Abstract
Pink1 and Parkin, identified through studies of hereditary early onset Parkinson's disease, are involved in mitochondria quality control. Parkin E3 ubiquitin ligase activity is activated by Pink1 kinase activity, although the mechanism is still elusive. Three recent reports uncover a surprising mechanism in which Pink1 directly phosphorylates ubiquitin to boost Parkin activity.
See also: LA Kane et al (April 2014), A Kazlauskaite et al (May 2014) and F Koyano et al (June 2014
Pink1, a mitochondrial kinase, and Parkin, a cytosolic E3 ubiquitin ligase, function in mitochondrial maintenance. Pink1 is normally imported into mitochondria, cleaved by PARL protease, and degraded by the proteasome. When mitochondria lose their membrane potential, Pink1 starts to accumulate on the mitochondrial outer surface, where it recruits and activates Parkin to trigger protein ubiquitination, eventually leading to mitochondrial autophagy (reviewed in Narendra et al, 2012). Parkin E3 ubiquitin ligase activity is inhibited by its N-terminal ubiquitin-like (UBL) domain through interaction with C-terminal RING-between-RING (RBR) E3 catalytic domain (Chaugule et al, 2011). Recombinant Pink1 can phosphorylate the UBL domain at Ser65 in vitro, and Parkin Ser65 phosphorylation is readily detected when mitochondria are depolarized (Kondapalli et al, 2012; Shiba-Fukushima et al, 2012). A simplified model is that Pink1 phosphorylation of Ser65 in the UBL domain releases Parkin autoinhibition. However, phospho-mimetic substitution (Ser65Glu) or even UBL domain deletion cannot bypass the requirement of Pink1 for Parkin activation (Sarraf et al, 2013). Are there any other Pink1 substrate(s) or Parkin phosphorylation sites involved in Parkin activation? Three recent reports provide an unexpected answer: Pink1 directly phosphorylates ubiquitin, and this is critical for Parkin activation (Kane et al, 2014; Kazlauskaite et al, 2014; Koyano et al, 2014). This discovery represents the first example of a functional phosphorylation on ubiquitin.
To identify the full spectrum of Pink1 substrates, proteomic approaches were used to compare the profile of mitochondria isolated from wild-type or Pink1-deficient cells after treatment by CCCP, a mitochondria-depolarizing drug (Kane et al, 2014; Kazlauskaite et al, 2014). Surprisingly, a phosphopeptide identified in mitochondria-associated proteins only in wild-type cells derives from ubiquitin, and phosphorylation occurs at the conserved Ser65 residue, which corresponds to Ser65 in the Parkin UBL. Based on structural similarity between ubiquitin and the UBL domain, Koyano et al (2014) hypothesized that Ser65 on ubiquitin may be phosphorylated by Pink1, and indeed ubiquitin in CCCP-treated lysates exhibited a phosphorylation-dependent electrophoresis band-shift, and the same phospho-Ser65 peptide could be identified by mass spectrometry. Quantification showed that phospho-ubiquitin represents 0.05% of total endogenous ubiquitin, although it is not known whether all the pSer65 ubiquitin molecules are associated with mitochondria. The kinase–substrate relationship was further established by in vitro kinase assays, in which recombinant Pink1 could phosphorylate wild-type but not S65A ubiquitin (Kane et al, 2014; Kazlauskaite et al, 2014; Koyano et al, 2014).
Does phospho-S65 ubiquitin play any role in Parkin regulation, and is there any interplay between Ser65 ubiquitin and Parkin UBL domain Ser65 phosphorylation? Using in vitro Parkin E3 activity assays, all three groups provide compelling evidence that both phosphorylations are involved in and required for full Parkin activation. Kazlauskaite et al found that substituting wild-type ubiquitin with S65A ubiquitin greatly diminished Parkin E3 activity, but Parkin with a UBL S65A mutation also had much lower activity compared to wild-type Parkin, indicating that both phosphorylations are required. Kane et al found that prephosphorylation of ubiquitin by recombinant Pink1 could significantly enhance Parkin-mediated protein ubiquitination in vitro. Using GFP-Parkin and purified depolarized mitochondria as a source of Pink1, Koyano et al found that phospho-mimetic S65D ubiquitin, but not WT or S65A ubiquitin, could stimulate autoubiquitination of GFP-S65E-Parkin bearing an additional phospho-mimetic substitution in the UBL domain, but observed no enhancement with GFP-WT-Parkin.
How does the phospho-Ser65 ubiquitin activate Parkin? Koyano et al showed that phospho-mimetic S65D ubiquitin with a C-terminal diglycine mutation, which abolishes its conjugation to target proteins, could still activate the E3 activity of Parkin, excluding the possibility that phospho-S65 ubiquitin is preferred by Parkin for conjugation. They further showed that phospho-mimetic S65D ubiquitin binds to S65E Parkin, but not WT Parkin. The activation mechanism appears to be allosteric and allows functional access of Parkin to its cognate ubiquitin-conjugating E2 enzyme, since phospho-S65 ubiquitin accelerates Parkin discharge of UBCH7˜ubiquitin (Kazlauskaite et al, 2014; Koyano et al, 2014). In summary, these studies uncover a surprising missing link in Parkin activation (Fig1).
