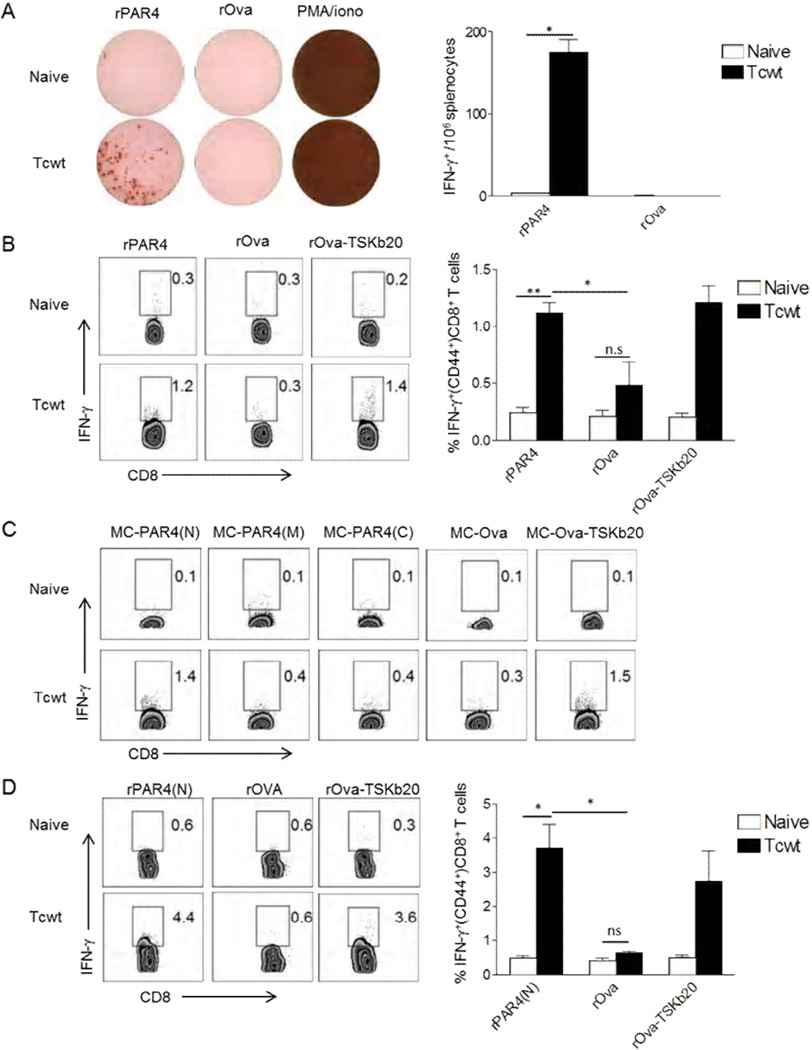
One critical factor in the generation of CD8+ T cell responses against T. cruzi proteins is the access of these proteins to the host cell cytoplasm (Garg et al., 1997). The observations of decomposition of the T. cruzi flagellum in host cells suggests that flagellar proteins from T. cruzi would be some of the first pathogen proteins available to host cell proteasome machinery for antigen processing in T. cruzi-infected host cells. Like other better-studied flagellar components, PAR4 is one of the more abundant proteins in T. cruzi trypomastigotes (Atwood et al., 2005) but has not previously been examined as a possible target of host immune responses. A distinct CD8+ T cell response to PAR4 was observed in T. cruzi (wild-type Brazil strain, Tcwt) infected mice (Figure 3A and B). An H2b MHC-l restricted epitope appeared to be present in the amino terminal region of PAR4 (PAR4(N)) (Figure 3C, Figure 2), with PAR4(N)-specific, functional CD8+ T cells being generated in T. cruzi infected C57Bl/6J mice (Figure 3D). Although clearly detectable, the PAR4-specific response is less potent than the highly dominant T. cruzi trans-sialidase derived, TSKb20 epitope-specific response (Martin et al., 2006). Thus, proteins in the T. cruzi flagellum released during amastigogenesis are processed and presented on MHC-I.
Figure 3. T. cruzi PAR4 induces CD8+ T cell response in T. cruzi-infected mice.
(A) Representative wells of IFN-γ ELISPOT assay with splenocytes from naïve or Tcwt infected (30 dpi) mice re-stimulated with rPAR4, rOVA or PMA/ionomycin for 18 hrs. Bar graph indicates the average number of spots per well for each group from an experiment with 3 mice/group. (B) Intracellular IFN-γ staining of splenocytes from naïve or Tcwt infected (180 dpi) mice restimulated with rPAR4, rOva or rOva-TSKb20 for 16hrs. The bar graph summarizes data from an experiment with 3 mice/ group. (C) Intracellular IFN-γ staining of splenocytes from naïve or Tcwt infected (48 dpi) mice, co-cultured for 16 hrs with MC-PAR4(N), MC-PAR4(M), MC-PAR4(C), MC-Ova or MC-Ova-TSKb20 cells. (D) Intracellular staining for IFN-γ in splenocytes from naïve or Tcwt infected (90 dpi) mice re-stimulated with rPAR4(N), rOva or rOva-TSKb20. The bar graph summarizes data from an experiment with 3 mice/ group. All histograms are gated on CD8+ CD44+ lymphocytes, with the inset numbers indicating the percentage of IFN-γproducing CD8+ T cells. All data representative of 2–4 similar experiments, with the bar graphs presenting data as mean ± SEM. **, * or n.s indicate p≤0.01, p≤ 0.05 or p>0.05 respectively, as determined by student t-test.
