Abstract
Mitochondrial F1Fo-ATP synthase generates the bulk of cellular ATP. This molecular machine assembles from nuclear- and mitochondria-encoded subunits. Whereas chaperones for formation of the matrix-exposed hexameric F1-ATPase core domain have been identified, insight into how the nuclear-encoded F1-domain assembles with the membrane-embedded Fo-region is lacking. Here we identified the INA complex (INAC) in the inner membrane of mitochondria as an assembly factor involved in this process. Ina22 and Ina17 are INAC constituents that physically associate with the F1-module and peripheral stalk, but not with the assembled F1Fo-ATP synthase. Our analyses show that loss of Ina22 and Ina17 specifically impairs formation of the peripheral stalk that connects the catalytic F1-module to the membrane embedded Fo-domain. We conclude that INAC represents a matrix-exposed inner membrane protein complex that facilitates peripheral stalk assembly and thus promotes a key step in the biogenesis of mitochondrial F1Fo-ATP synthase.
Keywords: assembly, F1Fo-ATP synthase, mitochondria, peripheral stalk
See also: DM Stroud & MT Ryan (August 2014)
Introduction
Exergonic hydrolysis of adenosine triphosphate (ATP) to adenosine diphospate (ADP) and inorganic phosphate (Pi) drives a large number of biochemical reactions in all living cells. Under aerobic conditions eukaryotic cells synthesize the vast majority of ATP through the activity of the F1Fo-ATP synthase complex that is located within the inner membrane of mitochondria. Here, redox-driven respiratory chain complexes expel protons from the mitochondrial matrix into the intermembrane space (IMS) compartment thereby generating a proton gradient across the inner membrane. Energy derived from this proton gradient is used by the ATP synthase to power ATP production from ADP and Pi on the matrix side of the inner mitochondrial membrane. This process is generally referred to as oxidative phosphorylation. F1Fo-ATP synthases from different organisms share a very similar overall architecture and are made up from essentially the same set of conserved core subunits, but may also contain specific accessory subunits that modulate stability and activity of the enzyme complex (Von Ballmoos et al, 2008).
In the baker's yeast Saccharomyces cerevisiae, the catalytic domain of F1Fo-ATP synthase is a hexameric complex composed of alternating Atp1 (F1α) and Atp2 (F1β) subunits that harbor the nucleotide-binding pockets of the enzyme (Stock et al, 1999; Von Ballmoos et al, 2008). Two structurally and functionally distinct stalk regions connect the catalytic domain to a membrane-embedded Fo-rotor domain formed by a ring-shaped decamer of Atp9 (subunit c) and a single copy of Atp6 (subunit a) that couples proton translocation to rotation of the Atp9 ring (Capaldi & Aggeler, 2002; Fillingame et al, 2003; Weber & Senior, 2003). The central stalk consists of the Atp3 (F1γ), Atp15 (F1ε) and Atp16 (F1δ) subunits and conveys rotational movements within the membrane-embedded domain to the nucleotide-binding sites by elastic power transmission (Noji et al, 1997; Junge et al, 2009). Catalytic domain and central stalk together form the F1-region of the ATP synthase. The Fo-domain of yeast mitochondrial ATP synthase additionally contains Atp8 and Atp17 (subunit f) that are essential for assembly and activity of the enzyme (Devenish et al, 2000). Rotation of the catalytic domain together with the Fo-rotor is prevented by the peripheral stalk (also termed stator stalk) that is made of the subunits Atp4 (subunit b), Atp5 (oligomycin-sensitivity-conferring protein, OSCP), Atp7 (subunt d), and Atp14 (subunit h) (Stock et al, 1999; Walker & Dickson, 2006). The small peripheral subunits Atp18 (subunit i), Atp19 (subunit k), Atp20 (subunit g), and Atp21 (subunit e) were shown to mediate the assembly and maturation of dimeric and oligomeric forms of the F1Fo-ATP synthase (Arnold et al, 1998; Arselin et al, 2003; Wagner et al, 2009, 2010). While oligomerization is not required for enzyme activity, such oligomers have been implicated in formation of the characteristic cristae domains of the inner mitochondrial membrane (Paumard et al, 2002; Dudkina et al, 2006; Davies et al, 2012).
Despite the tremendous physiological importance of F1Fo-ATP synthase complexes and the discovery of several inherited mitochondrial diseases caused by a partial ATP synthase deficiency (De Meirleir et al, 2004; Kucharczyk et al, 2009; Mayr et al, 2010; Spiegel et al, 2011; Torraco et al, 2012), still very little is known about the biogenesis of this fascinating enzyme. Two different genomes contribute to the formation of ATP synthase complexes: The Atp6, Atp8 and Atp9 proteins are encoded by mitochondrial DNA and inserted into the inner membrane from the matrix side (Devenish et al, 2000; Ott & Herrmann, 2010; Kehrein et al, 2013). All other components are nuclear-encoded and synthesized as precursors on cytosolic ribosomes with targeting information that directs them to mitochondria (Dolezal et al, 2006; Neupert & Herrmann, 2007; Chacinska et al, 2009). Some of these proteins, like Atp4 or the small peripheral subunits, are also integrated into the inner membrane upon import. All subunits of the F1-region and most proteins of the peripheral stalk must be fully translocated into the matrix. Thus, several protein biogenesis and sorting pathways contribute to formation of F1Fo-ATP synthase complexes, but mechanisms responsible for their coordination and mutual regulation are largely unexplored.
