Abstract
Much of the mechanism by which Wnt signaling drives proliferation during oncogenesis is attributed to its regulation of the cell cycle. Here, we show how Wnt/β-catenin signaling directs another hallmark of tumorigenesis, namely Warburg metabolism. Using biochemical assays and fluorescence lifetime imaging microscopy (FLIM) to probe metabolism in vitro and in living tumors, we observe that interference with Wnt signaling in colon cancer cells reduces glycolytic metabolism and results in small, poorly perfused tumors. We identify pyruvate dehydrogenase kinase 1 (PDK1) as an important direct target within a larger gene program for metabolism. PDK1 inhibits pyruvate flux to mitochondrial respiration and a rescue of its expression in Wnt-inhibited cancer cells rescues glycolysis as well as vessel growth in the tumor microenvironment. Thus, we identify an important mechanism by which Wnt-driven Warburg metabolism directs the use of glucose for cancer cell proliferation and links it to vessel delivery of oxygen and nutrients.
Keywords: angiogenesis, colon cancer, fluorescence lifetime imaging, metabolism, Wnt
See also: CB Thompson (July 2014)
Introduction
Normal patterns of Wnt signaling are necessary for tissue development and maintenance, but aberrant Wnt signaling is implicated in many cancers, especially colon cancer (Bienz & Clevers, 2000). Overactive Wnt signaling leads to constitutively active β-catenin, which via LEF/TCF (lymphoid enhancer factor/T-cell factor) transcription factors leads to inappropriate activation of Wnt target genes (Clevers, 2006; Klaus & Birchmeier, 2008). Previous studies have identified multiple functional outputs of oncogenic Wnt signaling, including proliferation, EMT induction, angiogenesis, migration, and cell survival (Brabletz et al, 2005). Here, we propose a novel function in the regulation of cancer metabolism.
Cancer metabolism is quickly regaining a forefront position in research as its role in epigenetics, proliferation, and survival is understood to be fundamentally connected. Otto Warburg first recognized that cancer cells ferment much of their glucose supply into lactate regardless of the presence of oxygen, a phenomenon termed the Warburg effect, or aerobic glycolysis (Warburg, 1956). It is now appreciated that cancer cells have different metabolic demands than normal cells and they therefore modify their use of metabolites to meet those demands. Instead of a dominant program for efficient production of ATP, proliferating tumor cells rely on a metabolic program of glycolysis to support anabolic production of biomass (DeBerardinis et al, 2008; Vander Heiden et al, 2009). An emphasis on glycolysis is thought to be driven not only from exposure to hypoxic conditions [mostly through stabilized HIF1α (Semenza, 2010)] but also by oncogenic signaling pathways, such as PI3K/AKT (Elstrom et al, 2004; Ward & Thompson, 2012). Although Wnt plays a well-known role in homeostatic liver metabolism (Liu et al, 2011) and can crosstalk with metabolic pathways in normal cells such as differentiating osteoblasts (Sethi & Vidal-Puig, 2010; Esen et al, 2013), there are little data implicating Wnt directly in the metabolic reprogramming of cancer.
Here, we probe the contribution of LEF/TCF/β-catenin activity to the metabolic programming of cancer cells. We block the downstream activity of Wnt through the disruption of β-catenin/LEF/TCF complexes by overexpressing dominant negative (dn)LEF/TCF isoforms that lack the β-catenin binding domain. We utilize both standard metabolomics analyses and a state-of-the-art microscopy technique for monitoring changes in the metabolism of living cells and tissues. The microscopy technique, called the phasor approach to fluorescence lifetime imaging microscopy (FLIM), utilizes two-photon microscopy for label-free detection of intrinsically autofluorescent molecules (Digman et al, 2008; Stringari et al, 2011). In our study, we used FLIM to monitor the metabolic coenzyme nicotinamide adenine dinucleotide (NADH), the principle electron acceptor in glycolysis and electron donor in oxidative phosphorylation. NADH FLIM is especially powerful as it provides a non-invasive, rapid, and sensitive readout of the metabolic status of single cells within their native microenvironment. We recently used FLIM analysis to discover that stem cells at the base of intestinal crypts are highly glycolytic and that a metabolic trajectory of glycolysis-to-oxidative phosphorylation tracks with a gradient of strong-to-weak Wnt signaling in intestinal crypts (Stringari et al, 2012). Here, we show through FLIM detection of free and bound NADH in vitro in living cancer cells and in vivo in living perfused tumors that blocking Wnt alters the metabolic program of cancer cells and leads to reduced use of aerobic glycolysis. We identify pyruvate dehydrogenase kinase (PDK1) as a novel Wnt target gene that promotes this effect, and we note that a cell-extrinsic effect of PDK1-driven glycolysis is enhanced tumor angiogenesis.
Results
Blocking Wnt alters the expression of metabolism-linked genes
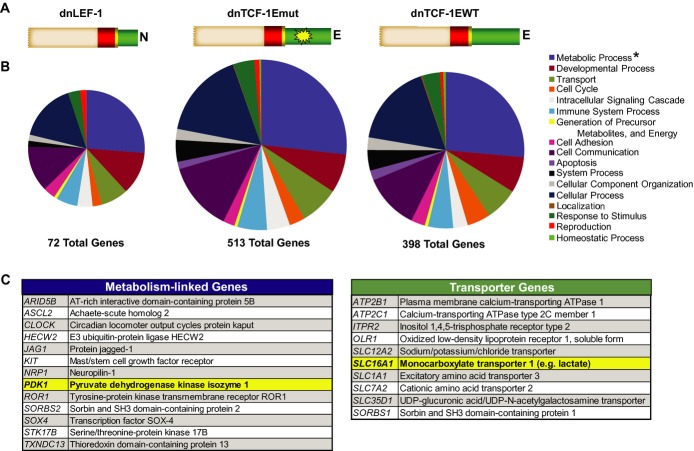
Although multiple roles of Wnt signaling in colon cancer have been well studied, such as proliferation via regulation of the G1 phase of the cell cycle (van de Wetering et al, 2002), there are other functions that have yet to be identified. To reveal new roles of Wnt in colon cancer, we performed Gene Ontology analysis of our recently published microarray data set (Hoverter et al, 2012). This data set reflected the change in global gene expression when three different dominant negative (dn) LEF/TCF isoforms were individually and rapidly induced with doxycycline in DLD-1 colon cancer cells (Fig1A). Induction of dnLEF/TCFs reduces Wnt signaling through interference with endogenous β-catenin/TCF and β-catenin/LEF complexes, and we therefore focused our analysis on genes that were downregulated within 8 and 22 h after induction. The ontology analysis was carried out using PANTHER software which classified the downregulated genes into different categories of biological processes (Thomas, 2003; Thomas et al, 2006). A consistently large subset of regulated genes fell under the category of metabolism (Fig1B and C; Supplementary Table S1). In fact, Panther binomial statistical analysis revealed that Wnt target genes connected to metabolism were the most highly overrepresented category compared to baseline representation of these genes in the human genome. These data, along with the notable downregulation of two key genes critical to cancer metabolism, pyruvate dehydrogenase kinase (PDK1) and the lactate transporter, MCT-1 (SLC16A1), led us to hypothesize that Wnt specifically and directly regulates a program of cellular metabolism in colon cancer cells.
Figure 1. Blocking Wnt alters the expression of metabolism-linked genes.
Panther Gene Ontology analysis of microarray data from DLD-1 cells overexpressing either dnLEF-1 (23 h), dnTCF-1Emut (8 h), or dnTCF-1EWT (8 h).
- Schematic of dnLEF/TCF isoforms expressed (beige box = context-dependent regulatory domain; red box = HMG DNA binding domain; yellow box = nuclear localization signal; green box = alternatively spliced C-terminal tails; yellow star = mutation in E-tail).
- Ontology analysis of dnLEF/TCF-downregulated genes reveals a large category of genes linked to metabolism. PANTHER binomial statistical analysis determined statistically significant overrepresentation of regulated genes within each category compared to representation in the human genome (*P-value < 0.01).
- List of metabolism-linked genes downregulated by all three dnLEF/TCF isoforms. List of transporter genes regulated by at least 2 dnLEF/TCF isoforms. For all genes on the list changes are P < 0.05, and range of fold changes is −1.3 to −6.0.
dnLEF-1 and dnTCF-1Emut do not alter the cell cycle or proliferation in vitro
To test this hypothesis, we used dnLEF/TCF overexpression to block Wnt target gene regulation in colon cancer cell lines and examined resultant changes in metabolism. Knowing that overexpression of the potent dnTCF-1E or dnTCF-4E isoforms halts cell cycle progression and cell proliferation (van de Wetering et al, 2002), while isoforms lacking the alternatively spliced C-terminal E-tail avoid this stall (Naishiro et al, 2001; Atcha et al, 2007), we induced the expression of dnLEF-1N which naturally lacks E-tail sequences. In parallel, we also induced the expression of dnTCF-1Emut, a mutated form of dnTCF-1E with a five-amino acid substitution in the auxiliary C-clamp DNA binding domain in the E-tail that eliminates the control of proliferation (Atcha et al, 2007). Use of these less potent dominant negative isoforms enabled an uncoupling of our test for effects on cell metabolism from effects on cycle progression. We utilized two different expression systems in two different colon cancer cell lines. In two independent clonal lines of DLD-1 colon cancer cells, we used a stably integrated doxycycline inducible system to express Flag-tagged dnLEF-1 [dnLEF-1(1) and dnLEF-1(2)]. In a second cell line (SW480), we used a lentiviral system to express physiological levels of Flag-dnLEF-1 or Flag-dnTCF-1Emut. In all cases, transgene expression was assessed by Western blot (Supplementary Fig S1A and E), and functional disruption of Wnt activity was confirmed by repression of the luciferase reporter Super TOPFlash (STOP) (Supplementary Fig S1B and F). We also confirmed that dnLEF-1 and dnTCF-1Emut had no impact on cell proliferation (Supplementary Fig S1C and G) or any parameter of the cell cycle profile (Supplementary Fig S1D and H). Cell cycle analysis performed at even longer time points of dnLEF/TCF expression also showed no change in the profile (data not shown). Overall, these data show that dnLEF-1N and dnTCF-1Emut expression have no effect on the cell cycle or the intrinsic ability of the cells to proliferate and can therefore reveal phenotypes of blocking Wnt that are uncoupled from these functions.