Over-expression of PAR4 in T. cruzi enhances PAR4-specific CD8+ T cell responses during infection
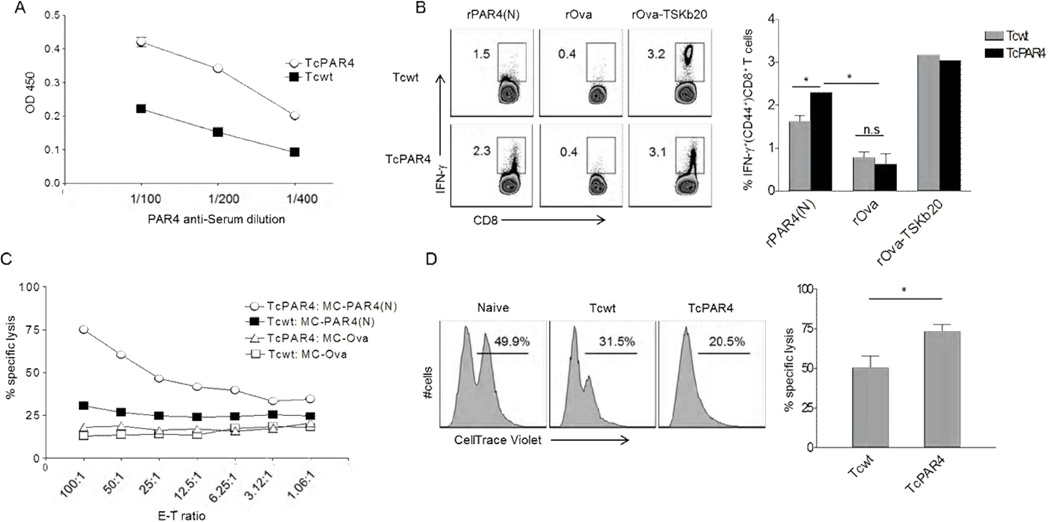
Although PAR4 is relatively abundant in T. cruzi trypomastigotes and elicits a CD8+ T cell response, we hypothesized that an increased expression of this protein would further promote its relative immunogenicity. T. cruzi over-expressing its native PAR4 (TcPAR4) protein were generated by single gene transgenesis, and were selected under strong drug pressure to achieve a ~2-fold increase in PAR4 expression (Figure 4A). TcPAR4 infection induced a significantly stronger PAR4-specific CD8+ T cell response compared to Tcwt infection, although still not at the level of the TSKb20-specific response (Figure 4B). Flagellar localization was critical to the enhanced PAR4-specific responses as T. cruzi over-expressing a truncated version of PAR4 lacking the flagellar localization signals (Tctr.PAR4) induced a comparable PAR4-specific CD8+ T cell response as Tcwt (Figure S5). Further, TcPAR4 infection also elicited greater in vitro (Figure 4C) and in vivo (Figure 4D) target cell lysis compared to infection with Tcwt. These results confirm that antigen abundance impacts the immunogenicity of proteins, even already abundant ones, in T. cruzi.
Figure 4. Over-expression of PAR4 in T. cruzi enhancements the generation of PAR4-specific CD8+ T cells.
(A) Relative expression levels of PAR4 in Tcwt and TcPAR4 trypomastigote stage (lysate) as determined by ELISA using serial dilutions of anti-PAR4 immune sera. (B) Intracellular IFN-γstaining of splenocytes derived from Tcwt or TcPAR4 infected (120 dpi) mice, restimulated with rPAR4(N), rOva or rOva-TSKb20 for 16hrs. Histograms are gated on CD8+ CD44+ lymphocytes, with the inset numbers indicating the percentage of IFN-γ producing CD8+ T cells. The bar graph summarizes data from an experiment with 3 mice/ group. (C) Cytolytic activity of splenocytes from TcPAR4 or Tcwt infected mice (96 dpi) against MC-PAR4(N) or MC-Ova target cells at different effector-target ratios. (D) Representative histograms comparing specific killing of CellTrace Violet (CTV) stained MC-PAR4(N) (CTVhi)/ MC-OVA (CTVlo) target cells inoculated into and recovered 18h later from the peritoneal cavity of naïve, Tcwt or TcPAR4 infected (300 dpi) mice. The inset numbers represent the relative proportions of recovered MC-PAR4(N) compared to MC-OVA. The bar graph shows the relative in vivo specific lysis of MC-PAR4(N) in Tcwt or TcPAR4 infected mice. Data represents an experiment with 3 mice/ group. All data representative of at least three similar experiments with the bar graphs presenting data as mean ± SEM. * or n.s indicate p≤ 0.05 or p>0.05 respectively, as determined by student t-test. See also figure S5
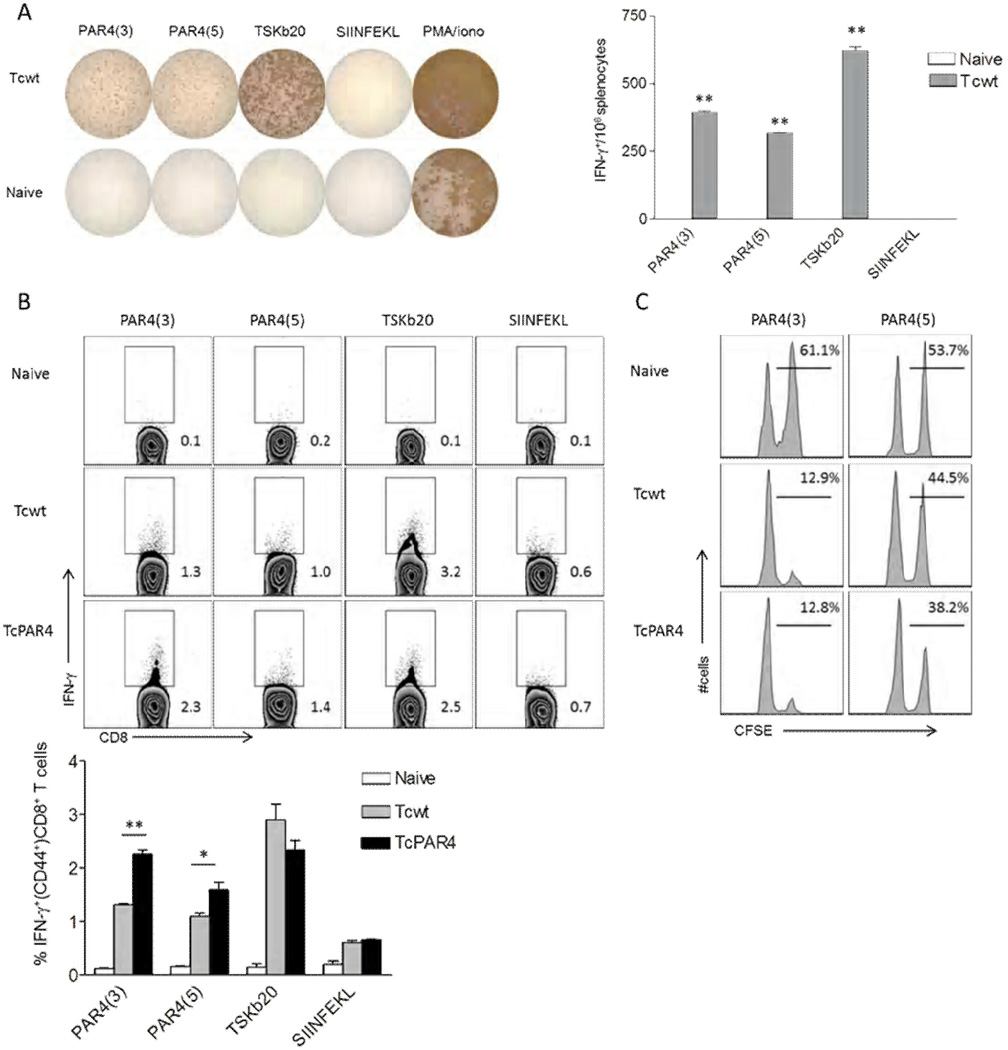
Based on the SYFPEITHI algorithm (Rammensee et al., 1999), we identified several potential CD8+ T cell epitopes from PAR4(N) for the H-2b haplotype and synthesized eight of these to test as possible CD8+ T cell targets. Two PAR4-derived H2Db haplotype-restricted epitopes: DSLLNEVSL (PAR4(3)) and KALSDEMEEM (PAR4(5)) were recognized by T cells from T. cruzi-infected C57BL/6 mice as reflected in both IFN-γ production (Figure 5 A, B) and in vivo cytolytic assays (Figure 5C). In agreement with our earlier observation, overexpression of PAR4 induced enhanced PAR4(3) and PAR4(5) specific CD8+ T cell responses (Figure 4B).