Current models suggest that assembly of the F1Fo-ATP synthase is a step-wise process that may include transient formation of discrete assembly intermediates (Rak et al, 2009, 2011). Several factors have been identified that are thought to facilitate assembly of distinct ATP synthase modules (Pícková et al, 2005; Ludlam et al, 2009). The matrix proteins Atp11, Atp12, and Fmc1 are required for biogenesis of the catalytic core of the F1-domain. Absence of any of these proteins leads to aggregation of the Atp1 and/or Atp2 subunits in the mitochondrial matrix (Ackerman & Tzagoloff, 1990; Wang et al, 2000; Lefebvre-Legendre et al, 2001). Yeast mutants lacking Atp11 or Atp12 were shown to be impaired in mitochondrial translation of ATP6 and ATP8 mRNAs (Rak & Tzagoloff, 2009). A number of further regulatory proteins have been suggested to support synthesis of mitochondrially encoded ATP synthase subunits and thus be required for production of functional enzyme complexes (summarized in Rak et al, 2009). The metalloprotease Atp23 is specifically required for maturation of the Atp6 subunit and directly implicated in assembly of the Fo-core domain together with Atp10 (Tzagoloff et al, 2004; Osman et al, 2007; Zeng et al, 2007). Notably, assembly factors specifically required for formation of the F1Fo peripheral stalk have not been identified so far.
Here we report on the identification of the inner membrane assembly complex (INAC) that promotes the biogenesis of mitochondrial F1Fo-ATP synthase. INAC consists of two so far uncharacterized mitochondrial inner membrane proteins, Ina22 and Ina17. Loss of INAC function causes dissociation of the F1-domain from the membrane-integral Fo-portion. This phenotype is explained by defects of the peripheral stalk segment, which is essential for the stable connection between F1 and Fo. Accordingly, we find that INAC facilitates assembly of the peripheral stalk and thus represents a critical assembly factor for the F1Fo-ATP synthase complex.
Results
Ina22, a mitochondrial protein required for respiratory growth
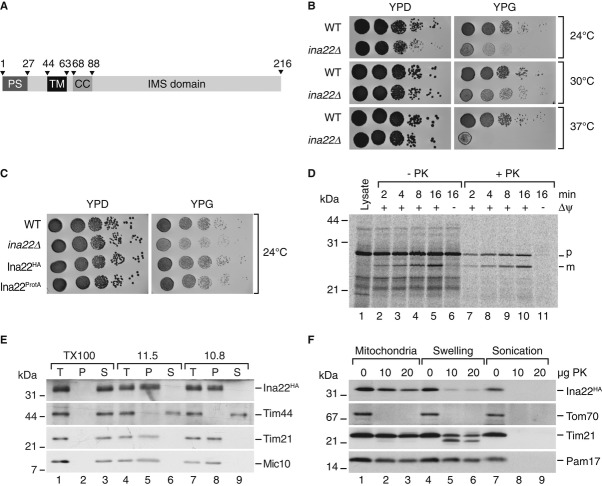
To identify novel proteins involved in the biogenesis of the mitochondrial oxidative phosphorylation machinery, we screened the yeast genome database for mutants affected in growth on non-fermentable carbon sources. Among the mutant strains, we selected yir024cΔ for further analyses that we later termed ina22Δ. INA22 (YIR024c) encodes an orphan protein with a predicted amino-terminal presequence for translocation across the inner mitochondrial membrane. Accordingly, Ina22 was previously identified in the yeast mitochondrial proteome (Sickmann et al, 2003). Moreover, Ina22 displays a single predicted transmembrane span, suggesting its localization in the inner mitochondrial membrane (Fig1A). In a high-throughput screen, absence of Ina22 was found to affect respiratory growth of yeast (Steinmetz et al, 2002; Qian et al, 2012). To verify this finding, we tested growth of the ina22Δ strain on fermentable (YPD) and non-fermentable (YPG) growth medium at different temperatures (Fig1B). The deletion strain was growing similar to wild-type on YPD medium at all tested temperatures. In contrast, on YPG medium growth of ina22Δ was significantly affected compared to the isogenic wild-type strain. This phenotype was exacerbated at lower and higher temperatures. For further analyses, we generated strains expressing Ina22HA and Ina22ProtA through chromosomal integration of the corresponding tag-encoding cassettes, allowing for expression of the fusion proteins from the authentic promoter. No growth difference between the wild-type and strains expressing Ina22HA or Ina22ProtA was observed (Fig1C). To assess translocation across the inner membrane and proteolytic removal of the presequence, we synthesized and radiolabeled Ina22 in a rabbit reticulocyte lysate and imported the precursor into purified mitochondria. Ina22 was imported to a protease-protected location in a membrane potential (Δψ)-dependent manner and processed to a faster migrating mature form indicative of inner membrane translocation (Fig1D). This finding supported the idea that Ina22 represented an inner membrane protein. Hence, we subjected mitochondria containing Ina22HA to alkaline extraction. As expected, Ina22 remained in the membrane pellet and was only released upon incubation with Triton X-100 (Fig1E). To assess the membrane topology of Ina22, we performed protease protection assays. The HA-tag was inaccessible to proteinase K (PK) treatment in intact mitochondria, but could be digested after the osmotic disruption of the outer mitochondrial membrane (swelling; Fig1F). In summary, these results confirmed that Ina22 was an integral inner mitochondrial membrane protein with its C-terminus exposed to the IMS.
Figure 1. Ina22 (Yir024c) is a mitochondrial inner membrane protein supporting respiratory growth.
- Predicted domain organization of S. cerevisiae Ina22. PS, presequence; TM, transmembrane domain; CC, coiled-coil domain; IMS, intermembrane space domain. Numbers indicate amino acid residues.
- Wild-type (BY4741) and ina22Δ yeast were spotted in serial 10-fold dilutions on YPD or YPG plates and grown at indicated temperatures for 2–5 days.
- As in (B), for indicated strains in YPH499 background.
- Radiolabeled Ina22 was imported into mitochondria for indicated times in the presence or absence of a membrane potential (Δψ) and subjected to treatment with proteinase K (PK) where indicated. p, precursor; m, mature.
- Membrane association of Ina22 assessed by carbonate extraction. T, total; P, pellet; S, soluble fraction.
- Submitochondrial localization of Ina22 was analyzed by protease protection. Indicated amounts of proteinase K (PK) were added to mitochondria in osmotically supporting SEM buffer (Mitochondria), hypotonic EM buffer (Swelling) or to mitochondria sonicated in the presence of 0.1% Triton X-100 (Sonication).