Blocking Wnt alters the metabolic program of colon cancer cells
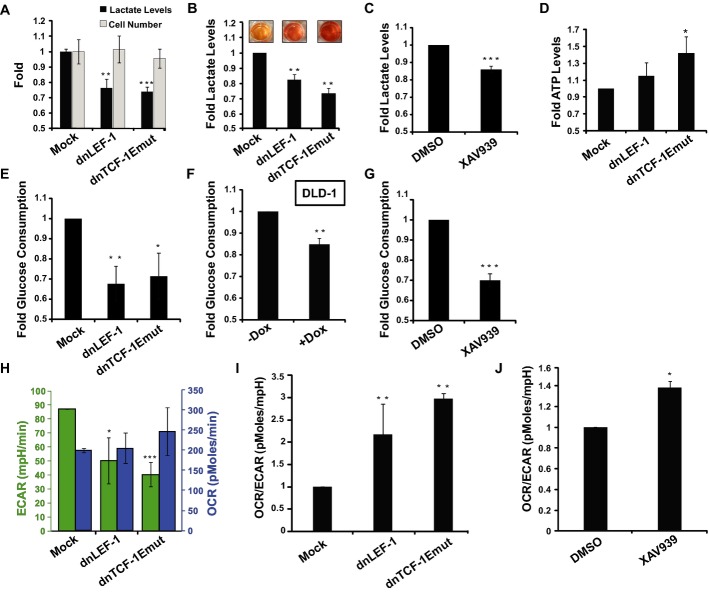
One of the most recognized hallmarks of cancer is Warburg metabolism, or a tendency for cancer cells to utilize glycolysis rather than oxidative phosphorylation regardless of the availability of oxygen (Warburg, 1956). We tested whether blocking Wnt alters carbohydrate metabolism by testing for changes in lactate (a byproduct of glycolysis) and ATP production (most efficiently produced via oxidative phosphorylation). A measure of lactate levels in the media of mock-infected SW480 cells compared to cells infected with lentivirus expressing dnLEF-1 or dnTCF-1Emut revealed a significant decrease in lactate production (Fig2A and B). This was true for both 2D conventional cultures and 3D suspension cultures in soft agar which were accompanied by a visual color change of the media (due to the secretion of excess lactic acid; Fig2B). The drop in lactate production occurred independently of cell number, which was not significantly altered because of the lack of any effect of dnLEF/TCF expression on proliferation (Fig2A). Similar analyses were performed in four different colon cancer cell lines (Supplementary Fig S2). We also interfered with Wnt by application of the drug XAV939. This small molecule inhibitor targets poly-ADP-ribosyltransferases tankyrase 1, 2 to destabilize β-catenin, an action that works even in colon cancer cells where its destruction complex components are defective (Huang et al, 2009). We observed similar decreases in lactate production upon XAV939 application, a change consistent with a decrease in Warburg-type metabolism (Fig2C). Another hallmark of Warburg is less efficient production of ATP (since oxidative phosphorylation produces more ATP per molecule of glucose than glycolysis). We therefore measured changes in ATP levels in SW480 cells infected with dnLEF/TCF-expressing lentivirus. We observed that any interference with Wnt signaling triggered increases in ATP production, suggesting an increased utilization of oxidative phosphorylation versus glycolysis (Fig2D). We also tested for rates of glucose consumption, and as expected, rates were lower when either dnLEF/TCFs or XAV939 was used to disrupt Wnt signaling (Fig2E–G). For more precise measurements, we used Seahorse technologies to measure the glycolytic flux (extracellular acidification rate or ECAR) and mitochondrial respiration (oxygen consumption rate or OCR) of colon cancer cells subjected to our conditions. We observed that expression of dnLEF/dnTCF in SW480 cells lowered the rate of glycolysis, but did not significantly affect the rates of respiration (Fig2H and I). XAV939 treatment induced a similar effect, which we report as a 30–40%increase in the rates of oxidative phosphorylation relative to glycolysis (OCR/ECAR ratio; Fig2J).
Figure 2. Blocking Wnt alters the metabolic program of colon cancer cells.
A Lactate levels are reduced with lentiviral transduction of dnLEF-1 or dnTCF-1Emut in SW480 cells growing under standard culture conditions for 10 days. Representative graph of three replicates is shown with error bars representing the SEM between three internal replicates.
B Fold change of lactate levels produced in 3D cultures. Images are of representative wells for each condition. Measurements performed on media collected from SW480 cells grown in soft agar after 22 days. Representative graph of three replicates is shown with error bars representing the SEM between three internal replicates.
C Fold change in lactate levels of SW480 cells treated with Wnt inhibitor XAV939 (10 μM) for a minimum of 4 days. Data represent the average of six independent trials (± SEM).
D ATP levels in SW480 cells collected 7 days post-transduction. Data represent the average of three independent trials (± SEM).
E–G Fold changes in glucose consumption in SW480 cells expressing dnLEF/TCFs (E), DLD-1 cells [dnLEF-1(2)] treated with doxycycline to induce dnLEF-1 expression (F), and SW480 cells treated with XAV939 (10 μM) (G). Data represent the average of four trials (± SEM).
H Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in SW480 cells transduced with MOCK, dnLEF-1, or dnTCF-1Emut virus. Data represent the average of three independent trials (± SD).
I Data from (H) represented as an OCR/ECAR ratio.
J OCR/ECAR ratio of SW480 cells treated with XAV939 (10 μM). Data represent the average of three independent trials (± SD).
Data information: *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001.
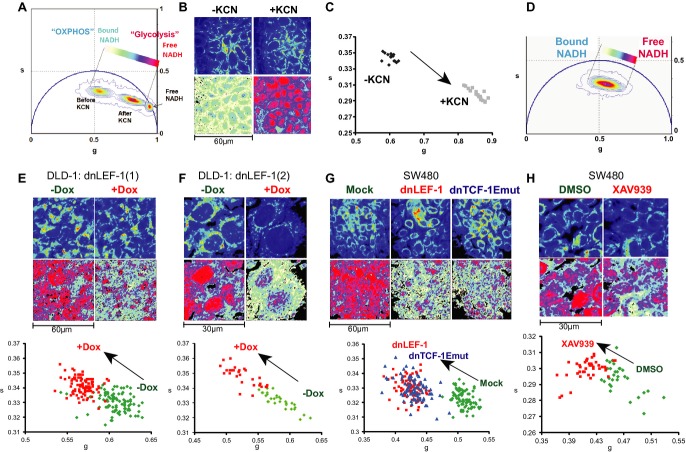
While the above technologies are quantitative, they are single end-point measurements of an entire population of cells. Recognizing that cancer metabolism can be heterogeneous and is best evaluated in living cells, we utilized the phasor approach to fluorescence lifetime imaging microscopy (FLIM) to evaluate metabolism. Specifically, we used FLIM to monitor dynamic shifts in patterns of aerobic glycolysis versus oxidative phosphorylation by following the signature of the metabolic and autofluorescent cofactor NADH. NADH autofluorescence can be excited at a specific wavelength (740 nm), and the pattern of the decay of this fluorescence differs depending on its bound or unbound state (Lakowicz et al, 1992). Glycolytic cells have a predominance of free, unbound NADH. Respiring cells have high rates of oxidative phosphorylation and a predominance of bound NADH (e.g., NADH bound to mitochondrial enzymes; Monici et al, 2003; Croce et al, 2004). The signature of fluorescence decay of NADH can be graphically represented on a 2D phasor plot where the decay rates for the pure free or bound species of NADH occupy very different positions (Becker et al, 2004; Colyer et al, 2008; Digman et al, 2008; Stringari et al, 2011). In a complex cellular environment where combinations of free and bound NADH co-exist, fluorescence signatures of decay map to experimental points between the extreme phasor plot positions of the pure free and bound species (Fig3A). In other words, experimental positions on the phasor plot indicate the relative ratio of free to bound NADH and therefore the relative levels of glycolysis and oxidative phosphorylation. In our analysis, increases in the free/bound NADH ratio (rightward and downward shift of experimental points in the phasor plot) indicate more glycolysis and an overall cancer type of metabolism (Bird et al, 2005; Skala et al, 2007; Yuan et al, 2007; Yu & Heikal, 2009).
Figure 3. Fluorescence lifetime analysis of NADH reveals shifts in glycolysis upon expression of dnLEF/TCF.
A Phasor plot of a FLIM analysis showing DLD-1 cells before and after a 1-min treatment with 4 mM potassium cyanide (KCN). The phasor position of pure free NADH is also shown.
B, C Top panel of images in (B) shows the autofluorescent intensity at 740 nm. Bottom panels in (B) correspond to the free/bound NADH coloring of the same field of cells according to the color map shown on the phasor plot in (A). Changes indicate an increase in free/bound NADH after KCN treatment. This is also shown in (C), a scatterplot analysis of average phasor positions of individual cells before and after KCN treatment.
D Phasor position of DLD-1/dnLEF-1(1) cells, and a color map to represent the gradient of relative levels of free and bound NADH.
E–H The top row of images show two-photon fluorescence intensity images excited at 740 nm for two clones of DLD-1 dnLEF-1 stable cells [dnLEF-1(1) and dnLEF-1(2)] and SW480 cells. The bottom row shows free/bound NADH color mapping as indicated by the color map in (D). The bottom panels show scatterplots where each point represents the average phasor position from one cell. Induction of dnLEF-1 or dnTCF-1Emut expression, or treatment with the Wnt inhibitor XAV939, results in a phasor shift toward bound NADH. All images and measurements were taken 5 days after seeding under confluent conditions. Representative data from single trials are shown from among at least three replicate experiments for each cell line. Each treatment resulted in a population on the scatterplot distinct from mock cells with P < 0.0001.
To demonstrate the relationship between a phasor shift and a change in metabolism, cells were treated with potassium cyanide (KCN) for 1 min to block mitochondrial respiration and trigger an increase in reduced, free NADH. FLIM analysis (using two-photon microscopy at 740 nm to excite bound and free NADH) before and after KCN treatment moved the phasor distribution closer to the pure free NADH position, indicating a release of NADH from mitochondria and an increase in the ratio of free to bound NADH (Fig3A). This result is also visually confirmed through a false-color mapping of free/bound NADH overlaid pixel-by-pixel on the original images taken of the cells, where pink/red and blue/white represent, respectively, higher and lower concentrations of free NADH relative to bound NADH (Fig3B). A scatterplot display of the data enables a comparison of the average position of individual cells for each set of conditions and therefore reveals the spread of the data between replicates of the same conditions as well as between different experimental conditions. In this case, the phasor positions of each pixel within one cell are averaged and plotted as a single point (Fig3C).
To assess the impact of blocking Wnt target gene activation on the ratio of free to bound NADH, we performed FLIM analysis on normoxic cells expressing dnLEF/TCFs or cells treated with the XAV939 inhibitor. Doxycycline induction of dnLEF-1N in two different clonal DLD-1 cell lines caused the average phasor position to shift away from free NADH to the upper left of the scatterplot, toward bound NADH (Fig3D, E and F). This shift represents a decrease in free/bound NADH, a change that was also evident in the color mapping of NADH states in the DLD-1 cells. Parental DLD-1 cells treated with the same concentration of doxycycline showed no significant change in the phasor positions, indicating that doxycycline alone does not alter the metabolic signature (Supplementary Fig S3D). To confirm this result in other colon cancer cells, we performed FLIM analysis on SW480 cells infected with empty virus or lentivirus carrying dnLEF-1N or dnTCF-1Emut expression cassettes (Fig3G; additional dnTCF isoforms in Supplementary Fig S3A and C), as well as analysis of cells treated overnight with XAV939, the Wnt signaling inhibitor (Fig3H). We also tested four other colon cancer cell lines that varied with respect to their endogenous level of Wnt signaling (Supplementary Fig S3C). Shifts in metabolism correlated directly with the ability to reduce Wnt signaling. These data suggest that blocking Wnt signaling in colon cancer cells results in a decrease in the ratio of free to bound NADH, which indicates a decrease in glycolysis. Taken together, our assessment of lactate production, ATP production, ECAR, OCR, glucose consumption, and FLIM signatures of NADH all indicates that oncogenic Wnt signaling promotes a glycolytic form of metabolism.