Figure 5. Target epitope identification in PAR4-specific CD8+ T cells.
(A) Representative wells of IFN-γ ELISPOT assay, where splenocytes from naïve or Tcwt infected (30 dpi) mice were restimulated for 18 hrs with the PAR4 peptides PAR4(3) or PAR4(5). T. cruzi trans-sialidase derived TSKb20, chicken ovalbumin derived SIINFEKL peptides or PMA/ionomycin served as controls. Bar graph indicates the average spots per well (±SEM) for each group from a representative of 3 trials with 3 mice/ group. ** indicates p≤0.01 comparing the indicated groups to SIINFEKL control by student t-test. (B) Representative intracellular IFN-γ staining of splenocytes derived from naïve, Tcwt or TcPAR4 infected (30 dpi) mice restimulated with PAR4(3), PAR4(5), TSKb20 or SIINFEKL for 5 hrs. Histograms are gated on CD8+ CD44+ lymphocytes, with the inset numbers indicating the percentage of IFN-γproducing CD8+ T cells. The bar graph summarizes data from an experiment with 3 mice/ group. (C) Representative histograms comparing specific killing of PAR4(3) or PAR4(5) (CFSEhi)/ SIINFEKL (CFSElo) peptide pulsed target splenocytes inoculated into and recovered 18h later from- naïve, Tcwt or TcPAR4 infected (300 dpi) mice. The inset numbers represent the percentage of PAR4(3) or PAR4(5) pulsed over total donor splenocytes recovered. Data represent an experiment with 3 mice/group. ** or * indicates p≤0.01 or ≤0.05 respectively by student t-test. All data are representative of at least 3 separate trials.
PAR4 transgenesis induces improved control of T. cruzi infection
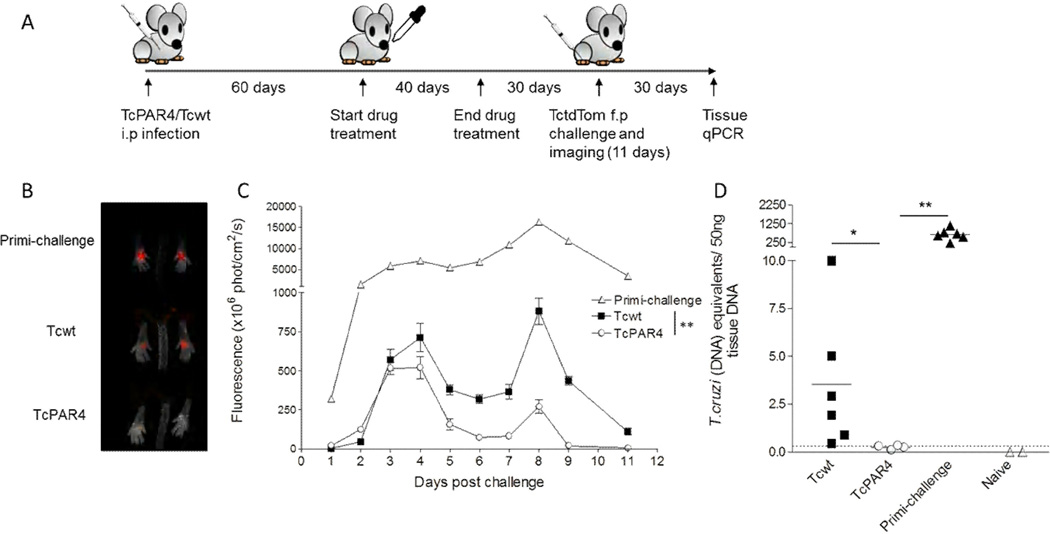
Given the heightened PAR4-specific CD8+ T cell response observed in infections with the PAR4-overexpressing TcPAR4, we hypothesized that immunization with TcPAR4 would provide a more potent and also cross-strain protective response against wild-type T. cruzi challenge relative to Tcwt. To test this hypothesis, we infected mice with TcPAR4 or Tcwt parasites and then drug-cured the infection using benznidazole to simulate vaccination (Bustamante et al., 2008). These mice were then challenged in each footpad with tdTomato fluorescent proteinexpressing T. cruzi (CL strain) (Canavaci et al., 2010), and parasite load at the site of infection was monitored by in vivo imaging (Collins et al., 2011) and by PCR for detection of parasite DNA in tissues (Fig 6A). By day 4 post-infectionTcPAR4-immunized mice displayed a significant decline in parasite numbers, as reflected in fluorescence signal, compared to Tcwt vaccinated or the control non-vaccinated (primi-infection) mice (Figure 6B,C). TcPAR4 immunized mice also exhibited significantly reduced parasite loads in skeletal muscle tissue at day 30 postchallenge, as compared to Tcwt immunized or non-immunized mice (Figure 6D). Thus, immunization with PAR4 over-expressing parasites stimulates a more effective adaptive immunity that better protects mice from challenge with WT T. cruzi.
Figure 6. TcPAR4 immunization enhances resistance to a T. cruzi challenge.
(A) Schematic showing the methodology of infection (intra-perotoneal, i.p), cure, challenge (in the superficial subcutaneous tissue of each footpad) and assessment of protection in TcPAR4 or Tcwt infected mice. (B) Representative image from day 4 post-challenge showing the acute control of T. cruzi at the site of infection in naive (primi-challenge), and Tcwt- or TcPAR4-vaccinated mice. The parasite load in foot pad, represented by the fluorescent signal of T. cruzi expressing tdTomato (TctdTom) is determined by in vivo imaging. (C) The control of TctdTom challenge based on mean fluorescent signal of all feet in primi-challenged, Tcwt or TcPAR4 immunized mice at indicated time points. Data are from an experiment with 3 mice/group, which is representative of 3 separate trials. (D) T. cruzi DNA in skeletal muscle tissue of C57BL/6 mice immunized with Tcwt or TcPAR4 and challenged with TctdTom as determined by quantitative real-time PCR, 30 days post challenge. Horizontal bars represent the mean. Unimmunized (primi-infection) or naïve mice served as controls. The dotted line represents the threshold of detection for the assay. Data represented as mean ± SEM, from one of 2 separate experiments with 3–6 mice/ group. ** or * indicates p≤0.01 or ≤0.05 respectively by student t-test. See also figure S6.