Loss of Ina22 function affects the F1Fo-ATP synthase
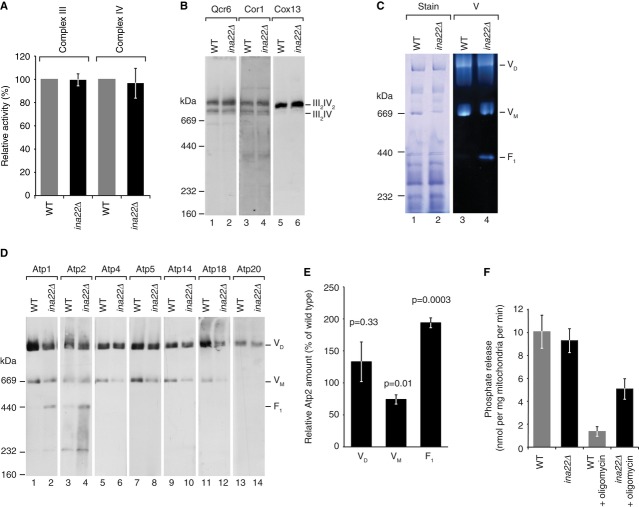
The respiratory growth defect of the ina22Δ strain led us to investigate a role of Ina22 in respiratory chain function. A steady state protein analysis of ina22Δ mutant mitochondria did not reveal obvious differences for any of the tested proteins (Supplementary Fig S1A). Thus, we assessed the activities of respiratory chain complexes III and IV directly to determine if their function was compromised (Fig2A). Both complex III and IV activities in ina22Δ mitochondria were indistinguishable from the wild-type control. As respiratory growth deficiency can also be caused by defective oligomerization of respiratory chain complexes (Chen et al, 2012; Strogolova et al, 2012; Vukotic et al, 2012), we analyzed supercomplex formation of complex III and IV by Blue Native (BN) PAGE. Western blot analyses revealed that III2/IV and III2/IV2 supercomplexes were similar in wild-type and ina22Δ mitochondria (Fig2B). Thus, no defects in respiratory chain complexes III or IV were apparent in the ina22Δ mutant mitochondria.
Figure 2. Deletion of INA22 affects the F1Fo-ATP synthase, but not respiratory chain complexes III and IV.
- Activities of respiratory chain complexes III and IV in wild-type and ina22Δ mitochondria as described in Materials and Methods. Activity of each complex in wild-type mitochondria was set to 100%. Data represent means ± standard error of the mean (s.e.m.), n ≥ 5.
- Wild-type and ina22Δ mitochondria were solubilized in 1% digitonin, analyzed by BN-PAGE with subsequent Western blotting using indicated antibodies against components of complexes III and IV.
- Wild-type and ina22Δ mitochondria were analyzed by BN-PAGE, Coomassie stained (Stain) or stained for in-gel F1Fo-ATP synthase activity (V).
- As in (B), but immunodecorated for proteins of complex V.
- Quantification of the Atp2 decoration in (D). Relative amount of Atp2 in dimer (VD), monomer (VM), and F1 in ina22Δ mitochondria compared to the wild-type (set to 100%) is shown. P-value based on two-sided t-test is indicated (n ≥ 5).
- In organello ATPase activity measurement was performed as described in Materials and Methods in wild-type and ina22Δ mitochondria, with or without addition of 50 μM oligomycin. Activity is presented in nmol of released phosphate per min per mg of mitochondrial protein (n = 5).
We speculated that the observed growth phenotype could be due to malfunction of the F1Fo-ATP synthase. First we analyzed the ATP-hydrolyzing activity of the catalytic domain by an in-gel assay (Fig2C). Interestingly, we observed dissociation of the F1-portion from the Fo-complex in ina22Δ mutant mitochondria (Fig2C). Compared to the wild-type control, a four to five fold increase in free F1-portion was observed in ina22Δ mitochondria. Immunoblotting with specific antibodies directed against ATP synthase subunits of different domains showed that the amounts of ATP synthase monomers were reduced in ina22Δ mitochondria and antibodies against Atp1 and Atp2 revealed the presence of free F1-subcomplexes not connected to Fo-domains (Fig2D and E). While the overall ATPase activity was similar in the presence or absence of Ina22, the F1Fo-ATP synthase of ina22Δ mutant mitochondria displayed a five fold higher ATP-hydrolyzing activity in the presence of the inhibitor oligomycin (Fig2F). Thus, lack of Ina22 considerably reduced the oligomycin sensitivity of ATP synthase. Based on these observations, we concluded that Ina22 was required for correct assembly of the functional F1Fo-ATP synthase.
Ina22 associates with selected subunits of F1Fo-ATP synthase
To address if Ina22 was a subunit of the F1Fo-ATP synthase, we isolated Ina22ProtA and associated proteins by IgG affinity chromatography from detergent-solubilized mitochondria. Purified proteins were separated by SDS-PAGE and the most abundant bands in a Coomassie-stained gel were analyzed by MALDI mass spectrometry (Fig3A). Along with the bait, the most abundant bands were found to contain the Atp1 and Atp2 subunits of the F1-portion. To obtain a full and unbiased view of the interaction network, we metabolically labeled cells using stable isotope labeling by amino acids in cell culture (SILAC; Ong et al, 2002) and isolated Ina22ProtA-containing complexes for characterization by quantitative mass spectrometry. Analysis of SILAC data from three independent replicates revealed significant enrichment of selected subunits of the F1Fo-ATP synthase (Fig3B and Supplementary Table S2). Notably, all of the identified proteins were subunits of the F1-domain or the peripheral stalk, whereas neither subunits of the Fo-part nor small accessory subunits required for dimerization of the enzyme were found enriched (Fig3C). Moreover, Cbp3, Cbp4, and Cbp6 (which participate in cytochrome b assembly; Gruschke et al, 2011, 2012) and the uncharacterized Ypl099c (Aim43), which we later termed Ina17, copurified with Ina22 (Fig3B). To support these mass spectrometric data we isolated Ina22ProtA from solubilized mitochondria and analyzed the precipitates by Western blotting. Indeed, co-isolation of Atp1, Atp2 of the F1-domain and Atp5 of the peripheral stalk could be confirmed with available antibodies. In contrast, the small dimer-promoting subunit Atp20 was not recovered (Fig3D). We were similarly able to confirm the co-isolation of Cbp3. Whereas the co-isolation of ATP synthase subunits with tagged Ina22 was in line with the observed defects of F1Fo-ATP synthase complexes, the presence of cytochrome b assembly factors was surprising. However, as the ina22Δ mutant did not display any defect in complex III activity or organization (Fig2A and B), we did not follow this aspect further in this study. In conclusion, the observed low efficiency of co-isolation of Ina22 with ATP synthase subunits and the fact that mitochondria-encoded subunits of the ATP synthase as well as the dimerization factors were not enriched in purified Ina22-containing complexes, indicate that Ina22 is not a stoichiometric subunit of the mature ATP synthase. We also assessed mitochondrial morphology by microscopy in comparison to atp20Δ mutant cells. While atp20Δ cells displayed defective mitochondrial morphology, ina22Δ mitochondria had only a mild defect (Supplementary Fig S2).