Blocking Wnt reduces PDK1 levels via regulation of transcription
We next asked whether the changes in metabolism induced by blocking Wnt could be attributed to any of the metabolism-linked target genes identified in the DLD-1 microarray experiments. One common target gene downregulated by all of the dnLEF/TCF proteins was pyruvate dehydrogenase kinase (PDK1). This kinase phosphorylates and inhibits the pyruvate dehydrogenase (PDH) complex in mitochondria (Roche et al, 2001). Inhibition of PDH, in turn, reduces the conversion of pyruvate to acetyl-CoA for entry into the TCA cycle and oxidative phosphorylation. Therefore, PDK1 plays an important modulatory role in the promotion of a glycolytic phenotype and is therefore not surprisingly found to be upregulated in a number of cancers (Koukourakis et al, 2006; Wigfield et al, 2008; Baumunk et al, 2012).
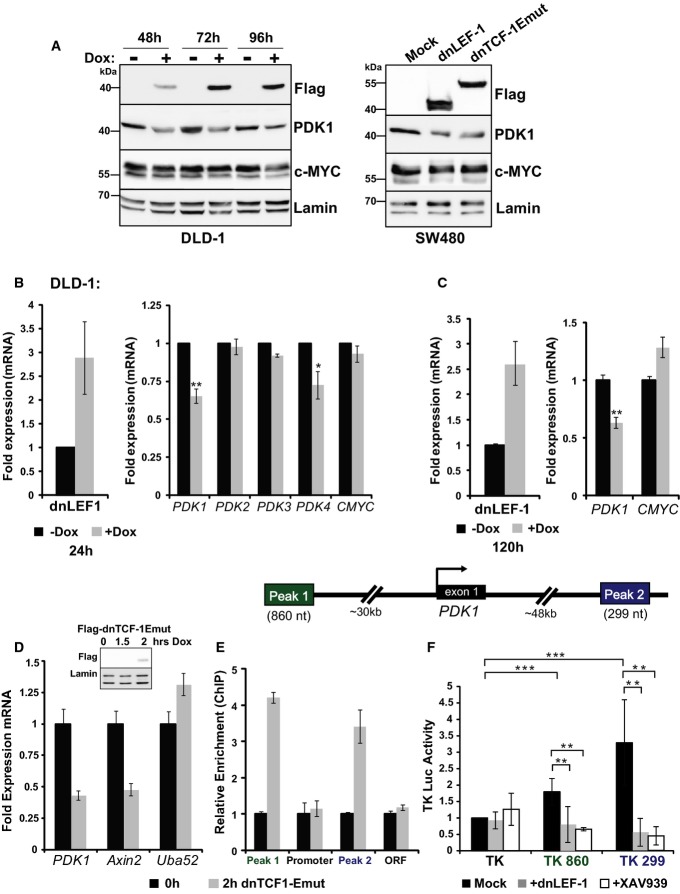
To validate the microarray data and test whether PDK1 levels are modulated by β-catenin and LEF/TCFs, Western blot analysis was performed for both DLD-1 and SW480 cells expressing dnLEF-1 or dnTCF-1Emut. PDK1 protein levels were reduced 50–75%for all cell lines (Fig4A). To confirm that this reduction is evident at the mRNA level, RT-qPCR analysis of PDK1 mRNA was performed for doxycycline-treated dnLEF-1 DLD-1 cells after 24 or 120 h of expression. At both time points, PDK1 mRNA levels were reduced to 60%, similar to the reduction observed at the protein level (Fig4B and C). Since there are four members of the PDK family (PDK1-4), we asked whether any of the additional PDK family members were also downregulated after blocking Wnt. RT-qPCR analysis of PDK2-4 levels in DLD-1 cells expressing dnLEF-1 indicates that neither PDK2 nor PDK3 levels were altered, while PDK4 showed a modest, but significant decrease from its already low basal level (Fig4B). Overall, these data suggest that PDK1 and possibly PDK4 are regulated by Wnt/β-catenin signaling in colon cancer cells.
Figure 4. Blocking Wnt directly reduces PDK1 levels via regulation of transcription.
A Whole-cell lysates from DLD-1 dnLEF-1(1) cells were collected 48, 72, and 96 h after 0.01 μg/ml doxycycline treatment and were probed with the antibodies shown. SW480 cells were harvested 48 h post-transduction.
B, C 9RT-qPCR analysis was performed on RNA collected from DLD-1 dnLEF-1(2) cells harvested 24 h (B) or 120 h (C) after the addition of doxycycline. Graphs shown represent the average of three trials (± SEM).
D RT-qPCR analysis was performed on 4-thiouridine-labeled RNA isolated from a 30-min pulse in the presence/absence of dnTCF-1Emut, induced by 2-h doxycycline treatment in DLD-1 cells. Known Wnt target genes Axin2 and Uba52 were used as positive and negative controls, respectively. A representative graph is shown of two replicates, with error bars representing the SD among three internal replicates.
E RT-qPCR analysis of chromatin immunoprecipitated from DLD-1 cells with or without induction of FLAG-dnTCF-1Emut, using anti-FLAG antibody. PCR primers designed to detect the indicated PDK1 genomic regions show that dnTCF-1Emut associates with distal regions that flank the PDK1 locus. A representative graph is shown of two replicates, with error bars representing the SD among three internal replicates.
F Luciferase reporter activity in SW480 cells shows that Peak 1 and Peak 2 regions confer elevated transcription activity to the heterologous thymidine kinase (TK) promoter. Expression of transduced dnLEF-1, or treatment with the Wnt inhibitor XAV939 (10 μM), eliminates the regulatory activity of these fragments. Graph shown represents the average of three independent replicates (± SD).
Data information: *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001).
Source data are available online for this figure.
A possible connection between Wnt and metabolism occurs through the Wnt target gene, c-Myc, and in fact, previous reports suggest that under certain conditions, c-Myc can potentiate the upregulation of PDK1 levels (Kim et al, 2007). We examined c-Myc levels after expression of dnLEF/TCF. With the exception of a slight decrease in c-Myc protein after 96 h of dnLEF-1 expression in DLD-1 cells (Fig4A), there is little to no difference in c-Myc mRNA or protein levels even after 120 h (Fig4A and C). This result also matches the previously published microarray, which showed no change in c-Myc mRNA after expression of dnLEF-1. Therefore, the change in PDK1 levels observed is independent of Wnt regulation of c-Myc.
To determine the kinetics of PDK1 transcription upon disruption of Wnt signaling, we used a 4-thiouridine labeling procedure to identify actively transcribed mRNAs (Dölken et al, 2008). After induction of dnTCF-1Emut for 2 h, DLD-1 cells were incubated with a pulse of 4-thiouridine for 30 min to incorporate the nucleotide label into nascently transcribed RNA. After the pulse, cells were harvested and labeled RNA was purified via a biotinylation/streptavidin pull-down procedure (Dölken et al, 2008). Semi-quantitative RT-PCR for PDK1 mRNA demonstrated that transcription of the PDK1 locus was immediately reduced upon the induction of dnTCF1E-mut expression (Fig4D). The known Wnt target gene AXIN2 served as a positive control, and constitutive, ubiquitous UBA52 served as a negative control. Induction of dnTCF-1Emut reduced transcription by at least 50%for both PDK1 and AXIN2, demonstrating that PDK1 is likely to be a direct Wnt target gene. We have also performed a genome-wide ChIP-seq study of dnTCF-1E binding (N.P. Hoverter, M.D. Zeller, M.M. McQuade, A. Garibaldi, A. Busch, K.J. Hertel, P. Baldi and M.L. Waterman, in preparation) and discovered in the genome data set that TCF-1 binds to distal upstream (‘Peak 1’) and downstream (‘Peak 2’) sites surrounding the PDK1 locus (Fig4E). Each genomic region of TCF-1 occupancy contains three putative Wnt response elements (Supplementary Fig S4). We designed PCR primers specific to these two regions as well as the PDK1 promoter and an internal site in the locus as a negative control. Using chromatin immunoprecipitation procedures, we determined that TCF-1 directly binds to the distal sites but not the promoter (Fig4E). To test whether these sites confer transcription regulation in colon cancer cells, we subcloned each distal region next to the heterologous thymidine kinase promoter and luciferase open reading frame. Transient transfection assays showed that both fragments increased promoter activity in SW480 colon cancer cells and induction of either dnLEF-1 or treatment with XAV939 eliminated this regulation (Fig4F). We also tested these fragments for their ability to regulate the PDK1 promoter (Supplementary Fig S4). While both regions conferred a distinct, modest level of activation, the PDK1 promoter itself was markedly sensitive to downregulation by dnLEF-1 (Supplementary Fig S4). These data demonstrate that PDK1 is a direct Wnt target gene and that regulation occurs through distal regulatory sites upstream and downstream of the locus.
PDK1 overexpression rescues the altered metabolic phenotype induced by blocking Wnt
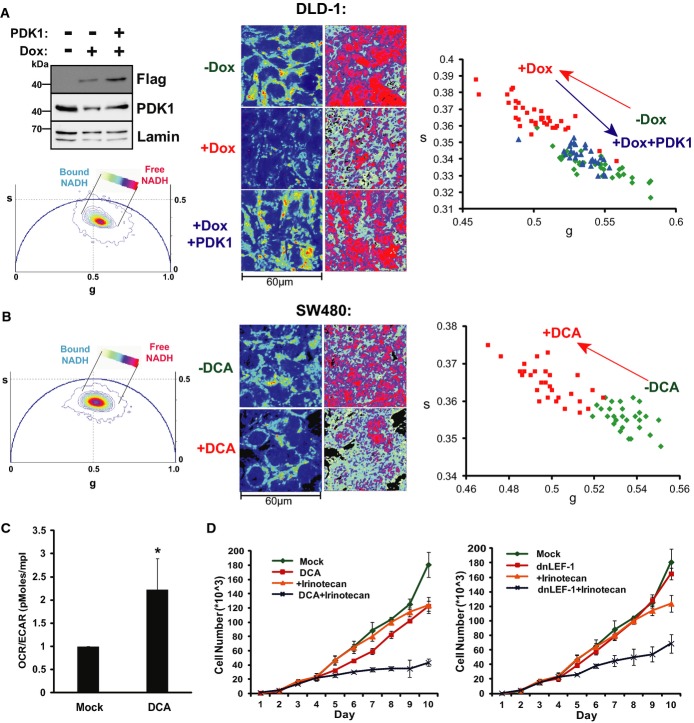
To determine how much of the Wnt-driven metabolic signature depends on PDK1, we asked whether restored expression could rescue the metabolic shift induced by dnLEF-1. We used lentiviral infection of DLD-1 cells to restore physiological levels of PDK1. As shown in Fig5A, dnLEF-1 expression alone led to a decrease in endogenous PDK1 levels and a decrease in glycolysis (a shift in the FLIM phasor position away from free NADH). In contrast, lentiviral restoration of normal PDK1 levels rescued the FLIM signature of glycolysis (a return of the phasor position back to that of cells not expressing dnLEF-1; Fig5A). This rescue is also evident in the free/bound NADH color mapping of the cells, showing similar levels of free/bound NADH in cells without doxycycline compared to cells with doxycycline and PDK1. When repeated in the second DLD-1 clonal line expressing dnLEF-1, a rescue of the FLIM signature was similar, indicating at least a partial rescue in the metabolic shift (Supplementary Fig S5). Taken together, these results suggest that reduction of PDK1 could be a major reason for the metabolic shift triggered by dnLEF/TCF interference with Wnt/β-catenin signaling.