Figure 1. Model of how Pink1-mediated ubiquitin and UBL phosphorylation may activate Parkin.
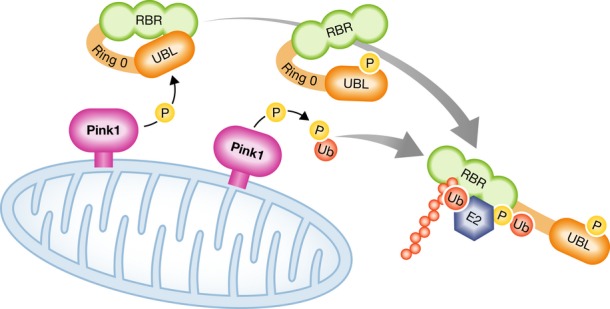
Pink1 phosphorylates both ubiquitin and the Parkin UBL domain at the Ser65 residue. Parkin UBL domain phosphorylation exposes a phospho-Ser65 ubiquitin binding site, and phospho-ubiquitin binding in turn activates Parkin E3 activity.
To understand how phospho-ubiquitin activates Parkin, the next step is to identify the exact binding site for phospho-Ser65-ubiquitin on Parkin, which was not characterized in the three recent studies. Interestingly, Wauer and Komander (2013) structurally identified a potential phosphopeptide binding site in Parkin's ‘Ring0’ domain, a highly conserved region between the UBL and the catalytic RBR domains. By determining crystal structures of Parkin, both Wauer and Komander (2013) and Trempe et al (2013) found a novel mechanism of Parkin regulation, in which the Ring0 domain inhibits Parkin E3 activity by blocking off the catalytic Cys431 in the RBR domain, such that activation of latent E3 activity of Parkin RBR domain in in vitro assays required removal of the Ring0 domain. Therefore, it is tempting to speculate that Ring0 represents a phospho-ubiquitin binding region involved in switching on Parkin ubiquitin ligase activity, although ultimately crystal structures of S65D ubiquitin bound to S65E Parkin will be necessary to validate such a molecular mechanism of activation.
Another major remaining question is whether UBL domain phosphorylation serves simply to relieve Parkin autoinhibition. Kazlauskaite et al showed that like phospho-Ser65-ubiquitin, the isolated phospho-Ser65-UBL domain can activate in vitro Parkin E3 activity in trans. ΔUBL-Parkin, though still able to translocate to depolarized mitochondria, is defective in mitochondria ubiquitination. Deleting the UBL domain greatly diminished ubiquitin trapping by Parkin bearing a serine substitution of its catalytic cysteine 431, also indicating a positive role for the UBL domain (Zheng & Hunter, 2013). Pink1 can stimulate Parkin self-association (Lazarou et al, 2013), and therefore, it is possible that the phospho-UBL domain may play a role similar to phospho-ubiquitin in activating additional Parkin molecules in trans.
Phosphorylation is generally counteracted by dephosphorylation. A mitochondrial phosphatase PGAM5 is a known binding partner of Pink1 (Imai et al, 2010), and in contrast to Pink1 being stabilized, PGAM5 cleavage is triggered by loss of mitochondrial membrane potential (Sekine et al, 2012). It will be interesting to test if PGAM5 can dephosphorylate p-Ser65-ubiquitin and p-Ser65-Parkin, and whether inhibition of PGAM5 could increase the efficiency of mitochondria clearance by Pink1/Parkin.
These findings also call attention to the function of phosphorylation of ubiquitin and other ubiquitin-like modifiers in general. Ubiquitin phosphorylation has been documented on S65, S57, T7, T12, Y59 in previous phospho-proteomic studies (referenced in Kane et al, 2014; Kazlauskaite et al, 2014), although the responsible kinases and especially their functional importance had remained elusive. Presumably, phosphorylation can change the binding properties of ubiquitin and ubiquitin chains, and phosphorylation occurring before conjugation may also affect linkage type or chain elongation.
Acknowledgments
The group of TH was supported by NIH grants (CA14195, CA80100 and CA82683). TH is a Frank and Else Schilling American Cancer Society Professor, and the Renato Dulbecco Chair in Cancer Biology. XZ is supported by a Salk Pioneer Fund Postdoctoral Scholar Award.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4:a011338. doi: 10.1101/cshperspect.a011338. pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Kanamaru Y, Koike M, Nishihara A, Okada M, Kinoshita H, Kamiyama M, Maruyama J, Uchiyama Y, Ishihara N, Takeda K, Ichijo H. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J Biol Chem. 2012;287:34635–34645. doi: 10.1074/jbc.M112.357509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauvé V, Grenier K, Seirafi M, Tang MY, Ménade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]