Figure 3. Ina22 interacts physically with a subset of F1Fo-ATP synthase subunits.
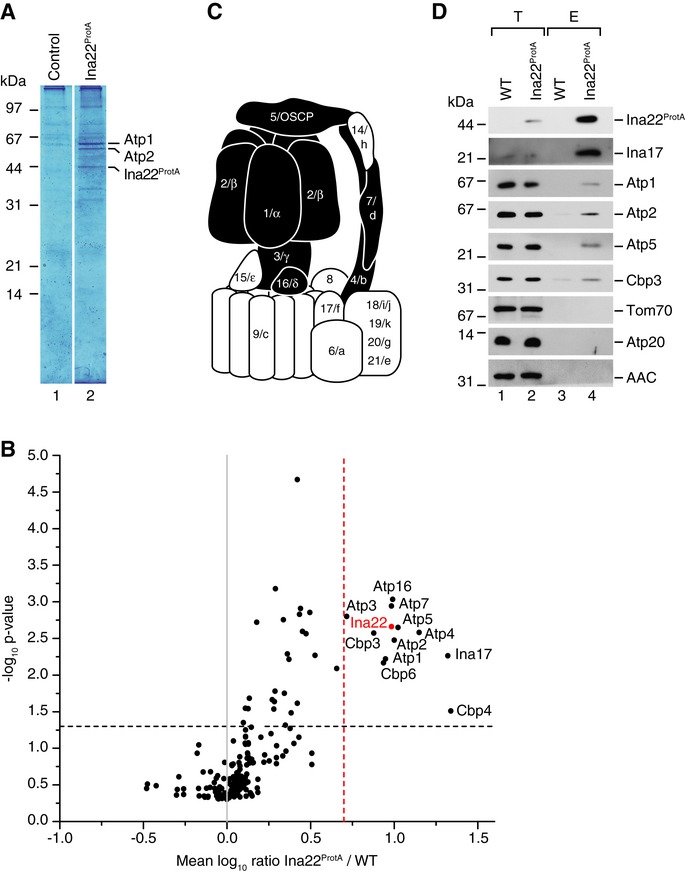
- IgG affinity purification from wild-type and Ina22ProtA mitochondria. Selected bands were identified by MALDI mass-spectrometry.
- Quantitative mass spectrometric analysis of protein complexes purified from the Ina22ProtA mitochondria after SILAC. Proteins enriched in Ina22ProtA purifications compared to control purifications from wild-type mitochondria with a mean log10 ratio over 0.7 and P-value < 0.05 are indicated (n = 3).
- Schematic representation of the F1Fo-ATP synthase. Subunits enriched in (B) are shown in black.
- Protein complexes were purified from wild-type and Ina22ProtA mitochondria by IgG affinity chromatography and analyzed by SDS-PAGE and Western blotting. Elution (E), 100%; total (T), 0.5%.
The inner membrane protein Ina17 affects ATP synthase organization
The mass spectrometric analyses revealed significant enrichment of Ina17 in the Ina22 complex purification (Fig3B) and we confirmed this copurification by Western blotting (Fig3D). Ina17 is a protein with a predicted presequence, a single hydrophobic amino acid stretch sufficient in length to form a transmembrane span, a predicted coiled-coil, and a calculated molecular weight of 21.8 kDa for the precursor (Fig4A). Previous analyses had identified Ina17 as mitochondrial protein (Reinders & Zahedi, 2006; Hess et al, 2009; Gebert et al, 2012). Upon incubation of radiolabeled Ina17 with purified mitochondria, the precursor protein was imported in a Δψ-dependent manner and processed to a faster migrating mature form suggesting transport across the inner membrane (Fig4B). To address if Ina17 was a mitochondrial membrane protein, we subjected purified mitochondria to alkaline extraction. Ina17 was resistant to alkaline treatment and was only released by detergent solubilization indicating that it represented an integral membrane protein (Fig4C). In order to determine the topology and submitochondrial localization of Ina17, we performed protease protection experiments. Ina17 was resistant to protease treatment in intact mitochondria but became accessible upon rupture of the outer membrane by hypoosmotic treatment. Since the antiserum against Ina17 was directed against a C-terminal peptide, we concluded that the epitope is exposed to the IMS (Fig4D).
Figure 4. Ina17 is a mitochondrial inner membrane protein.
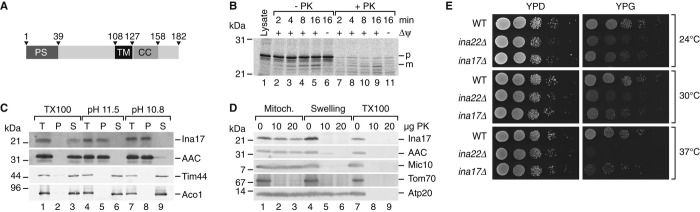
- Schematic representation of Ina17. PS, predicted presequence. TM, transmembrane span. CC, predicted coiled coil domain. Numbers indicate amino acid residues.
- In organello import of Ina17. In vitro translated [35S]-labeled Ina17 was imported into wild-type mitochondria for indicated times, in the presence or absence of a membrane potential (Δψ), with or without subsequent proteinase K (PK) treatment.