Figure 5. PDK1 overexpression rescues the altered metabolic phenotype induced by blocking Wnt.
- Western blot analysis of lysates collected from DLD-1 dnLEF-1(2) cells treated with or without 0.01 μg/ml doxycycline for 24 h and with or without PDK1 lentivirus for 72 h. FLIM imaging was performed at confluency (96 h doxycycline and 7 days post-transduction). Intensity images are shown on the left. FLIM results, shown through the free/bound NADH color mapping (right), as well as the scatterplot, show that doxycycline induction of dnLEF-1 shifted the phasor toward bound NADH, while PDK1 rescue shifted it back to its original position (P < 0.0001 comparing −Dox to +Dox and comparing +Dox to +Dox+PDK1).
- FLIM analysis of SW480 cells treated with 50 mM DCA for 48 h shows a phasor shift toward bound NADH (P < 0.0001 comparing +DCA to mock).
- OCR/ECAR ratio of DCA (10 mM)-treated SW480 cells shows that blocking PDK1 activity doubles the oxygen consumption rate (mitochondrial activity) relative to the extracellular acidification rate (glycolysis-produced lactate). Data shown represent the average of three independent trials (± SD; *P-value < 0.05).
- Sulforhodamine B cell proliferation assay of SW480 cells treated with or without 2.5 μM irinotecan, 20 mM DCA, and dnLEF-1 lentivirus shows an increased sensitivity to irinotecan when treated with DCA or dnLEF-1. A representative graph of two trials is shown. Error bars represent the standard deviation between eight internal replicates.
Source data are available online for this figure.
Given that we propose PDK1 expression as a major Wnt target for glycolysis, we compared the effects of dnLEF/TCF to dichloroacetate (DCA), a well-known small molecule inhibitor of PDK1 (Whitehouse et al, 1974). A 48-h treatment of DCA led to the same characteristic shift in the phasor plot toward bound NADH that was seen with expression of dnLEF/TCF, again indicating a decrease in a glycolytic phenotype (Fig5B). The FLIM shift corresponded to an increase in the ratio of oxygen consumption (oxidative phosphorylation) relative to the extracellular acidification rate (OCR/ECAR, Fig5C). These changes in metabolic metrics match previous reports that DCA treatment decreases lactate excretion (Bonnet et al, 2007) and increases ATP levels (Sun et al, 2011) similar to our measurements with dnLEF/TCF expression. Another reported phenotype of DCA treatment in cancer cells is an increase in sensitivity to chemotherapy treatment (Cairns et al, 2007; Stockwin et al, 2010; Tong et al, 2011). Therefore, we examined whether dnLEF-1 forced similar increases in sensitivity to the chemotherapeutic agent irinotecan. We treated SW480 cells with DCA alone, dnLEF-1 alone, DCA with irinotecan, or dnLEF-1 with irinotecan. Neither 2.5 μM irinotecan nor dnLEF-1 alone had much effect on cell proliferation and 20 mM DCA had only a very slight impact. In contrast, both DCA and dnLEF-1 greatly enhanced the sensitivity of the cells to irinotecan, showing a synergistic decrease in the proliferation rate (Fig5D). These data suggest that dnLEF/TCF inhibition of PDK1 in many ways mimics the effects of DCA. However, while there was congruence between dnLEF/TCF and the DCA phenotypes, DCA targets all four members of the PDK family, not just PDK1. We therefore used lentiviral shRNA knockdown to test for the specific significance of PDK1 (Supplementary Fig S5B and C). Western blot analysis of multiple knockdown lines revealed reduced PDK1 protein levels and significant reduction of phosphorylation of its main substrate, mitochondrial pyruvate dehydrogenase (PDHpSer293, Supplementary Fig S5B). FLIM analysis of each knockdown cell line revealed significant shifts in the free/bound NADH signature that mimic the decrease in glycolysis observed with dnLEF1 or dnTCF1 expression or DCA treatment (Supplementary Fig S5C). These results indicate that PDK1 is an important Wnt target gene and that it contributes major activities to glycolysis. However, we also observed the knockdown cultures to revert to a glycolysis mode of metabolism several days later. Western blot analysis of the reverted cultures showed that while PDK1 protein levels remained low, phosphorylation of the target substrate pyruvate dehydrogenase had recovered, a sign of compensatory rescue via other PDK family members or other kinases (data not shown). Thus, the more stable shift in metabolism observed with DCA treatment or dnLEF1/dnTCF expression suggests that total cellular PDK activity is targeted by Wnt signaling beyond single, selective regulation of PDK1. We conclude that PDK1 is a major Wnt target gene, but that it is coordinately regulated within an entire gene program for glycolysis.
Blocking Wnt reduces in vivo tumor growth
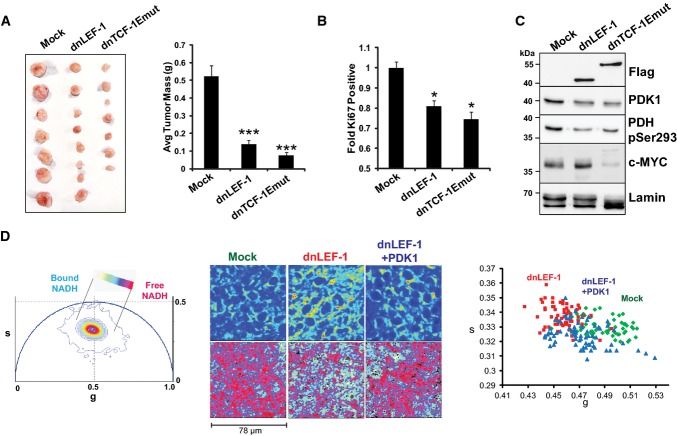
Since growing cells in vitro (on plastic) does not accurately mimic the metabolic demands of an in vivo tumor, we tested the impact of dnLEF/TCF expression on cancer cells in a xenograft tumor model. As shown in Fig6A, expression of dnLEF-1 or dnTCF-1Emut in SW480 cells injected into immune-deficient NOG mice drastically decreased tumor growth by up to 90%. The tumors created from cells expressing dnLEF/TCF had 20–30%fewer proliferating cells as measured by Ki67 staining (Fig6B). To compare the changes in protein expression in vivo with the previous results in vitro, we used Western blot analysis to examine levels of PDK1 and c-Myc. PDK1 protein was reduced in both dnLEF/TCF tumor types. Levels of c-Myc protein were variable but overall similar between the dnLEF-1-expressing tumors compared to mock, while c-Myc protein was significantly reduced in the dnTCF-1Emut tumors (Fig6C). These results are consistent with the previously described microarray which showed a decrease in c-MYC RNA levels with dnTCF-1Emut, but not dnLEF-1 (Hoverter et al, 2012). Western blot analysis also revealed stable reduction of pyruvate dehydrogenase phosphorylation in both dnLEF/TCF tumor types (PDHpSer293; Fig6C). Overall, the tumor analysis suggests that while tumors expressing dnLEF/TCF show similar reductions in PDK1 as cells in vitro, the expression of dnLEF/TCF has a much different effect on cell growth: no change in vitro but a strong, negative effect on growth in vivo.
Figure 6. Blocking Wnt reduces in vivo tumor growth.
- Expression of dnLEF-1 or dnTCF-1Emut in SW480 cells results in smaller xenograft tumors. Images of tumors are shown with quantification of the tumor mass and volume at the time of harvest. Data include measurements of eight tumors for each condition. Error bars represent the SEM among eight replicates (***P-value < 0.001).
- Ki67 staining of paraffin-embedded sections from xenograft tumors shows fewer Ki67-positive cells with dnLEF-1/dnTCF-Emut expression. Data shown represent the average of counts from at least eight fields. Error bars represent the SEM among at least eight replicates (*P-value < 0.05).
- Western blot was performed on protein lysates prepared from freshly extracted xenograft tumors. Western blot analysis of endogenous PDK1 and its target, pyruvate dehydrogenase (pSer293-PDH), shows decreases when dnLEF-1 or dnTCF-1Emut tumors are expressed. cMYC expression shows variable levels of expression.
- Phasor plot representation for the color mapping of in vivo tumor FLIM analysis, fluorescence intensity (top image panels), FLIM color mapping (bottom panels), and scatterplot analysis are as described in Fig5 except that these analyses were performed on living, surgically exposed, yet still actively perfused, xenograft tumors. Both the free/bound NADH color mapping (bottom row) and scatterplot (average phasor position of individual cells within each tumor) show a shift in the phasor position toward bound NADH with dnLEF-1 expression and a return to free NADH with PDK1 overexpression (P < 0.0001 comparing dnLEF-1 to mock and comparing dnLEF-1+PDK1 to dnLEF-1). A minimum of three fields of view per tumor were analyzed. Data shown are from one mouse representative of eight replicates (additional tumor analysis in Supplementary Fig S7).
Source data are available online for this figure.
To test whether the same change in metabolism that we detected in vitro by FLIM analysis is also detected in vivo, we adapted FLIM analysis for living, actively perfused xenograft tumors. For all tumors, tail vein injection of fluorescent dextran (FITC dextran) was used to continuously image live blood cell flow through the vasculature of the tumor. Mice were anesthetized and intact xenograft tumors were exposed (with feeder vessels preserved) followed by immediate stabilization of the mice over the microscope objective in a temperature-controlled environment. Non-invasive, confocal FLIM imaging was performed at depths between 50 and 200 μm around the subcapsular region of the tumors (see Supplementary Fig S6 for a schematic of in vivo FLIM). A similar metabolic response to Wnt signaling inhibition was observed with these analyses. That is, dnLEF/dnTCF1-expressing tumors were less glycolytic (phasor position shifted toward bound NADH; Fig6D). A notable difference compared to cells grown in monoculture in vitro was that xenograft tumors had a greater degree of variability in FLIM profile, reflecting the existence of a more complex tumor microenvironment (additional replicates shown in Supplementary Fig S7). To test whether PDK1 expression could rescue this effect, we imaged xenograft tumors of dnLEF-1 cells that had been lentivirally transduced with a PDK1 expression vector prior to injection into the mice. PDK1 expression rescued the metabolic signature, shifting the phasor plot back toward the free NADH position (Fig6D; Supplementary Fig S7).