Rapid detection of T. cruzi infection by PAR4 specific CD8+ T cells
T. cruzi infection induces very robust CD8+ cell responses directed at epitopes in highly immunodominant trans-sialidase family of proteins, with upward of 30% of the entire mouse CD8+ T cell compartment being specific for these epitopes at the peak of the response (Martin et al., 2006). Therefore it was somewhat surprising that a relatively modest increase in T. cruzi specific CD8+ T cells provoked by the overexpression of PAR4 would impact the course of a challenge infection as observed here. Since T. cruzi sacrifices its flagellum within a few hours of host cell invasion, we hypothesized that T. cruzi-infected cells may be detected by PAR4-specific CD8+ T cells relatively quickly as well. To test this hypothesis, murine fibroblasts infected with T. cruzi for various lengths of time were co-cultured with PAR4(3)- or TSKb20-primed CD8+ T cells and monitored for recognition of their respective epitopes. Whereas PAR4(3)+CD8+ T cells recognized host cells infected with T. cruzi within the first few hours postinfection, T cells specific for the immunodominant TSKb20 epitope did not recognize T. cruzi infected host cells until >24 hrs post-infection (Figure 7A,B).
Figure 7. Rapid detection of T. cruzi infected cells by PAR4-specific CD8+ T cells.
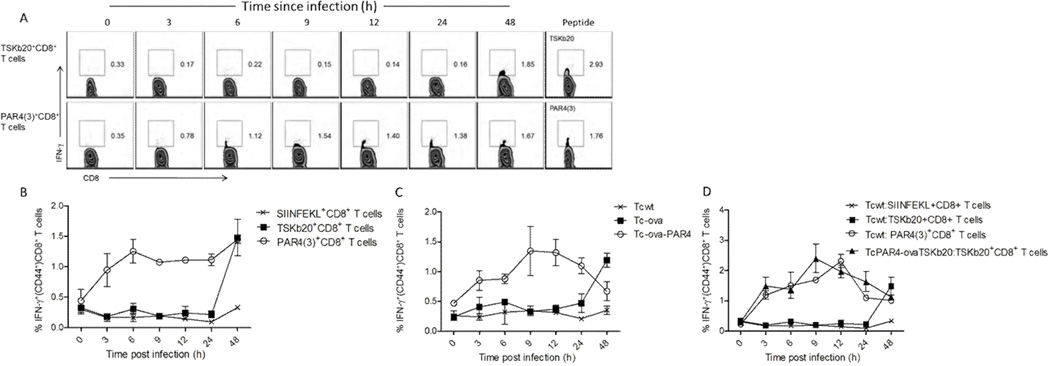
Representative flow plots (A) and line graph (B) showing the timing of IFN-γ response of TSKb20- or PAR4-specific splenic CD8+ T cells restimulated with murine fibroblasts infected with T. cruzi for the indicated lengths of time. Restimulation with the corresponding peptide epitopes served as controls. All histograms are gated on CD8+ CD44+ lymphocytes, with the inset numbers indicating the percentage of IFN-γ producing cells. SIINFEKL specific CD8+ T cells served as controls. (C)Timing of IFN-γ response in OVA specific CD8+ lymphocytes derived from Lm-ova infected mice co-incubated with murine fibroblasts infected with Tcwt, Tc-ova or Tc-ova-PAR4 for the indicated lengths of time. (D) Timing of IFN-γ response in OVA/TSKb20/PAR4(3) specific CD8+ splenic lymphocytes co-incubated with murine fibroblasts infected with Tcwt or TcPAR4-ovaTSKb20 for the indicated lengths of time. The line graphs present data as mean ± SEM from one of three separate experiments. See also figure S7.
To control for possible differences in T cell populations and variable epitope affinity for MHC, we engineered T. cruzi to express the model antigen ovalbumin (OVA) as a secreted protein (Tcova) or linked to PAR4 (Tc-ova-PAR4) as well as a version of OVA in which the SIINFEKL epitope had been replaced with the T. cruzi TSKb20 epitope (TcPAR4-ovaTSKb20) and assessed the timing of presentation of the respective epitopes in infected host cells. In agreement with the results of recognition of the endogenous trans-sialidase- and PAR4-specific T cells, the OVA SIINFEKL epitope was detectable within hours of host cell infection by Tc-ova-PAR4 parasites while presentation of the secreted OVA took > 1 day (Figure 7C). Likewise, TSKb20-specific T cells recognized host cells within hours when the TSKb20 epitope was physically linked to PAR4 (Figure 7D). Thus the ability of PAR4-specific T cells to detect parasite infection earlier in the infection cycle may be integral to the more potent effector activity of these cells relative to the abundant trans-sialidase-specific T cells.
DISCUSSION
Though the life history of T. cruzi has been systematically elucidated over more than one hundred years (Brener, 1973; de Souza, 1984; Tyler and Engman, 2001), some aspects of it still remain a mystery. One such intriguing feature is the cell biology of intracellular amastigogenesis, where the elongated, flagellated trypomastigotes remodel into oval amastigotes with no external flagellum. The T. cruzi ubiquitin-proteasome system has been implicated in this morphogenesis, as being responsible for recycling of now unneeded trypomastigote structures (Gonzalez et al., 1996). However, some key structural proteins have remained unaccounted for in this process. Notably absent among parasite proteasome targets during amastigogenesis are components of T. cruzi paraflagellar rod proteins (de Diego et al., 2001), suggesting an alternate mechanism of dealing with the flagella during remodeling. This, along with the observation of CD8+ T cells specific to flagellar structural proteins in T. cruzi infection (Egui et al., 2012; Michailowsky et al., 2003; Wrightsman et al., 2002) led us to hypothesize that the T. cruzi flagellum is ‘lost’ into the host cell cytosol during amastigogenesis. In this study, we make the observation of the release of the detached T. cruzi flagellum into the cytosol of the host cell during amastigogenesis as a product of an asymmetric division of T. cruzi.