- Membrane association of Ina17 was assessed by carbonate extraction, as in Fig1E.
- Proteinase K protection assay was performed as in Fig1F.
- Growth test of ina22Δ and ina17Δ strains compared to wild-type. 10-fold serial dilutions of each strain were grown on YPG or YPD at indicated temperatures for 2–5 days.
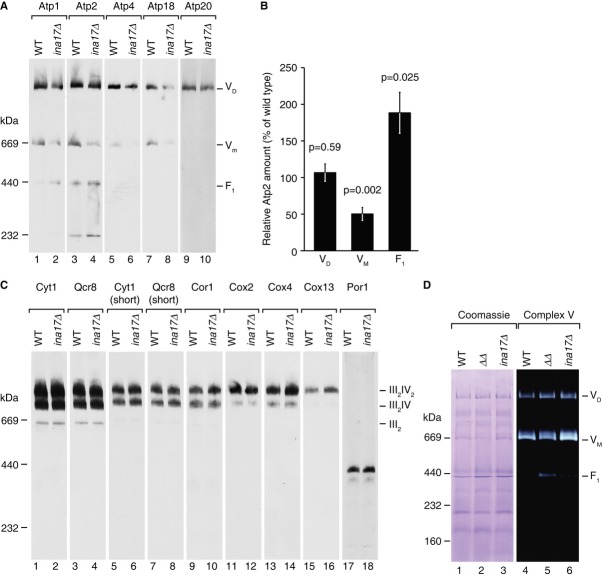
Because ina22Δ cells displayed respiratory defects, we then analyzed the growth phenotype of ina17Δ mutant cells on fermentable and non-fermentable medium. ina17Δ mutant cells displayed a clear growth defect compared to wild-type cells on non-fermentable medium. This phenotype was exacerbated at higher temperature (Fig4E). Thus, ina17Δ mutant cells displayed a similar growth defect as observed for the ina22Δ mutant. For further analyses, mitochondria were isolated from ina22Δ and ina17Δ mutant cells and initially analyzed for steady state levels of selected mitochondrial proteins. As expected, these analyses confirmed the ina17Δ mutant, as the Ina17 protein was not detected. However, none of the tested proteins was altered in abundance compared to the wild-type control (selected proteins are shown in Supplementary Fig S1B). Thus, we analyzed F1Fo-ATP synthase complexes by BN-PAGE. Similar to the phenotype observed for the F1Fo-ATP synthase in ina22Δ mutant mitochondria, ina17Δ mitochondria displayed a partial dissociation of ATP synthase with a significant amount of free F1-domain and reduced amounts of the monomeric form (Fig5A and quantification in 5B). In contrast to the F1Fo-ATP synthase complexes, III2IV2 supercomplexes of the respiratory chain remained stable (Fig5C). The partial disconnection of the F1-portion from the Fo-domain was also apparent by an in–gel activity assay (Fig5D). For comparison, we generated an ina17Δ/ina22Δ double mutant (Supplementary Fig S3) and found that destabilization of ATP synthase was exacerbated in the double mutant mitochondria (Fig5D). In the ina17Δ/ina22Δ double mutant the amount of free F1-portion was four fold increased compared to the ina17Δ and eight fold in comparison to wild-type.
Figure 5. Deletion of INA17 causes defects in the F1Fo-ATP synthase.
- 10 μg of wild-type and ina17Δ mitochondria were analyzed by BN-PAGE and Western blotting with indicated antisera.
- Quantification of the Atp2 signal in (A). Atp2 amount in the ATP synthase dimer (VD), monomer (VM), and F1 in ina17Δ relative to wild-type (set to 100%) is shown. P-value based on a two-sided t-test is indicated (n = 6).
- Same as in (A), but using antisera against respiratory chain complexes III and IV.
- Solubilized mitochondria were analyzed by BN-PAGE and Coomassie staining or F1Fo-ATP synthase in-gel activity staining.
Ina22 and Ina17 form a complex in the inner membrane
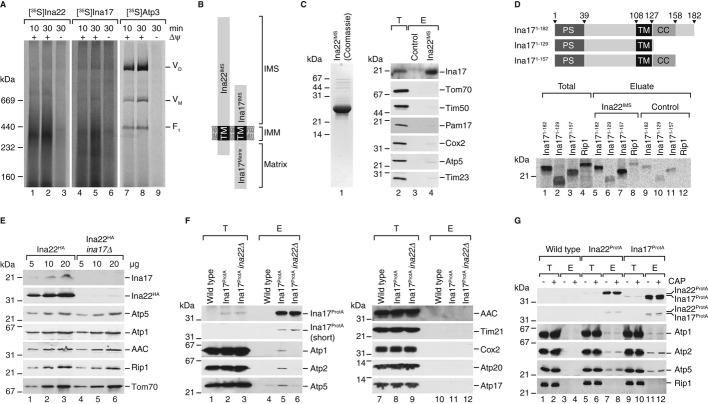
Our analyses had revealed that Ina17 interacts with Ina22 and that both Ina17- and Ina22-deficient mutants were affected in the organization of the F1Fo-ATP synthase. To assess if both proteins indeed formed complexes in mitochondria, we performed import assays with the radiolabeled precursors of Ina22, Ina17 and Atp3. Subsequently, mitochondria were solubilized and protein complexes separated by BN-PAGE. Protein complexes containing the imported proteins were detected by digital autoradiography. While Atp3 efficiently assembled into F1Fo-ATP synthase complexes, Ina22 and Ina17 assembled predominantly into co-migrating complexes with an apparent molecular weight of ˜300 kDa (Fig6A). (Ina17 also formed a faster migrating complex in the range of ˜200 kDa, which was not seen for Ina22.)
Figure 6. Ina22 and Ina17 form a complex in the inner membrane.
- Full-length radiolabeled Ina22, Ina17 and Atp3 were imported into wild-type mitochondria for indicated times in the presence or absence of a membrane potential (Δψ), with subsequent BN-PAGE analysis and detection of the radioactive signal by digital autoradiography.
- Schematic presentation of the membrane topology of Ina17 and Ina22. IMS, intermembrane space; IMM, inner mitochondrial membrane; TM, transmembrane domain.