Blocking Wnt reduces tumor vessel density, while reintroduction of PDK1 restores it
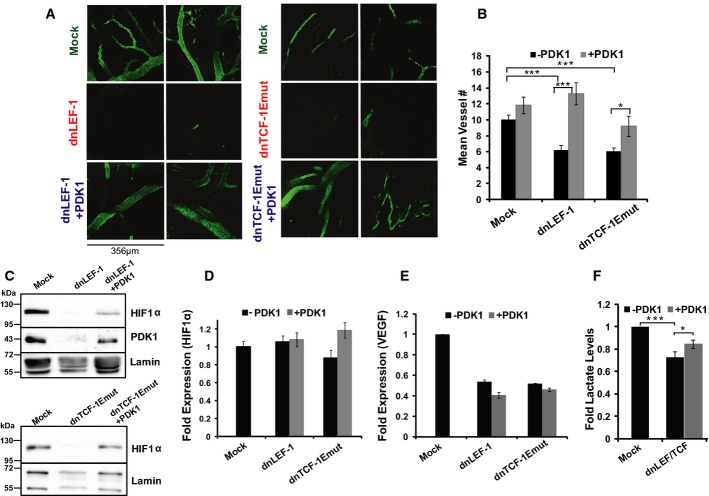
Much of the cost and benefit analysis of glycolytic metabolism has focused on intracellular changes in metabolites and biosynthetic intermediates. However, imaging live tumors highlighted a striking extrinsic benefit of glycolytic metabolism on the tumor microenvironment. FITC dextran labeling of the vasculature revealed that the density of blood vessels was greatly diminished in dnLEF/TCF-expressing tumors (Fig7A). Thus, one important reason these dnLEF/dnTCF-expressing tumors were reduced in size was because of poor nutrient delivery. Interestingly, re-expression of PDK1 strictly in the injected tumor cells appeared to restore vessel density to levels equivalent to that of the mock tumors (Fig7A). To quantitate these changes, we stained tumor sections for the endothelial marker CD31 and counted vessel density in the same subcapsular region where the FLIM analysis was performed. Vessel density was greatly diminished in the tumors expressing dnLEF-1 or dnTCF-1Emut, but rescue expression of PDK1 returned vessel density back to the original level observed in the mock tumors (Fig7B). A few studies have shown that enhanced glycolysis can promote angiogenesis through the stabilization of HIF1α (Hunt et al, 2007; Milovanova et al, 2008; Sutendra et al, 2012). We therefore evaluated HIF1α protein levels in the xenograft tumors. Interestingly, HIF1α protein levels were reduced upon dnLEF/TCF expression, and levels were at least partially restored with reintroduction of PDK1 (Fig7C). HIF1α mRNA levels were unaffected and stable in all conditions (Fig7D), suggesting that regulation occurs at a post-transcriptional step. We also note that while the well-known angiogenic factor VEGF was downregulated in the dnLEF/TCF-expressing tumors, addition of PDK1 did not restore these levels (Fig7E), suggesting that the PDK1 rescue of angiogenesis is VEGF independent. Overall, dnLEF/TCF expression in vivo reduced tumor growth, reduced PDK1 levels, reduced the glycolytic metabolic signature (including tumor lactate levels, Fig7F), and reduced the density of blood vessels feeding the tumor. Restoration of PDK1 expression rescued both the change in metabolism and the reduction in vessel density, a striking demonstration of cell-autonomous and non-autonomous effects of a metabolic enzyme.
Figure 7. Blocking Wnt reduces tumor vessel density, while reintroduction of PDK1 restores it.
A Tumor vessel density within the subcapsular region is greatly diminished with dnLEF/TCF, but is restored with PDK1 expression. Images show FITC dextran-labeled vasculature in the xenograft tumors (˜ 50–200 μm deep).
B Quantification of vessel density derived from the CD31 staining of paraffin-embedded sections of each tumor. For quantification, the number of vessels per field of view (40× objective) was averaged from every cross-section (at least 20 for each condition) within the subcapsular region for each tumor. Error bars represent the SEM among at least 20 replicate fields.
C Western blot analysis was performed on protein lysates prepared from freshly extracted xenograft tumors.
D, E RT-qPCR analysis of (D) HIF-1α and (E) VEGF expression in xenograft tumor RNA. HIF-1α mRNA levels do not change among the various tumors (normalized to GAPDH), but VEGF expression levels are downregulated in the dnLEF-1 and dnTCF-1Emut tumors. However, PDK1 rescue expression does not change the levels of either HIF-1α or VEGF mRNA. RT-qPCR data shown are representative of four replicate tumor sets. Error bars represent the standard error between three internal replicates.
F Lactate levels in dnLEF/TCF-expressing tumors are reduced compared to MOCK levels and are partially restored with PDK1 rescue expression. Data show the average fold lactate levels (compared to mock) in five tumors for each condition, with error bars representing the SEM (*P-value < 0.05; ***P-value < 0.001).
Source data are available online for this figure.
Discussion
Here, we report that Wnt/β-catenin signaling directs a metabolic program of glycolysis in colon cancer cells, a common cancer phenotype known as the Warburg effect. This metabolic change is accompanied by non-autonomous affects on the microenvironment in the form of increased vessel development. A direct Wnt target gene, PDK1, is identified to participate in both of these Wnt-driven, cancer-supporting phenotypes (model in Fig8A). We also describe for the first time the use of FLIM imaging to detect metabolic changes in living, actively perfused xenograft tumors. We suggest that effects on metabolism are a common, core function of all LEF/TCF family members and isoforms, even those isoforms previously understudied due to their lack of regulation of proliferation and the cell cycle.
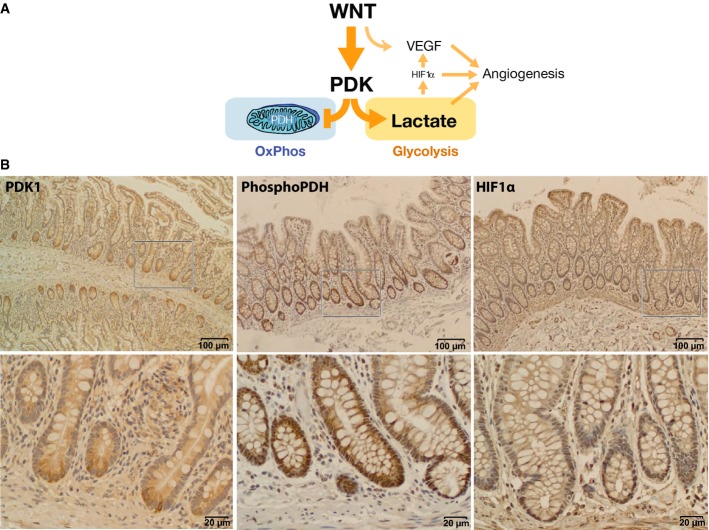
Figure 8. Wnt promotes aerobic glycolysis through PDK1.
- Model of Wnt regulation of cancer metabolism and angiogenesis through PDK1.
- Immunohistochemical staining of normal human small intestine (PDK1) and normal human colon (phospho-PDH and HIF1α) shows correlations between high levels of Wnt signaling, PDK1 activity, and HIF1α levels. Bottom row shows higher-power images of boxed portions in the top row.
A prevailing theme in the field of cancer metabolism is that the same oncogenic pathways that drive transformation must also favor metabolic pathways that support this process (Ward & Thompson, 2012). In fact, given that Wnt signaling plays a critical involvement in embryonic development, tissue remodeling, and cancer biology, and given that the success of these processes depends on tight coordination with the metabolic pathways that support them, it has been suggested that Wnt is a prime candidate for directing this coordination (Sethi & Vidal-Puig, 2010). Our study supports this paradigm as we demonstrate that blocking Wnt target gene activation alters cancer cell metabolism by reducing aerobic glycolysis. Two recent studies also support this conclusion. One study in breast cancer cells found that activation of Wnt signaling enhanced glycolysis through indirect actions of the transcriptional repressor Snail on cytochrome c oxidase, a component of the electron transport chain (Lee et al, 2012). A second, more recent study found that Wnt stimulates a fast-acting, short-term Wnt-Lrp5-Rac1-mTORC2-Akt signal to upregulate protein and activity levels of glycolytic enzymes, including PDK1 (Esen et al, 2013). Regulation occurred within 10–20 min of Wnt stimulation and was independent of β-catenin, LEF/TCFs, and transcription. Our findings contrast with this system because we find that β-catenin and LEF/TCF are required to directly target PDK1 transcription. Indeed, our test for a connection to mTORC2 was negative as inhibition of β-catenin with either dnLEF/TCF expression or XAV939 treatment for as long as 3 h had no effect on mTORC2 activity in DLD-1 and SW480 cells and inhibition of mTORC2 for 24 h with the specific mTOR inhibitor PP242 had no effect on PDK1 mRNA levels (Supplementary Fig S9).
We propose that Wnt signals have short-term and long-term modes of metabolic regulation. Short-term modes trigger an mTOR kinase cascade that affects protein stability of glycolytic enzymes. Long-term modes use β-catenin to change gene expression of at least some of these enzymes as well as other metabolically relevant components (Fig1C, Supplementary Materials and Methods). This latter mode is of primary importance in cancers that harbor aberrant, chronic, elevated levels of Wnt signaling with or without Wnt ligands. Chronic Wnt signaling could transform the metabolic potential of cancer cells by elevating the overall levels of key components for glycolysis and thereby set the stage for robust Warburg metabolism in the face of whatever environment the transformed cells encounter.
PDK1 is a well-known, key metabolic regulator of glycolysis. This kinase phosphorylates the pyruvate dehydrogenase (PDH) complex to inhibit the first step of converting pyruvate to acetyl-CoA (Roche et al, 2001), an important regulatory nexus between glycolysis and respiration. PDK1 phosphorylation of PDH suppresses entry of fuel into mitochondria and instead promotes glucose fermentation to lactate in the cytoplasm, an important activity for promoting the Warburg effect in cancer. Indeed, PDK1 is upregulated in numerous cancers, including colon cancer (Koukourakis et al, 2006; Wigfield et al, 2008; Baumunk et al, 2012). Here, we propose that cancer relevant levels of Wnt signaling use LEF/TCFs for direct regulation of PDK1. PDK1 mRNA was identified through microarray analysis as downregulated after blocking Wnt in colon cancer cells (Fig1). Our chromatin immunoprecipitation, 4-thiouridine labeling, and transient transfection analyses show that this downregulation is direct (Fig4D–F, Supplementary Fig S4). Genome-wide ChIP-seq studies by others (ENCODE; Kent et al, 2002) also show that the PDK1 gene locus is occupied by TCF-4 (TCF7L2) in multiple cancer cell lines (Supplementary Fig S8). Interestingly, peaks of occupancy are also present in the promoter regions of the other PDK family members, especially for PDK2 and PDK4. It is possible that multiple PDK isoforms have the capacity to respond to Wnt signaling, a possibility underscored by our observation of compensatory recovery of PDH phosphorylation when PDK1 was selectively targeted for knockdown by shRNAs.
Our experiments demonstrate that PDK1 is a pivotal mediator of Wnt effects on metabolism as rescue of its expression restored glycolysis and vessel density in tumors. However, we expect that PDK1 is only one target of an entire gene program driven by Wnt to promote cancer-supporting metabolism. For example, while PDK1 knockdown caused a significant shift of metabolism in cultures, the shift was not as stable as the dnLEF/TCF treatments. Also, PDK1 rescue expression did not entirely restore tumor mass. It is possible that knockdown of a single Wnt target gene out of the context of an entire gene program is not sufficient to shift metabolism and tumor proliferation phenotypes. Ontology analysis identified additional metabolism-linked genes that were downregulated by dnLEF/TCFs (Fig1 and Supplementary Table S1). This includes genes that encode nutrient and small molecule transporter genes that affect metabolic pathways. For example, the lactate transporter MCT-1 (monocarboxylate transporter-1 or SLC16A1) is a critical player in tumor metabolism and is upregulated in colon cancer (Pinheiro et al, 2008). MCT-1 was downregulated with dnLEF/TCF expression, both by microarray analysis and by RT-qPCR analysis of xenograft tumors (Supplementary Fig S8). The MCT-1 promoter is occupied by TCF-4, especially in HCT116 colon cancer cells (Supplementary Fig S8), implicating MCT-1 as a Wnt target gene. Previous studies have identified other metabolism-connected Wnt target genes. For example, the well-known Wnt target gene c-Myc plays pivotal roles in cancer metabolism, driving both aerobic glycolysis and glutaminolysis (Osthus et al, 2000; Wise et al, 2008; Dang, 2010). In fact, c-Myc has been shown to enhance HIF-1α-mediated regulation of PDK1 (Kim et al, 2007). We note here, however, that c-Myc is not required in our system to mediate changes in metabolism as its levels are not altered with the expression of dnLEF-1 (supported by our microarray, Western blot, and RT-qPCR analyses). However, as Wnt/β-catenin signaling is the main driver of colon cancer, we expect that the Wnt target c-Myc should cooperate with the genes identified in our study to collectively drive strong, glycolytic signatures of Warburg metabolism. Additional Wnt target genes linked to glycolysis have also been identified in the liver. For example, Chafey et al show that Wnt augments LDH (lactate dehydrogenase) activity and downregulates two mitochondrial ATPase subunits in hepatocytes, thus driving glycolysis at the expense of oxidative phosphorylation (Chafey et al, 2009). While it is not clear whether these effects are direct or indirect, these data along with the other evidence above suggest that Wnt signaling promotes a gene program that supports a cancer-promoting metabolic profile of enhanced aerobic glycolysis.