The transition of trypomastigote to amastigote in T. cruzi takes place without nuclear division and progresses through an intermediate stage (2K1N) with duplicate flagella, basal body and kinetoplast, but only a single nucleus. Cytokinesis then generates a daughter cell (1K1N) with a short flagellum and a second daughter (1K0N) with a long flagellum and associated basal body and kinetoplast, but with no evidence of a nucleus. Based upon previous studies documenting the presence of amastigotes-like forms within the LAMP+ lysosomal vacuole, (Nagajyothi et al., 2011), it is likely that this early division process is also taking place within the vacuole and the 2 daughters are then released into the cytoplasm upon dissolution of the vacuolar membrane (Figure S7). An analogous asymmetric division process resulting in long flagellum and short flagellum daughters is also observed in T. brucei (Sharma et al., 2008; Van Den Abbeele et al., 1999). During stage transition in T. brucei procyclics, the resultant short-flagellated daughter epimastigote continues through the cell-division, while the longer flagellated daughter epimastigote does not and appears to die (Sharma et al., 2009; Sharma et al., 2008; Van Den Abbeele et al., 1999). In the tsetse fly salivary glands asymmetric division results in the production of progeny with distinct flagella, including trypomastigotes pre-adapted for infection of mammals (Rotureau et al., 2012). In T. cruzi the IK1N daughter presumably spawns the amastigotes that will eventually fill the host cell, while the 1K0N daughter appears to be degraded in the host cell. 1K0N cells termed ‘zoids’, that retain their flagellum and basal body-kinetoplast complex, have also been reported in T. brucei, (Robinson et al., 1995). In T. cruzi, the intact flagella of the 1K0N daughter, initially evident in host cells 2–6 hrs post-infection were undetectable by 12 hours post-infection, however HA-tagged (PAR4) particles are visible soon thereafter. These observations, in addition to providing critical insights into the biology of T. cruzi, also shed light on the hitherto unclear mechanism by which CD8+ T cell responses could be generated against the unique structural components in T. cruzi (Miller et al., 1996; Wrightsman et al., 2002; Wrightsman et al., 1995) or Leishmania spp (Maga et al., 1999; Saravia et al., 2005) flagella that are absent from these pathogens’ predominant intracellular stages (Portman and Gull, 2010).
CD8+ T cells are critical to the adaptive immune control of most intracellular pathogens owing to their ability to produce a variety of cytokines, and to directly target infected host cells for destruction. In its cytoplasmic niche, T. cruzi releases various proteins that are processed and presented on class I MHC molecules (Garg et al., 1997), and may be detected by specific CD8+ T cells possessing cognate T cell receptors. However, unlike the genetically simpler viral or bacterial model pathogens, protozoans, including T. cruzi, potentially elicit CD8+ T cell responses against a large numbers of epitopes, spanning multiple proteins (Doolan et al., 2003; Frickel et al., 2008; Martin et al., 2006). These responses also appear in a focused, reproducible hierarchy, in which T cell clones of certain epitope specificities are represented in greater quantities (dominant) than others (subdominant) - in a phenomenon termed immunodominance (Yewdell and Bennink, 1999). Given the complexity of the T. cruzi genome, with hugely expanded sets of variant gene families encoding various surface-expressed and secreted proteins (El-Sayed et al., 2005), multiple clones of CD8+ T cells form the immunodominance hierarchy, led by those specific to the trans-sialidase family of proteins (Martin et al., 2006; Rosenberg et al., 2010). Trans-sialidases are part of an enormous gene- family in T. cruzi, encoded by >3000 genes (Weatherly et al., unpublished), and are also the targets of the majority of T. cruzi specific CD8+ T cells in mice, with the responses to other proteins appearing significantly subdominant. Though T. cruzi trans-sialidases have been used as vaccine candidates (Costa et al., 1998; Fontanella et al., 2008), their diversity and strain-variant nature (Martin et al., 2006) make them unreliable targets for immunization against T. cruzi. For this reason, we have focused on the identification of other sub-dominant, strain-invariant epitopes from non-large gene-family proteins as potential vaccine candidates.
A number of studies in viral models have reported a lack of correlation between pathogen control and the general magnitude or breadth of T cell responses (Addo et al., 2003; Betts et al., 2001). In contrast, the timing of expression of an epitope by a pathogen can frequently be linked to the efficiency of T cell responses and to correlate with protection (Adnan et al., 2006; Yates et al., 2011). Hence, it is logical that even though strong, immunodominant CD8+ T cell responses are generated during T. cruzi infection, the relatively delayed expression on infected host cells of epitopes recognized by these T cells may make these responses less effective, and allow for parasite persistence despite the strength of this response.
The shedding of the trypomastigote flagellum soon after host cell entry places epitopes in flagellar proteins among the first signals identifying T. cruzi-infected cells to host T cells. Further, the earlier and progressively better control of a T. cruzi challenge infection in mice with enhanced numbers of PAR4-specific CD8+ T cells suggests that this ability to recognize infected cells earlier in the infection cycle is highly beneficial in terms of infection control.
In addition to the timing of presentation of flagella-derived peptides, a number of other characteristics make proteins in the T. cruzi flagellum particularly attractive as immunological targets. These are among the most abundant proteins in the parasite, are encoded by single genes and appear largely invariant among T. cruzi strains. On the negative side, the flagellar proteins may only be present in the host cells ephemerally, although PAR4-specific T cells recognize infected host cells until at least 48 hrs post infection (Figure 7). However, since production of trypomastigotes and reinvasion of new host cells is occurring continuously during infection, flagellar proteins are available to prime and boost T cell responses and to serve as targets for CD8+ T cells throughout the infection.
Given the apparent negative consequences to T. cruzi of shedding the flagellum upon entering host cells (i.e. providing early notification to the immune system that the host cell is infected), it is surprising that this route of disposal has survived natural selection. Possibly, this is the best option for jettisoning such a large and complex structure which might be difficult for the parasite to efficiently resorb and otherwise recycle. Also, unlike flagella in mammalian or algal cells, the flagellum in T. cruzi trypomastigotes is attached to the cell body along most its length. Adsorption of such a structure may be more disruptive than asymmetric division. Notably, the T cell responses induced by PAR4 are subordinate in terms of frequency to trans-sialidase-specific responses. Perhaps this is one way that the massive ts family of proteins serves to generate immune evasion, by deflecting the attention of the dominant T cell responses away from highly conserved proteins exposed early in the infection process and toward a set of related epitopes that are capable of constant change and are variable among isolates.