- Pull-down assay with immobilized Ina22IMS. Purified Ina22IMS (Coomassie-stained SDS gel is shown in the left panel) was immobilized on sepharose beads and incubated with solubilized wild-type mitochondria. Bound proteins were eluted and analyzed by SDS-PAGE and Western blotting with indicated antisera. E, eluate, 100%; T, total, 1%.
- Pull-down assay as in (C) with Ina22IMS-coupled sepharose and the indicated [35S]-labeled proteins synthesized in vitro. Eluate, 100%; total, 10%. Schematic representation of Ina17 truncation constructs in the top panel as in Fig4A.
- Steady-state protein levels in mitochondria from Ina22HA and Ina22HA/ina17Δ strains assessed by SDS-PAGE and Western blotting with indicated antisera.
- IgG affinity purification from Ina17ProtA, Ina17ProtA/ina22Δ and isogenic wild-type mitochondria. Purified proteins were analyzed by SDS-PAGE and Western blotting. E, eluate, 100%; T, total, 0.7%.
- Affinity purification as in (F) from the indicated strains treated with chloramphenicol (CAP) prior to isolation of mitochondria. Long and short exposures of Protein A decorations are shown in top two panels, respectively.
While Ina22 is largely exposed to the IMS, Ina17 exposes domains into the matrix and the IMS (Fig6B). Based on this topology, we asked if both proteins interacted through their IMS-domains. Therefore, we expressed and purified the recombinantly produced Ina22IMS-domain from E. coli cells. The purified domain was immobilized on CNBr-activated Sepharose columns and incubated with mitochondrial extracts obtained after solubilization in Triton X-100-containing buffer. Upon extensive washing bound proteins were analyzed by Western blotting. While control inner membrane proteins were not recovered in significant amounts, Ina17 was efficiently bound by the Ina22IMS-domain, indicating that both proteins indeed associate via their IMS-domains (Fig6C). To address if Ina22 and Ina17 interacted through their predicted coiled-coil domains, we expressed and radiolabelled full-length Ina17 and two truncations in reticulocyte lysates. All tested constructs containing the coiled-coil region specifically bound to Ina22IMS. In contrast, binding of Ina171–129, lacking the coiled coil, to Ina22IMS was significantly less efficient (Fig6D).
Our previous experiments showed that Ina17 was present in wild-type-like amounts in the absence of Ina22 (Supplementary Fig S1). We generated an ina17Δ mutant strain expressing a HA-tagged version of Ina22 to assess if the presence of Ina22 depended on Ina17. Surprisingly, mitochondria isolated from this strain contained drastically reduced amounts of Ina22 (Fig6E). Hence, the presence of Ina22 in mitochondria apparently depended on Ina17, while vice versa this was not the case.
Because the F1-portion of the F1Fo-ATP synthase is predominantly exposed to the mitochondrial matrix, we speculated that the observed association of Ina22 with the ATP synthase occurred in complex with its partner Ina17. To address this hypothesis, we tagged Ina17 with Protein A at the C-terminus by chromosomal integration of the corresponding Protein A-encoding cassette. Ina17ProtA-containing mitochondria were isolated, solubilized, and Ina17ProtA together with bound proteins isolated via IgG-chromatography. Similar to Ina22, subunits of the F1-portion and peripheral stalk such as Atp1, Atp2, Atp5 co-isolated with Ina17ProtA, whereas components of the Fo-domain (e.g. Atp17) and the dimeric ATP synthase complex (Atp20) or other abundant inner membrane proteins (e.g. AAC, Cox2) were not recovered in the eluate (Fig6F). Thus, Ina17 displayed a selective association with the same F1Fo-ATP synthase subdomains that were observed to bind to Ina22.
Since Ina22-levels were reduced in the absence of Ina17, we assessed the ATP synthase association of Ina17 in the absence of Ina22. Therefore, we generated an ina22Δ yeast strain expressing Ina17ProtA for complex isolation. When Ina17ProtA was isolated from ina22Δ mitochondria, the association of Ina17 with the F1-portion and the peripheral stalk of the F1Fo-ATP synthase was lost (Fig6F, lane 5 and 6). Accordingly, complex formation between Ina17 and Ina22 is a prerequisite for F1-association of Ina17. Based on the phenotypes of the mutant cells and the complex formation between Ina22 and Ina17, we termed this complex INAC.
We asked, if the association of INAC with the F1-portion and peripheral stalk was dependent on the core subunits Atp6, Atp8, and Atp9 of the Fo-part. Therefore, we blocked the synthesis of mitochondria-encoded proteins by chloramphenicol treatment and isolated Ina22ProtA- and Ina17ProtA-containing complexes from purified mitochondria. Compared to the untreated control, the association of the F1-portion and peripheral stalk were not decreased upon block of mitochondrial translation (Fig6G). As a control, we assessed the association of the cytochrome c oxidase assembly factor, Coa3, with the translational activator Mss51 under these conditions (Mick et al, 2010). As reported previously the association was significantly reduced in the presence of chloramphenicol (Supplementary Fig S4A). We also assessed the amount of mitochondria-encoded proteins that associated with INAC. To this end we radiolabelled mitochondrial translation products in isolated mitochondria prior to Ina17ProtA isolation. While Western blotting revealed subunits of the F1-portion in the eluate as expected, only very low levels of radiolabelled mitochondria-encoded Fo-subunits were recovered (Supplementary Fig S4B). Accordingly, INAC interacts with the F1-portion and peripheral stalk of the ATP synthase independent of newly synthesized Fo subunits Atp6, Atp8 and Atp9 and does not associate to a significant extent with the Fo-portion.