Given the strong evidence for direct regulation of PDK1 transcription, we asked whether Wnt and glycolysis gene programs were correlated in primary human tumors. We used publically available gene expression data from The Cancer Genome Atlas Data Portal to test for correlations between PDK1 expression and signatures of overactive Wnt signaling (TCGA, https://tcga-data.nci.nih.gov/tcga/). No correlation was identified, a finding consistent with recent studies of large tumor data sets (> 2,500) that found confounding levels of heterogeneity for gene signatures of metabolism, and consistent with our observation of compensatory PDK activities in PDK1 shRNA-knockdown lines (Hu et al, 2013). Nevertheless, a different method of analysis detected links between Wnt signaling and glycolysis in tumors (Supplementary Fig S10). In this analysis, a comparison of the expression level of Wnt and glycolysis genes (defined by Kyoto Encyclopedia of Genes and Genomes (KEGG)) was used for unsupervised clustering of 238 human tumors into nine tumor groups with one group of 21 tumors strongly clustered (group 4, Supplementary Fig S10A). This same cluster was detected when gene signatures were compared via distance measures within the cluster versus the other eight tumor groups (Supplementary Fig S10B). Strong similarities (correlations and anti-correlations) between Wnt signaling components and glycolysis genes were detected within this group, relationships that distinguished the cluster from the remaining clusters. Importantly, PDK1 ranked as a highly correlated gene in the 21 tumors, its expression most often having negative, anti-correlations with Wnt components (see Supplementary Materials and Methods). The majority of anti-correlating genes encode antagonists of Wnt signaling or components of non-canonical Wnt signaling, and interestingly, the STRING algorithm for protein interaction networks revealed that most of these anti-correlating components engage in direct or indirect interactions with the Wnt component Dishevelled (Supplementary Fig S10C; Szklarczyk et al, 2011). These data are compelling because any kind of clustering of colon tumors on the basis of gene expression suggests a promising approach to probe differences between tumors for diagnosis and/or prognosis and they most certainly warrant further investigation and validation.
Most tumorigenic actions of signaling pathways like Wnt represent distorted use of normal functions crucial to cellular homeostasis. How might Wnt promotion of aerobic glycolysis be beneficial to normal tissues? One organ that illustrates this benefit is the liver where Wnt signaling regulates liver function, including functional and anatomical liver zonation, hepatocyte maturation, and glycolytic metabolism (Tan et al, 2006; Thompson & Monga, 2007; Colletti et al, 2009; Gebhardt & Hovhannisyan, 2010). The pancreas is also central to whole-body glucose homeostasis, a tissue that exhibits important links between Wnt signaling and disease. In fact, polymorphisms in the TCF7L2 (TCF-4) locus encode the strongest links to risk of diabetes of any SNP (Grant et al, 2006; Lyssenko et al, 2007; Cadigan & Waterman, 2012). Most relevant to our study is that Wnt regulation of glycolysis is important to normal intestinal tissue. We recently used FLIM analysis to identify a metabolic gradient along the crypt-villus axis of the mouse small intestine (Stringari et al, 2012). Consistent with our finding that Wnt signals drive a glycolytic phenotype, we observed the strongest glycolytic signatures at the base of the crypt and lowest at the top of the villi, correlating with high and low Wnt activity, respectively. According to our model (Fig8A), Wnt promotes a glycolytic signature through the regulation of PDK1, which then promotes HIF1α stabilization. Consistent with this model, we observe high levels of PDK1 protein and its target, phospho-PDH, at the bottom of human intestinal crypts (Fig8B). Stabilized HIF1α is also detected in crypt bases (in addition to the hypoxic region at the tops of crypts as has been reported previously) (Giles et al, 2006; Fig8B). Wnt-stimulated HIF1α stabilization could contribute to a positive feedback loop to support glycolysis in the intestinal stem cell niche as HIF1α targets several components of glycolysis. Wnt also plays an important direct role in the maintenance of stem cells (Korinek et al, 1998; Nusse, 2008), which utilize aerobic glycolysis as a metabolic resource (Suda et al, 2011; Zhou et al, 2012). The benefits of this type of metabolism in stem cells include support for proliferation as well as reduction in ROS (reactive oxygen species) production to protect the integrity of the genome (Kondoh et al, 2007; Prigione et al, 2010).
In the present study, we identify a new link between Wnt signaling and tumor angiogenesis. Although Wnt has been connected to angiogenesis through direct regulation of VEGF expression (Zhang et al, 2001), we report for the first time the role of Wnt-directed PDK1 expression in this process. Indeed, PDK1 and PDK2 have recently been shown to enhance angiogenesis (McFate et al, 2008; Sutendra et al, 2012). We suggest that one mechanism for this phenomenon is increased lactate production because levels of this metabolite were increased in PDK1 rescue tumors (Fig7F). Lactate has been associated with increased angiogenesis via reduced ADP ribosylation and stabilization of HIF-1α protein (Lu et al, 2002; Hunt et al, 2007; Milovanova et al, 2008). Stabilized HIF-1α could enhance angiogenesis at least partly through upregulation of VEGF (Forsythe et al, 1996; Pugh & Ratcliffe, 2003) and SDF-1 (stromal cell-derived factor-1; Karshovska et al, 2007). While we observe changes in HIF1α levels that correlate with tumor vessel density, we did not observe concomitant changes in VEGF levels, suggesting that other angiogenic factors such as SDF-1 might be relevant (Fig7C–E). Even though the exact mechanism linking Wnt and PDK1 to angiogenesis is not known, our discovery that PDK1 rescues vascularization highlights how the metabolic status of cancer cells communicates to the tumor microenvironment.
Consistent with our finding that oncogenic Wnt regulates PDK1, blocking Wnt mimics the effects of small molecule inhibition of the PDK family of kinases through DCA. DCA treatment reduces glycolysis, increases oxidative phosphorylation, decreases lactate production, increases cancer cell sensitivity to chemotherapy drugs, and reduces xenograft tumor mass and vascularity (Bonnet et al, 2007; Papandreou et al, 2011). A DCA trial in human glioblastoma patients resulted in promising tumor regression, as well as enhanced tumor apoptosis, and to note: decreased tumor vascularity (Michelakis et al, 2010). Collectively, results from multiple studies suggest that the effects of DCA are much more robust in vivo than in vitro (Papandreou et al, 2011), mirroring our lack of growth phenotype when blocking Wnt in culture, as opposed to the drastic reduction in xenograft tumor size in vivo. This could be attributed to the excess nutrients available in in vitro culture which can mask dependencies on specific metabolic pathways. More intriguing is the possibility that DCA and dnLEF-1 expression are more effective in vivo because blocking aerobic glycolysis has profound effects on the tumor microenvironment, such as reduced vascularity. Our study therefore has important implications for the development of cancer therapies targeting the Wnt pathway and for testing Wnt inhibitors in vivo, as inhibitors that do not block proliferation or cell cycle progression in artificial culture conditions in vitro may still be effective at limiting tumor growth in vivo via their effects on the more sensitive Wnt metabolism program. In fact, partial interference with oncogenic Wnt signaling may be an effective treatment strategy as it might normalize metabolic activity in tumor cells while preserving essential Wnt signaling functions in normal cells such as cell cycle and cell differentiation programs.
Materials and Methods
Cell lines/constructs
Colon cancer cell lines, SW480 and DLD-1, were cultured in either Dulbecco’s modified Eagle’s medium (DMEM; Gibco 11960) or RPMI-1640 medium (Cellgro 15-040), supplemented with 10%fetal bovine serum (FBS) and 2 mM glutamine. Unless otherwise stated, experiments were performed in the same media. DLD-1 dnLEF-1 and dnTCF-1Emut stable cells were created by transfecting Tet-inducible dnLEF-1N or dnTCF-1Emut into DLD-1 TR7 cells (a generous gift from M. van de Wetering and H. Clevers) as previously described (Yokoyama et al, 2010; Hoverter et al, 2012). Cells were maintained in 500 μg/ml zeocin and 10 μg/ml blasticidin. However, it was found that dnLEF/TCF expression in the presence of zeocin reduces cell proliferation (Yokoyama et al, 2010, and data not shown). Therefore, in order to avoid any contribution from zeocin, all experiments were performed in the absence of antibiotics. Induction of dnLEF-1 or dnTCF1-Emut was achieved through the addition of 0.01 or 1.0 μg/ml doxycycline to the media, respectively. Lentiviral constructs were cloned via Cold Fusion (System Biosciences) by inserting coding sequences for Flag-tagged dnLEF-1N, Flag-tagged dnTCF-1Emut (Atcha et al, 2007), or Flag-tagged PDK1 [a gift of Jean Zhao (Boehm et al, 2007); Addgene plasmid #20564] into pCDH lentivector (System Biosciences; SBI #CD533A-2). See Supplementary Materials and Methods for cloning information regarding TK and PDK1 reporters.
Lentiviral preparation and infection
Lentivirus was prepared and target cells were transduced using System Biosciences lentivirus technology according to their specifications (see Supplementary Materials and Methods for details). Cells were harvested for subsequent assays 72 h post-transduction unless otherwise stated.
Lactate assay
Lactate measurements were performed on media collected from both cells grown in suspension (soft agar) and cells grown on solid support (plastic). SW480 cells were embedded in soft agar 72 h post-transduction at a concentration of 1,000 cells in 2.5 ml of 0.3%agar (in DMEM with 20%FBS). Fresh media were added once a week. After 22 days of growth (5 days since last media change), images of wells were taken and media were collected. For cells grown on plastic, SW480 cells (72 h post-transduction) were seeded in a 96-well plate at 5,000 cells per well. Media were changed after 72 h and then left unchanged until media were collected after 7 days. Media were collected from three triplicate wells, while the number of cells in three wells was determined using the SRB assay (described below). SW480 cells treated with XAV939 (Sigma X3004) were treated with either 10 μm XAV939 or DMSO for at least 4 days prior to media collection. For lactate measurements of xenograft tumors, pre-weighed flash-frozen xenograft tumors were homogenized in a Precellys 24 homogenizer in the presence of 10 μl HBSS (Hank’s balanced salt solution) per mg tissue at 6,000 rpm, twice, for 15 s each. Tissue extraction and lactate assay were performed according to L-Lactate Assay kit (Eton Bioscience #1200011002). All measurements were performed in triplicate.