Conserved and abundant flagellar proteins, particularly ones like PAR4 that lack homologues in mammals, should be further investigated as potential vaccine targets for flagellated intracellular pathogens like Leishmania spp. and T. cruzi. We have explored this possibility with the intention of engineering attenuated strains of T. cruzi to be more immunogenic and immuneprotective. Such lines have promise as animal vaccines to reduce infection in companion animals and thus transmission to humans (Cohen and Gurtler, 2001). But these flagellar proteins may also have potential as conventional vaccines. Establishing a robust T cell response to these conserved proteins before the exposure to ts epitopes might short-circuit this potential evasion mechanism and result in much enhanced parasite control. The approach of letting the life history of a pathogen instruct on what the immune system should be responding to, rather than observing what it naturally responds to - perhaps misdirected to – in the course of infection might indeed be useful in vaccine development for other complex pathogens with vast numbers of potential vaccine targets.
EXPERIMENTAL PROCEDURES
Mice, parasites and infections
Female C57BL/6 mice were purchased from The Jackson Laboratory or bred and maintained in our animal facility under specific pathogen-free conditions. T. cruzi epimastigotes (Brazil strain) were transfected as described previously (Garg et al., 1997) with pTREX plasmid (Lorenzi et al., 2003) containing the coding sequence of full length (1743 bp) or truncated (1693 bp) T.cruzi paraflagellar rod protein 4 (PAR4) gene (TriTrypDB accession no: TcCLB.510353.30), with or without fusion to an upstream td-Tomato gene or influenza haemagglutinin (HA)-tag, to generate transgenic T. cruzi: TcPAR4. Unless otherwise indicated, all infections were initiated by inoculating vero cell culture passaged trypomastigote stage T. cruzi, intra-peritoneally (i.p) (104 parasites) or subcutaneously in the foot pad (f.p) (104 parasites). All animal protocols were approved by the University of Georgia Institutional Animal Care and Use Committee.
Microscopy
Immunofluorescence microscopy was performed to determine presence of the PAR4 or tr.PAR4 in epimastigote, trypomastigote and intracellular and extracellular amastigote stages of T. cruzi, modifying the protocol described previously (Agrawal et al., 2009), using TcPAR4, TctdTom (T. cruzi (CL strain) expressing the tdTomato protein (Canavaci et al., 2010)) or Tcwt strains to infect Vero or human foreskin fibroblast (HFF) cells. Extracellular amastigogensis was achieved by axenic conversion as described in detail before (Tomlinson et al., 1995). Briefly, T. cruzi trypomastigotes were incubated in RPMI media with 10% fetal calf serum, pH 5.0, at 37°C/5%CO2 to induce their conversion into amastigotes. The initial, intermediate and terminal morphological stages in amastigogenesis were observed by fluorescence microscopy and quantified by sampling at various time points during this process. Anti-PAR4 polyclonal serum from in mice, td-Tomato or anti-HA (Roche) antibodies were used to track PAR4 or tr.PAR4. Propidium Iodide or DAPI indicated DNA and anti-centrin (Chlamydomonas reinhardtii) (Millipore) distinguished T. cruzi basal body. T. cruzi plasma membrane localizing phosphatidyl inositol phospholipase C (anti-TcPiPLC, a gift from Dr. Roberto Docampo) or filipin (binding to cholesterol) were used to mark the parasite surface. Antibody to the kinetoplast associated protein (KAP)3 (anti-KAP3, a gift from Dr. Stenio Fragoso) marked the kinetoplast and MitoTracker Red (Invitrogen) that would accumulate only in active mitochondria delineated a functional kinetoplast. The presence of active DNA replication that is associated with nuclear division in the host cell or T. cruzi was determined by addition of BrdU (BD Biosciences). First and subsequent generations of amastigotes were observed by paraformaldehyde fixation of infected host cells before 12 hrs or after 24 hrs of infection respectively. Images were acquired with an Applied Precision Delta Vision microscope and were deconvolved and adjusted for contrast using its Softworx software.
In vivo cytotoxicity assay
Equal numbers of MC-PAR4(N) or MC-Ova were labeled with different concentrations of CellTrace Violet (CTV) (Invitrogen) to produce CTV hi MC-PAR4(N) and CTV lo MC-Ova. Equal numbers of either were then transferred i.p into recipients, and after 18 h, reisolated by peritoneal lavage to detect CTV stained cells by flow cytometry. Similarly, spleen cells from naive mice incubated for 1 h at 37°C with 10 µM PAR4 (3), PAR4(5), or SIINFEKL peptides were labeled with different concentrations of CFSE (Molecular Probes) as described before (Martin et al., 2006) to produce CFSE high and low populations. Equal numbers of CFSElabeled cells were transferred i.v into recipients, and after 18 h, splenocytes were isolated and CFSE-labeled cells were detected by flow cytometry. Percentage of specific lysis was determined in either case using the equation: 1 − [(% CTV or CFSElo naive/% CTV or CFSEhi naive)/(% CTV or CFSElo infected/CTV or CFSEhi infected)] × 100%.
Assessing protection
The acute control of T. cruzi challenge was determined as described previously (Collins et al., 2011). 2×105 TctdTom parasites were subcutaneously inoculated into both footpads of naïve mice, or mice drug-cured (Bustamante et al., 2008) from TcPAR4 or Tcwt infections (104 i.p), 30 days after the end of drug treatment. Mouse feet were imaged every day using the Maestro 2 in vivo imaging system (Caliper Life Sciences) with the green set of filters (acquisition settings: 560 to 750 in 10-nm steps; exposure time of 88.18 ms and 2 × 2 binning) for the indicated time period. The total fluorescent signal was quantified and the values represented as photons/cm2/second. Skeletal muscle tissue from challenged mice were analyzed at 30 dpi, by real-time PCR for the presence of T. cruzi (DNA) as described before (Cummings and Tarleton, 2003).
Assessing presentation of CD8+ T cell epitopes from T. cruzi infected cells
PAR4(3), TSKb20 or SIINFEKL specific CD8+ T cells were generated by dendritic cell (DC) vaccination as described previously (Padilla et al., 2009). In short, C57Bl/6 mice were vaccinated with bone-marrow derived DCs loaded with PAR4(3), TSKb20 or SIINFEKL peptides and boosted 5 days later with syngenic splenocytes loaded with the respective peptides. OVA specific CD8+ T cells were generated by infection with Listeria monocytogenes expressing ova (Lm-ova), as described previously (Berg et al., 2003). Splenocytes (106) collected 21 days post DC vaccination or 14 days post Lm-ova infection were co-incubated for 9–12 hrs with 105 murine fibroblast (MC57G fibrosarcoma cell line, ATCC) cells that have been infected for various lengths of time with Tcwt; or with the corresponding peptide epitopes as positive controls. For infecting MC57G cells, 106 T. cruzi trypomastigotes were added to 105 cells and washed off after 3 hrs. At the end of co-incubation with infected fibroblasts or the peptide epitopes, CD8+ CD44+ T cells were evaluated for intracellular IFN-γ as described above.