The INA complex facilitates the assembly of the peripheral stalk
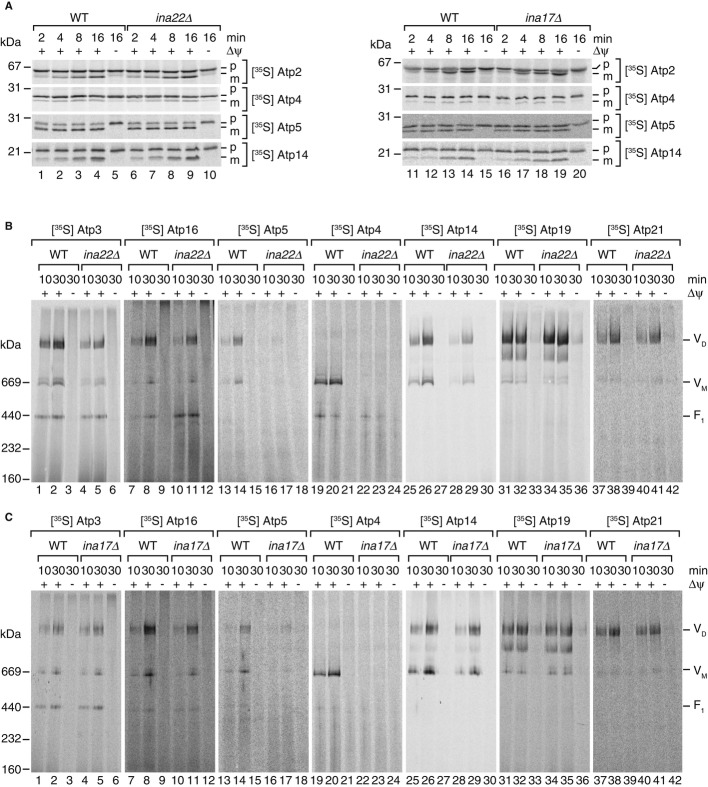
In ina22Δ and ina17Δ mutant cells the F1Fo-ATP synthase displayed partial dissociation. Characteristic for both mutant phenotypes was the accumulation of an increased free pool of the F1-portion. At the same time both proteins interacted with a fraction of the F1-domain and peripheral stalk at steady state. Based on these observations, we speculated that the INA complex formed by Ina22 and Ina17 was involved in the formation of a stable connection between the F1- and Fo-domains of the ATP synthase, a process that requires an intact peripheral stalk. To address this hypothesis directly, we expressed and radiolabeled selected precursor proteins of the different subdomains of ATP synthase in rabbit reticulocyte lysate and imported them into wild-type and INAC mutant mitochondria. All of the tested proteins were efficiently imported into wild-type and mutant mitochondria and processed to a faster migrating mature form (selected precursor proteins are shown in Fig7A). In order to assess the incorporation of imported subunits into ATP synthase complexes, we performed BN-PAGE analysis of selected import reactions. Atp3, a subunit of the central stalk, assembled into monomeric and dimeric ATPase complexes, but was also recovered in a free F1 pool. Compared to wild-type, assembly of Atp3 into the ATPase monomer and dimer was reduced (Fig7B, lanes 4 & 5). Similarly, slight reduction was observed for assembly of Atp16 of the central stalk (Fig7B, lanes 10 & 11). We then analyzed subunits of the peripheral stalk. A striking assembly defect was observed for Atp5 (oligomycin-sensitivity-conferring protein, OSCP) and Atp4 (Fig7B, lanes 16 & 17, 22 & 23). In addition, an obvious, but less pronounced assembly defect was observed for Atp14 (Fig7B, lanes 28 & 29). As a control, we assessed assembly of the dimerization factors Atp19 and 21. Both proteins assembled with similar efficiency into ATP synthase complexes in mutant and wild-type mitochondria (Fig7B, lanes 31–42). These experiments demonstrated that Ina22 was primarily required for assembly of peripheral stalk subunits to the F1Fo-ATP synthase. Thus, we performed similar assembly analyses in ina17Δ mitochondria. Again, severe assembly defects were observed for peripheral stalk subunits Atp5 and Atp4 and to a lesser, but still noticeable, extent for Atp14 (Fig7C). Moreover, similar to what was observed in ina22Δ mitochondria, assembly of Atp16 was reduced compared to wild-type while assembly of Atp3 was less affected in ina17Δ than in the absence of Ina22. In contrast, no defect was observed for assembly of Atp19 and Atp21 (Fig7C). In summary, both INA complex constituents, Ina22 and Ina17, are required for efficient assembly of peripheral stalk constituents into the F1Fo-ATP synthase complex. Since the peripheral stalk stably connects F1- and Fo-domains of ATP synthase, this finding explained the apparent instability of the enzyme complex in INAC mutant mitochondria.
Figure 7. Assembly of F1Fo-ATP synthase peripheral stalk subunits is impaired in ina22Δ and ina17Δ.
A [35S]-labeled ATP synthase subunits were imported into isolated mitochondria from wild-type, ina22Δ (left panel), or ina17Δ (right panel) for indicated times and analyzed by SDS-PAGE and digital autoradiography.
B, C After import of radiolabeled F1Fo-ATP synthase subunits into wild-type and ina22Δ (B) or ina17Δ (C) mitochondria, non-imported proteins were digested by proteinase K and samples analyzed by BN-PAGE followed by digital autoradiography.
Discussion
Mitochondrial protein complexes of the oxidative phosphorylation system, namely complexes I, III, IV and the F1Fo-ATP synthase, assemble from subunits of dual genetic origin. Hence, mitochondria are challenged by the task to assemble sophisticated nano-machines from proteins that are imported into mitochondria from the cytoplasm and those that are inserted into the inner membrane from the matrix in a co-translational manner (Rak et al, 2009; Soto et al, 2011; Smith et al, 2012). In case of the F1Fo-ATP synthase, the core subunits of the proton-conducting Fo-domain are mitochondria-encoded, while the catalytic F1-ATPase domain is built from imported subunits. Several chaperone-like factors have been described that are selectively involved in the biogenesis of either the F1-portion (Atp11, Atp12, Fmc1) or the Fo-sector (Atp10, Atp23). However, very little has been known about a critical later step in the F1Fo-ATP synthase assembly pathway: the connection of the soluble F1-domain to the membrane-embedded Fo-domain through the peripheral stalk segment (Prescott et al, 1994; Rak et al, 2009; Pagliarani et al, 2013). With the discovery of INAC as a protein complex that ensures stable coupling of the two main modules of F1Fo-ATP synthase by supporting peripheral stalk formation, we now shed first light on this terra incognita.