Sulforhodamine B cell growth (SRB) assay
Cells were seeded in 96-well plates at 5,000 cells per well with eight replicates for each condition and time point. Media were refreshed every day (with or without 0.01 μg/ml doxycycline, 2.5 μM irinotecan, or 20 mM DCA as needed). Cells were fixed and stained according to published protocols (Skehan et al, 1990) for a 10-day period. Optical density readings were performed at 450 nm.
ATP assay
SW480 cells were seeded at 125,000 cells per well (12-well plate) 72 h post-transduction and harvested 96 h after seeding. ATP measurements were performed according to the ENLITEN ATP Assay System (Promega #FF2000). All measurements were performed in triplicate.
Glucose consumption assay
Cells were seeded at 5,000 cells per well in a 96-well plate and media were harvested 6 d later. Low glucose media (DMEM or RPMI with 5.5 mM glucose) were added 24 h (SW480) or 12 h (DLD-1) prior to harvest. XAV939 (10 μM) treatment was performed for 4 days, while doxycycline was added throughout the experiment to induce dnLEF-1 in DLD-1 cells. Glucose concentration in the media was determined according to Glucose (HK) kit (Sigma GAHK20).
XF24 Extracellular Flux Analyser
Metabolic rates of oxygen consumption (OCR) and extracellular acidification (ECAR) were measured using an XF24 Extracellular Flux Analyser (Seahorse Bioscience) as described previously (Zhang et al, 2012). Cells were plated at a density of 100,000 cells per well in a XF24 Cell Culture Microplate (Seahorse #100777-004). One hour before the assay, growth media were replaced by XF Assay Medium (Seahorse #102365-100) supplemented with 5.5 mM glucose and adjusted to pH 7.4. Oligomycin, rotenone, and antimycin A were prepared for final concentrations of 1 μM, and FCCP was prepared for a final concentration of 200 nM. Inhibitors were injected during the measurements: 2 min of mixing, 2 min of incubation, and 4 min of measurement. After the assay, protein was collected and measured with BCA Protein Assay Reagent (Thermo # 23225) for data normalization. XAV939 and DCA were added 16 h before assay at concentrations of 10 μM and 10 mM, respectively, and were maintained in the XF Assay Medium during the measurements. To exclude non-mitochondrial oxygen consumption, OCR values were calculated by taking the difference between the OCR before the addition of inhibitors and the OCR after the addition of rotenone and antimycin A.
Fluorescence lifetime imaging microscopy (FLIM) of cells in vitro
Cells were seeded at 150,000 cells per plate (35-mm glass bottom dishes). Infection with lentivirus was performed 72 h prior to seeding. Treatment with 0.01 μg/ml doxycycline was performed starting at the time of seeding. FLIM imaging was performed 5 days after seeding (under confluent conditions). Both XAV939 (10 μM) and DCA (50 mM) were added 48 h prior to imaging. For KCN treatment, cells were treated with 4 mM KCN 24 h after seeding. Cells were imaged by FLIM before the addition of KCN, as well as 1 min after treatment. Fluorescence lifetime images were acquired with a two-photon microscope coupled with a Becker and Hickl 830 card (Becker and Hickl, Berlin). See Supplementary Materials and Methods for additional description of FLIM hardware, software, and analysis.
Western blot analysis
Thirty micrograms of lysate was analyzed by Western blot using the following antibodies: Flag (1:1,000; Cell Signaling #2368), lamin (1:1,000 Cell Signaling #2032), PDHK1 (1:1,000 Cell Signaling #3820), c-Myc (1:1,000 Cell Signaling #5605), PDHpSer293 (1:1,000 Calbiochem #AP1062), HIF1α (1:1,000 Genetex #GTX127309), AKT (1:1,000 Cell Signaling #9272), phospho-Akt (Ser473; 1:1,000 Cell Signaling #9271), and secondary antibody (1:15,000 anti-rabbit IgG-horseradish peroxidase; Amersham). Blots were imaged on the Fujifilm LAS4000 Imaging System. For the preparation of xenograft lysates, tumors were homogenized in a Precellys 24 homogenizer in the presence of lysis buffer at 6,000 rpm, twice, for 15 s each.
Real-time PCR
Total RNA was isolated with Trizol from DLD-1 cells after treatment with doxycycline for either 24 h or 120 h. Xenograft tumors were homogenized in a Precellys 24 homogenizer in the presence of Trizol at 6,000 rpm, twice, for 15 s each. A total of 2 μg of RNA was reverse-transcribed using random primers according to the High Capacity cDNA Reverse Transcription kit (Invitrogen #4374966). Real-time quantitative PCR (RT-qPCR) was performed with Maxima SYBR Green/ROX qPCR Master Mix (Fermentas #K0222). Relative change in gene expression was calculated using the ΔΔCt method using GAPDH expression for normalization. See Supplementary Materials and Methods for a list of primer sequences.
4′ Thiouridine labeling and isolation of nascent RNA
4′ Thiouridine labeling was performed in duplicate following 2 h of mock (no doxycycline) or 1.0 μg/ml doxycycline treatment of DLD-1 colon cancer cells. 500 μM 4′ thiouridine (Sigma) was added to cells for 30 min at 37°C. Collected cells were resuspended in 4 ml Trizol reagent and total RNA purified. Labeled RNA was chemically biotinylated and purified using streptavidin-coated magnetic beads as described (Dölken et al, 2008). Briefly, 4′ thiouridine-labeled RNA was biotinylated with 2 μl biotin-HPDP [Pierce: 1 mg/ml dissolved in dimethylformamide]/1 μg RNA added to 1 μl 10× biotinylation buffer (100 mM Tris pH 7.4, 10 mM EDTA) and 7 μl water] for 1.5 h at room temperature with rotation followed by RNA precipitation. Biotinylated RNA was separated from bulk, unlabeled RNA using streptavidin-coated magnetic beads (μMacs Streptavidin kit).
Chromatin Immunoprecipitation (ChIP)
DLD-1 cells with or without 2 h doxycycline treatment (to induce expression of FLAG-dnTCF-1Emut) were cross-linked with 1%formaldehyde in 1× PBS for 12 min at room temperature. Cross-linking was quenched with 125 mM glycine for 5 min, and recovered cells were washed with 1× PBS. Cellular lysates were balanced for immunoprecipitation by Bradford assay (500 arbitrary units (AU) per sample) and pre-cleared with 30 μl His-magnetic beads (Invitrogen 10103D) for 30 min prior to the addition of 50 μl of FLAG antibody-conjugated magnetic beads (Sigma A2220). FLAG antibody beads were pre-blocked by 3 washes with 1 ml 1× PBS/BSA solution (5 mg/ml BSA fraction V in 1× PBS). Immunoprecipitations were carried out overnight at 4°C followed by magnetic separation and two 5-min washes with 1 ml LiCL buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1%NP-40, 1%sodium deoxycholate). Beads were washed twice with cold 1× TBS before suspension in proteinase K buffer [30 mM Tris–HCl pH 8.0; 5 μl of proteinase K (20 mg/ml)] and 300 mM NaCl for an overnight incubation at 65°C. ChIP DNA was recovered with the Fermentas GeneJET PCR Purification Kit in preparation for PCR. Real-time PCR (RT-PCR) was used to quantify enrichment of PDK1 gene locus regions. Primers specific to the genomic regions are provided in Supplementary Materials and Methods.
Luciferase assay
Cells were seeded at 2.5 × 105 per 6-well 24 h prior to transfection. Each well was transfected with 0.1 μg Super8xTopflash (kind gift from Dr RT Moon) or TK or PDK1 luciferase reporters and 0.1 μg thymidine kinase β-galactosidase plasmid using BioT transfection reagent (Bioland Scientific). Cells were treated with 0.01 μg/ml doxycycline or 10 μM XAV939 at the time of transfection where indicated. Cells were harvested 24 h post-transfection and assayed for luciferase activity and β-galactosidase activity (for normalization).
Xenograft tumors
SW480 stable transductants for xenograft injection were prepared through lentiviral infection with pCDH vector alone, dnLEF-1 or dnTCF-1Emut, with or without PDK1, followed by selection with 500 μg/ml G418. Cells (2.5 × 106) were injected subcutaneously into immune-deficient NOG mice. Tumors were removed and measured 4 weeks after injection. Xenograft tumors for FLIM analysis were injected at 5 × 106 cells per tumor and allowed to grow for 3 weeks.
Fluorescence lifetime imaging microscopy (FLIM) of in vivo xenograft tumors
All animal procedures were approved by the UC Irvine IACUC. Nine-week-old NOG mice (Jackson Labs) with 3-week-old xenograft tumors were anesthetized with 100 mg/kg ketamine/10 mg/kg xylazine. FITC or TRITC dextran (155 kDa; 12.5 mg/ml) was injected into the tail vein to label the tumor vasculature. Xenografts were exposed via a skin flap cut so as to avoid severing the xenografts’ feeder vessels. The mice were positioned on the stage in an environment chamber maintained at a constant 27°C. Images were acquired at an average depth of 50–200 μm, using FITC/TRITC labeling of vessels to distinguish tumor cells from metabolically distinct endothelial and blood cells. At least three 78-μm fields of view were imaged for each tumor. Tumors were examined from eight mice. Fluorescence lifetime images were acquired with a Zeiss 710 microscope (see Supplementary Materials and Methods for additional description of FLIM hardware, software, and analysis).
Immunohistochemistry
For Ki67 staining, deparaffinized 3.5-μm sections of formalin-fixed paraffin-embedded (FFPE) tumor xenografts were stained for Ki67 (Dako #M7240, 1:400 dilution) using a Ventana Benchmark Ultra autostainer with peroxidase-based detection. For each section, the number of positive nuclei per 8–10 fields (at a 20× magnification) was manually enumerated and the mean ± SEM was calculated. For CD31 staining, sections of FFPE tumor xenografts were blocked in avidin-biotin and a Mouse-On-Mouse kit (Vector Labs, Burlingame, CA) and subjected to antigen retrieval (citrate buffer pH 6.0 with steaming) and then incubated in a 1:50 dilution of anti-mouse CD31 (Dianova rat anti-mouse CD-31, #DIA-310), followed by peroxidase development. Enumeration of vessel density was done by counting vessels in every cross-section within the subcapsular region of each tumor sample and calculating mean ± SEM. For PDK staining in small intestine and phospho-PDH and HIF-1α staining in colon, following pressure cooker antigen retrieval in citrate buffer, sections were blocked in 3%H2O2, goat serum, and avidin-biotin-blocking reagent (Vector Labs). Sections were incubated in primary antibodies such as anti-PDK (Santa Cruz sc-28783, 1:1,000), anti-PDHpSer293 (Calbiochem #AP1062, 1:200), and anti-HIF-1α (Thermo PA1-16601, 1:500), followed by biotinylated secondary antibodies and visualization using a peroxidase-conjugated avidin-based Vectastain protocol. Slides were then counterstained with hematoxylin and mounted.
Bioinformatic analysis of human adenocarcinoma gene expression
Analysis of Wnt signaling and glycolysis pathway gene expression in human colon cancer used the publicly available colon adenocarcinoma (COAD) mRNA expression data from The Cancer Genome Atlas (TCGA) Data Portal (https://tcga-data.nci.nih.gov/tcga/). Gene memberships of the Wnt signaling and glycolysis pathways were used as defined by Kyoto Encyclopedia of Genes and Genomes (KEGG). Hierarchical clustering of log2-transformed normalized expression of 238 tumors was used to identify groups of tumor samples showing similar expression profiles for the 185 analyzed genes as well as direct tumor-to-tumor correlations for the expression of pathway components. STRING analysis (http://string-db.org; Szklarczyk et al, 2011) was used to display interactions among the PDK1 correlating genes. For additional details, see Supplementary Materials and Methods.