Statistical analysis
Data are presented as the mean plus/ minus the standard error of mean (SEM). Statistical analyses compared the groups with a two-tailed student t-test. Only p values of less than or equal to 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Angel Padilla, Dr. Juan Bustamante, Dr. Demba Sarr, Gretchen Cooley, Duo Peng and Bharath Kumar Bolla for technical assistance and all the members of Tarleton Research Group for helpful suggestions throughout this work. We acknowledge the insightful suggestions provided by Drs. Boris Striepen (UGA) and Philip Bastin (Pasteur institute) for this study and thank Drs. Stenio Fragoso and Roberto Docampo for providing anti-KAP(3) and anti-TcPiPLC sera respectively. We also thank Julie Nelson and Muthugapatti Kandasamy of the Center for Tropical and Emerging Global Diseases Flow Cytometry and Biomedical Microscopy Core facilities at the University of Georgia and the staff at the Coverdell Center rodent vivarium for their assistance. This work was supported by NIH Grants AI-22070 and AI-089952 to R.L.T
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
SPK designed and conducted the experiments and wrote the manuscript; RLT designed the experiments and wrote the manuscript.
REFERENCES
- Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnan S, Balamurugan A, Trocha A, Bennett MS, Ng HL, Ali A, Brander C, Yang OO. Nef interference with HIV-1-specific CTL antiviral activity is epitope specific. Blood. 2006;108:3414–3419. doi: 10.1182/blood-2006-06-030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NW, Hong KS, Robbins ES, Nussenzweig V. Stage-specific surface antigens expressed during the morphogenesis of vertebrate forms of Trypanosoma cruzi. Exp Parasitol. 1987;64:474–484. doi: 10.1016/0014-4894(87)90062-2. [DOI] [PubMed] [Google Scholar]
- Atwood JA, 3rd, Weatherly DB, Minning TA, Bundy B, Cavola C, Opperdoes FR, Orlando R, Tarleton RL. The Trypanosoma cruzi proteome. Science. 2005;309:473–476. doi: 10.1126/science.1110289. [DOI] [PubMed] [Google Scholar]
- Bastin P, MacRae TH, Francis SB, Matthews KR, Gull K. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol Cell Biol. 1999;19:8191–8200. doi: 10.1128/mcb.19.12.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Matthews KR, Gull K. The paraflagellar rod of kinetoplastida: solved and unsolved questions. Parasitol Today. 1996;12:302–307. doi: 10.1016/0169-4758(96)10031-4. [DOI] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, Forman J. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brener Z. Biology of Trypanosoma cruzi. Annual review of microbiology. 1973;27:347–382. doi: 10.1146/annurev.mi.27.100173.002023. [DOI] [PubMed] [Google Scholar]
- Bustamante JM, Bixby LM, Tarleton RL. Drug-induced cure drives conversion to a stable and protective CD8+ T central memory response in chronic Chagas disease. Nat Med. 2008;14:542–550. doi: 10.1038/nm1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavaci AM, Bustamante JM, Padilla AM, Perez Brandan CM, Simpson LJ, Xu D, Boehlke CL, Tarleton RL. In vitro and in vivo high-throughput assays for the testing of anti-Trypanosoma cruzi compounds. PLoS Negl Trop Dis. 2010;4:e740. doi: 10.1371/journal.pntd.0000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JE, Gurtler RE. Modeling household transmission of American trypanosomiasis. Science. 2001;293:694–698. doi: 10.1126/science.1060638. [DOI] [PubMed] [Google Scholar]
- Collins MH, Craft JM, Bustamante JM, Tarleton RL. Oral exposure to Trypanosoma cruzi elicits a systemic CD8(+) T cell response and protection against heterotopic challenge. Infect Immun. 2011;79:3397–3406. doi: 10.1128/IAI.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Franchin G, Pereira-Chioccola VL, Ribeirao M, Schenkman S, Rodrigues MM. Immunization with a plasmid DNA containing the gene of trans-sialidase reduces Trypanosoma cruzi infection in mice. Vaccine. 1998;16:768–774. doi: 10.1016/s0264-410x(97)00277-6. [DOI] [PubMed] [Google Scholar]
- Cummings KL, Tarleton RL. Rapid quantitation of Trypanosoma cruzi in host tissue by real-time PCR. Mol Biochem Parasitol. 2003;129:53–59. doi: 10.1016/s0166-6851(03)00093-8. [DOI] [PubMed] [Google Scholar]
- de Diego JL, Katz JM, Marshall P, Gutierrez B, Manning JE, Nussenzweig V, Gonzalez J. The ubiquitin-proteasome pathway plays an essential role in proteolysis during Trypanosoma cruzi remodeling. Biochemistry. 2001;40:1053–1062. doi: 10.1021/bi001659k. [DOI] [PubMed] [Google Scholar]
- de Souza W. Cell biology of Trypanosoma cruzi. International review of cytology. 1984;86:197–283. doi: 10.1016/s0074-7696(08)60180-1. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egui A, Thomas MC, Morell M, Maranon C, Carrilero B, Segovia M, Puerta CJ, Pinazo MJ, Rosas F, Gascon J, et al. Trypanosoma cruzi paraflagellar rod proteins 2 and 3 contain immunodominant CD8(+) T-cell epitopes that are recognized by cytotoxic T cells from Chagas disease patients. Mol Immunol. 2012;52:289–298. doi: 10.1016/j.molimm.2012.05.021. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Feldman AM, McNamara D. Myocarditis. The New England journal of medicine. 2000;343:1388–1398. doi: 10.1056/NEJM200011093431908. [DOI] [PubMed] [Google Scholar]
- Fontanella GH, De Vusser K, Laroy W, Daurelio L, Nocito AL, Revelli S, Contreras R. Immunization with an engineered mutant trans-sialidase highly protects mice from experimental Trypanosoma cruzi infection: a vaccine candidate. Vaccine. 2008;26:2322–2334. doi: 10.1016/j.vaccine.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Frickel EM, Sahoo N, Hopp J, Gubbels MJ, Craver MP, Knoll LJ, Ploegh HL, Grotenbreg GM. Parasite stage-specific recognition of endogenous Toxoplasma gondii-derived CD8+ T cell epitopes. J Infect Dis. 2008;198:1625–1633. doi: 10.1086/593019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Nunes MP, Tarleton RL. Delivery by Trypanosoma cruzi of proteins into the MHC class I antigen processing and presentation pathway. J Immunol. 1997;158:3293–3302. [PubMed] [Google Scholar]