Statistical analysis
Statistical evaluation was performed by Student’s unpaired t-test. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Josh Teer and Kevin Huang for initial work on PDK1 reporter plasmids. We thank Jean Zhao for providing the PDK1 expression vector and Drs. Eric Stanbridge, Peter Donovan, David Fruman, and Aimee Edinger for their advice, critique, and reagents, and Dr. Klemens Hertel and Angela Garibaldi for assistance with the 4-thiouridine labeling experiment. The work of K.T.P and M.L.W. was supported by NIH Grants CA096878, CA108697, and a P30CA062203 to the Chao Family Comprehensive Cancer Center. K.W. and R.A.E. were supported by P30CA062203, and C.S., M.A.D., and E.G. were supported in part by NIH Grants P41-RRO3155, P41 GM103540, and P50-GM076516. T.T. was supported by NIH NRSA T32GM007185, and M.T. was supported by NIH Grants R01CA90571, R01CA156674, R01CA185189, R01GM073981, and P01GM081621. C.G. was supported by NIH Grant U01 AA021838 and the Chao Family Comprehensive Cancer Center Support Grant, CA-62203.
Author contributions
KTP and MLW conceived of the project. MLW, KTP, CS, EG, TT, MT, and RAE designed experiments, with CS and EG providing direction on FLIM and TT and MT providing Seahorse expertise. CG performed the bioinformatics analysis. KTP carried out experiments on all aspects of the project with assistance from SS-T, NH, and MM. CS performed FLIM experiments and analysis. KW and RAE performed mouse husbandry and mouse manipulation for FLIM. TT carried out the Seahorse experiments. KTP and MLW wrote the manuscript. KTP, CS, RAE, EG, TT, SS-T, MM, NH, MT, and MLW provided critical editing of manuscript. EG, RAE, MT, and MLW supervised research.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information for this article is available online: http://emboj.embopress.org
References
- Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting J-HT, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumunk D, Reichelt U, Hildebrandt J, Krause H, Ebbing J, Cash H, Miller K, Schostak M, Weikert S. Expression parameters of the metabolic pathway genes pyruvate dehydrogenase kinase-1 (PDK-1) and DJ-1/PARK7 in renal cell carcinoma (RCC) World J Urol. 2012;31:1191–1196. doi: 10.1007/s00345-012-0874-5. [DOI] [PubMed] [Google Scholar]
- Becker W, Bergmann A, Hink MA, König K, Benndorf K, Biskup C. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc Res Tech. 2004;63:58–66. doi: 10.1002/jemt.10421. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling review. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, White JG, Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, Harry G, Hashimoto K, Porter CJ, Andrade MA, Thebaud B, Michelakis ED. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Hlubek F, Spaderna S, Schmalhofer O, Hiendlmeyer E, Jung A, Kirchner T. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 2005;179:56–65. doi: 10.1159/000084509. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Symp Quant Biol. 2012;4:1–22. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Papandreou I, Sutphin PD, Denko NC. Metabolic targeting of hypoxia and HIF1 in solid tumors can enhance cytotoxic chemotherapy. Proc Natl Acad Sci USA. 2007;104:9445–9450. doi: 10.1073/pnas.0611662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafey P, Finzi L, Boisgard R, Caüzac M, Clary G, Broussard C, Pégorier J-P, Guillonneau F, Mayeux P, Camoin L, Tavitian B, Colnot S, Perret C. Proteomic analysis of beta-catenin activation in mouse liver by DIGE analysis identifies glucose metabolism as a new target of the Wnt pathway. Proteomics. 2009;9:3889–3900. doi: 10.1002/pmic.200800609. [DOI] [PubMed] [Google Scholar]
- Clevers H. Review Wnt/β-Catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E, Tripodi M, Amicone L. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–672. doi: 10.1053/j.gastro.2009.05.038. [DOI] [PubMed] [Google Scholar]
- Colyer RA, Lee C, Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc Res Tech. 2008;71:201–213. doi: 10.1002/jemt.20540. [DOI] [PubMed] [Google Scholar]
- Croce AC, Ferrigno A, Vairetti M, Bertone R, Freitas I, Bottiroli G. Autofluorescence properties of isolated rat hepatocytes under different metabolic conditions. Photochem Photobiol Sci. 2004;3:920–926. doi: 10.1039/b407358d. [DOI] [PubMed] [Google Scholar]
- Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Digman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013;17:745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: the role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- Giles RH, Lolkema MP, Snijckers CM, Belderbos M, van der Groep P, Mans DA, van Beest M, van Noort M, Goldschmeding R, van Diest PJ, Clevers H, Voest EE. Interplay between VHL/HIF1alpha and Wnt/beta-catenin pathways during colorectal tumorigenesis. Oncogene. 2006;25:3065–3070. doi: 10.1038/sj.onc.1209330. [DOI] [PubMed] [Google Scholar]
- Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Hoverter NP, Ting J-H, Sundaresh S, Baldi P, Waterman ML. WNT/p21 circuit directed by the C-clamp, a sequence-specific DNA binding domain in TCFs. Mol Cell Biol. 2012;32:3648–3662. doi: 10.1128/MCB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Hunt TK, Aslam RS, Beckert S, Wagner S, Ghani QP, Hussain MZ, Roy S, Sen CK. Aerobically derived lactate stimulates revascularization and tissue repair via redox mechanisms. Antioxid Redox Signal. 2007;9:1115–1124. doi: 10.1089/ars.2007.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karshovska E, Zernecke A, Sevilmis G, Millet A, Hristov M, Cohen CD, Schmid H, Krotz F, Sohn H-Y, Klauss V, Weber C, Schober A. Expression of HIF-1alpha in injured arteries controls SDF-1alpha mediated neointima formation in apolipoprotein E deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:2540–2547. doi: 10.1161/ATVBAHA.107.151050. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gao P, Liu Y-C, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006;66:632–637. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci USA. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Jeon HM, Ju MK, Kim CH, Yoon G, Han SI, Park HG, Kang HS. Wnt/Snail signaling regulates cytochrome c oxidase and glucose metabolism. Cancer Res. 2012;72:3607–3617. doi: 10.1158/0008-5472.CAN-12-0006. [DOI] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Wu JJ, Rovira II, Liu J, Gavrilova O, Lu T, Bao J, Han D, Sack MN, Finkel T. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4:ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem. 2002;277:23111–23115. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjögren M, Ling C, Eriksson K-F, Lethagen A-L, Mancarella R, Berglund G, Tuomi T, Nilsson P, Del Prato S, Groop L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, Wu H, Schell MJ, Tsang TM, Teahan O, Zhou S, Califano JA, Jeoung NH, Harris RA, Verma A. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer T-L, Mackey JR, Fulton D, Abdulkarim B, McMurtry MS, Petruk KC. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2:31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- Milovanova TN, Bhopale VM, Sorokina EM, Moore JS, Hunt TK, Hauer-Jensen M, Velazquez OC, Thom SR. Lactate stimulates vasculogenic stem cells via the thioredoxin system and engages an autocrine activation loop involving hypoxia-inducible factor 1. Mol Cell Biol. 2008;28:6248–6261. doi: 10.1128/MCB.00795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monici M, Agati G, Fusi F, Pratesi R, Paglierani M, Santini V, Bernabei PA. Dependence of leukemic cell autofluorescence patterns on the degree of differentiation. Photochem Photobiol Sci. 2003;2:981. doi: 10.1039/b306276g. [DOI] [PubMed] [Google Scholar]
- Naishiro Y, Yamada T, Takaoka AS, Hayashi R, Hasegawa F, Imai K, Hirohashi S. Restoration of epithelial cell polarity in a colorectal cancer cell line by suppression of beta-catenin/T-cell factor 4-mediated gene transactivation. Cancer Res. 2001;61:2751–2758. [PubMed] [Google Scholar]
- Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Goliasova T, Denko NC. Anticancer drugs that target metabolism: is dichloroacetate the new paradigm? Int J Cancer. 2011;128:1001–1008. doi: 10.1002/ijc.25728. [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Longatto-Filho A, Scapulatempo C, Ferreira L, Martins S, Pellerin L, Rodrigues M, Alves VAF, Schmitt F, Baltazar F. Increased expression of monocarboxylate transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch. 2008;452:139–146. doi: 10.1007/s00428-007-0558-5. [DOI] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- Roche TE, Baker JC, Yan YH, Hiromasa Y, Gong XM, Peng T, Dong JC, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi JK, Vidal-Puig A. Wnt signalling and the control of cellular metabolism. Biochem J. 2010;427:1–17. doi: 10.1042/BJ20091866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci USA. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Stockwin LH, Yu SX, Borgel S, Hancock C, Wolfe TL, Phillips LR, Hollingshead MG, Newton DL. Sodium dichloroacetate selectively targets cells with defects in the mitochondrial ETC. Int J Cancer. 2010;127:2510–2519. doi: 10.1002/ijc.25499. [DOI] [PubMed] [Google Scholar]
- Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci Rep. 2012;2:568. doi: 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Sun RC, Board PG, Blackburn AC. Targeting metabolism with arsenic trioxide and dichloroacetate in breast cancer cells. Mol Cancer. 2011;10:142. doi: 10.1186/1476-4598-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Dromparis P, Kinnaird A, Stenson TH, Haromy A, Parker JMR, McMurtry MS, Michelakis ED. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene. 2012;32:1638–1650. doi: 10.1038/onc.2012.198. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SPS. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- Thomas PD. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MD, Monga SPS. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- Tong J, Xie G, He J, Li J, Pan F, Liang H. Synergistic antitumor effect of dichloroacetate in combination with 5-fluorouracil in colorectal cancer. J Biomed Biotechnol. 2011;2011:740564. doi: 10.1155/2011/740564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB, Heiden MGV. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de LauW, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Whitehouse S, Cooper RH, Randle PJ. Mechanism of activation of pyruvate dehydrogenase by dichloroacetate and other halogenated carboxylic acids. Biochem J. 1974;141:761–774. doi: 10.1042/bj1410761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML, Harris AL. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br J Cancer. 2008;98:1975–1984. doi: 10.1038/sj.bjc.6604356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang X-Y, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama NN, Pate KT, Sprowl S, Waterman ML. A role for YY1 in repression of dominant negative LEF-1 expression in colon cancer. Nucleic Acids Res. 2010;38:6375–6388. doi: 10.1093/nar/gkq492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Heikal AA. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B. 2009;95:46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Li Y, Cameron BD, Relue P. Fluorescence anisotropy of cellular NADH as a tool to study different metabolic properties of human melanocytes and melanoma cells. IEEE J Sel Top Quantum Electron. 2007;13:1671–1679. [Google Scholar]
- Zhang X, Gaspard JP, Chung DC. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001;61:6050–6054. [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Wisidagama DRR, Setoguchi K, Hong JS, Van Horn CM, Imam SS, Vergnes L, Malone CS, Koehler CM, Teitell MA. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc. 2012;7:1068–1085. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, Ruohola-Baker H. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.