- Gonzalez J, Ramalho-Pinto FJ, Frevert U, Ghiso J, Tomlinson S, Scharfstein J, Corey EJ, Nussenzweig V. Proteasome activity is required for the stage-specific transformation of a protozoan parasite. J Exp Med. 1996;184:1909–1918. doi: 10.1084/jem.184.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi HA, Vazquez MP, Levin MJ. Integration of expression vectors into the ribosomal locus of Trypanosoma cruzi. Gene. 2003;310:91–99. doi: 10.1016/s0378-1119(03)00502-x. [DOI] [PubMed] [Google Scholar]
- Luhrs KA, Fouts DL, Manning JE. Immunization with recombinant paraflagellar rod protein induces protective immunity against Trypanosoma cruzi infection. Vaccine. 2003;21:3058–3069. doi: 10.1016/s0264-410x(03)00108-7. [DOI] [PubMed] [Google Scholar]
- Maga JA, LeBowitz JH. Unravelling the kinetoplastid paraflagellar rod. Trends in cell biology. 1999;9:409–413. doi: 10.1016/s0962-8924(99)01635-9. [DOI] [PubMed] [Google Scholar]
- Maga JA, Sherwin T, Francis S, Gull K, LeBowitz JH. Genetic dissection of the Leishmania paraflagellar rod, a unique flagellar cytoskeleton structure. Journal of cell science. 1999;112(Pt 16):2753–2763. doi: 10.1242/jcs.112.16.2753. [DOI] [PubMed] [Google Scholar]
- Martin DL, Weatherly DB, Laucella SA, Cabinian MA, Crim MT, Sullivan S, Heiges M, Craven SH, Rosenberg CS, Collins MH, et al. CD8+ T-Cell responses to Trypanosoma cruzi are highly focused on strain-variant trans-sialidase epitopes. PLoS Pathog. 2006;2:e77. doi: 10.1371/journal.ppat.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailowsky V, Luhrs K, Rocha MO, Fouts D, Gazzinelli RT, Manning JE. Humoral and cellular immune responses to Trypanosoma cruzi-derived paraflagellar rod proteins in patients with Chagas' disease. Infect Immun. 2003;71:3165–3171. doi: 10.1128/IAI.71.6.3165-3171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Wrightsman RA, Manning JE. Trypanosoma cruzi: protective immunity in mice immunized with paraflagellar rod proteins is associated with a T-helper type 1 response. Exp Parasitol. 1996;84:156–167. doi: 10.1006/expr.1996.0101. [DOI] [PubMed] [Google Scholar]
- Nagajyothi F, Weiss LM, Silver DL, Desruisseaux MS, Scherer PE, Herz J, Tanowitz HB. Trypanosoma cruzi utilizes the host low density lipoprotein receptor in invasion. PLoS Negl Trop Dis. 2011;5:e953. doi: 10.1371/journal.pntd.0000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla AM, Simpson LJ, Tarleton RL. Insufficient TLR activation contributes to the slow development of CD8+ T cell responses in Trypanosoma cruzi infection. J Immunol. 2009;183:1245–1252. doi: 10.4049/jimmunol.0901178. [DOI] [PubMed] [Google Scholar]
- Portman N, Gull K. The paraflagellar rod of kinetoplastid parasites: from structure to components and function. Int J Parasitol. 2010;40:135–148. doi: 10.1016/j.ijpara.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. The Journal of cell biology. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg CS, Martin DL, Tarleton RL. CD8+ T cells specific for immunodominant trans-sialidase epitopes contribute to control of Trypanosoma cruzi infection but are not required for resistance. J Immunol. 2010;185:560–568. doi: 10.4049/jimmunol.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotureau B, Subota I, Buisson J, Bastin P. A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development. 2012;139:1842–1850. doi: 10.1242/dev.072611. [DOI] [PubMed] [Google Scholar]
- Saravia NG, Hazbon MH, Osorio Y, Valderrama L, Walker J, Santrich C, Cortazar T, Lebowitz JH, Travi BL. Protective immunogenicity of the paraflagellar rod protein 2 of Leishmania mexicana. Vaccine. 2005;23:984–995. doi: 10.1016/j.vaccine.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Sharma R, Gluenz E, Peacock L, Gibson W, Gull K, Carrington M. The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009;25:517–524. doi: 10.1016/j.pt.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Peacock L, Gluenz E, Gull K, Gibson W, Carrington M. Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes. Protist. 2008;159:137–151. doi: 10.1016/j.protis.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Tarleton RL, Koller BH, Latour A, Postan M. Susceptibility of beta 2-microglobulin-deficient mice to Trypanosoma cruzi infection. Nature. 1992;356:338–340. doi: 10.1038/356338a0. [DOI] [PubMed] [Google Scholar]
- Tarleton RL, Sun J, Zhang L, Postan M. Depletion of T-cell subpopulations results in exacerbation of myocarditis and parasitism in experimental Chagas' disease. Infect Immun. 1994;62:1820–1829. doi: 10.1128/iai.62.5.1820-1829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson S, Vandekerckhove F, Frevert U, Nussenzweig V. The induction of Trypanosoma cruzi trypomastigote to amastigote transformation by low pH. Parasitology. 1995;110(Pt 5):547–554. doi: 10.1017/s0031182000065264. [DOI] [PubMed] [Google Scholar]
- Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. Int J Parasitol. 2001;31:472–481. doi: 10.1016/s0020-7519(01)00153-9. [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele J, Claes Y, van Bockstaele D, Le Ray D, Coosemans M. Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology. 1999;118(Pt 5):469–478. doi: 10.1017/s0031182099004217. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, McElhaugh MJ, Eisenlohr LC. Generation of CD8(+) T cell memory in response to low, high, and excessive levels of epitope. J Immunol. 2002;168:4455–4461. doi: 10.4049/jimmunol.168.9.4455. [DOI] [PubMed] [Google Scholar]
- Wrightsman RA, Luhrs KA, Fouts D, Manning JE. Paraflagellar rod protein-specific CD8+ cytotoxic T lymphocytes target Trypanosoma cruzi-infected host cells. Parasite Immunol. 2002;24:401–412. doi: 10.1046/j.1365-3024.2002.00479.x. [DOI] [PubMed] [Google Scholar]
- Wrightsman RA, Miller MJ, Saborio JL, Manning JE. Pure paraflagellar rod protein protects mice against Trypanosoma cruzi infection. Infect Immun. 1995;63:122–125. doi: 10.1128/iai.63.1.122-125.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AJ, Van Baalen M, Antia R. Virus replication strategies and the critical CTL numbers required for the control of infection. PLoS computational biology. 2011;7:e1002274. doi: 10.1371/journal.pcbi.1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tarleton RL. Parasite persistence correlates with disease severity and localization in chronic Chagas' disease. J Infect Dis. 1999;180:480–486. doi: 10.1086/314889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.