The inner mitochondrial membrane proteins Ina17 and Ina22 are constituents of INAC. Although obvious homologous of Ina17 and Ina22 were only found in the fungal kingdom, the fact that a number of distinct assembly intermediates of the F1-portion have been identified in human mitochondria suggests that proteins with similar functions could assist the assembly of mammalian F1Fo-ATP synthase (Fernández-Vizarra et al, 2009). Both proteins expose domains to the IMS and matrix side. Interestingly, we find that INAC specifically interacts with the F1-portion and peripheral stalk segment of the F1Fo-ATP synthase. Genetic interaction between INAC constituents and ATP18, YME1, MGE1 or prohibitins have been reported. Although we did not detect a physical interaction of INAC with the encoded proteins, the high throughput analysis is in agreement with a role of Ina17 and Ina22 in mitochondrial biogenesis and/or membrane organization. We find that the lack of Ina17 or Ina22 does not lead to a loss of the F1Fo-ATP synthase but causes dissociation of the F1-portion from the Fo-membrane unit. When we assessed the assembly of nuclear-encoded, imported subunits into the F1Fo-ATP synthase, we found that subunits of the peripheral stalk (Atp4, Atp5, Atp14) displayed severe assembly defects in ina17Δ or ina22Δ mitochondria. Especially the incorporation of Atp4, which is the central structural component of the peripheral stalk, into the F1Fo-ATP synthase was impaired. Although we cannot formally exclude a role of INAC in the dynamic regulation of the mature ATP synthase complex assembly state, these data strongly support the idea that INAC assists in the biogenesis of F1Fo-ATP synthase by facilitating the assembly of newly imported subunits into the peripheral stalk segment. Our analyses further showed that INAC is not a stoichiometric subunit of the active F1Fo-ATP synthase. This finding agrees with previous reports on the abundance of cellular proteins, which showed that the cellular amount of Ina22 represent < 0.5% of Atp2 (Ghaemmaghami et al, 2003). This ratio, together with the specific association of INAC with the F1- and peripheral stalk portions but not Fo, and the INAC requirement for incorporation of peripheral stalk subunits indicate a transient association of INAC with F1Fo-ATP synthase modules. Taking the localization of INAC into consideration, it is likely that the association with the F1- and peripheral stalk portions occurs at the inner membrane. A lack of INAC function does not affect the formation of the catalytic α3β3 oligomer. Hence, the ATPase activity of the F1-ATPase is not reduced under standard conditions in ina17Δ or ina22Δ mitochondria. However, in the presence of oligomycin the mutant mitochondria displayed increased resistance to the antibiotic treatment, which suggests a defect in Fo assembly or its association with the F1-portion27,53. This matches the observed phenotype of INAC mutants.
Our data suggest that Ina22 and Ina17 interact with each other through their IMS domains. While Ina22 exposes only a small domain of 17 amino acids into the mitochondrial matrix, Ina17 possesses a larger (˜8 kDa) matrix-segment. Ina22 is destabilized in the absence of Ina17, but this is not the case in the reverse situation. However, the association of Ina17 with the F1-portion depends on Ina22, indicating that both proteins are required for the interaction. It is a generally accepted concept that the assembly of the ATP synthase occurs in a modular manner (Rak et al, 2011; Fox, 2012). In this process, the mitochondria-encoded Atp6 and Atp8 of the Fo-portion associate in the inner membrane. In parallel, a ring of mitochondria-derived Atp9 subunits is formed to which the fully assembled nuclear-encoded catalytic core, composed of Atp1, 2, 3, 15 and 16, is thought to associate. The assembly of the lateral stalk, of which the Atp4 subunit represents the only inner membrane protein, is thought to initially engage with the Atp6/Atp8 complex and subsequently becomes linked to the α, β ring through Atp5. In contrast to this, the identification of INAC leads us to speculate that a full F1/peripheral stalk-assembly intermediate exists at the inner membrane that lacks any of the mitochondria-encoded subunits. In agreement with this, the association of INAC with the F1 portion and the peripheral stalk occurs in the absence of newly synthesized Atp6, Atp8, and Atp9. Thus, we conclude that INAC assists in the process of linking this fully formed complex with the membrane integral module. The function of INAC is not essential for the assembly process per se, as a mature F1Fo-ATP synthase is formed in its absence. Hence, alternative assembly pathways appear to exist. However, in the absence of INAC the stability of the F1Fo-ATP synthase is affected and the assembly process, as indicated by our assays, appears to be significantly less efficient. Thus, we conclude that INAC promotes the correct association of the F1-portion to the Fo-segment of the ATP synthase and that a lack of INAC weakens the association through improper molecular interactions.
Through the identification of a novel assembly factor complex, this study considerably advances the knowledge about the biogenesis of a key player in cellular energy metabolism, the mitochondrial F1Fo-ATP synthase. Our characterization of INAC-deficient mutant mitochondria allowed a first glance at the crucial process of peripheral stalk assembly. Further work will however be required to understand the molecular mechanism of INAC function and its interplay with other known assembly factors.
Materials and Methods
Yeast strains and growth conditions
Yeast strains used in this study are listed in Supplementary Table S1. Wild-type strains YPH499 and BY4741 were used (Sikorski & Hieter, 1989; Brachmann et al, 1998). Deletion strains ina22Δ (yir024cΔ), ina17Δ (aim43Δ), and the corresponding wild-type strain BY4741 were obtained from the Euroscarf collection. All other strains were generated by homologous recombination of PCR derived cassettes from plasmids pYM10, pYM2 and pFA6a–HIS3MX6, as previously described (Janke et al, 2004). All yeast strains were grown on non-fermentable YPG (1% yeast extract, 2% peptone, 3% glycerol) or fermentable YPD medium (1% yeast extract, 2% peptone, 2% glucose), unless otherwise indicated. Mitochondria were isolated from yeast grown at 30°C on YPG according to standard procedures (Meisinger et al, 2006) and stored in single-use aliquots at −80°C. For mitochondrial translation inhibitor treatment, yeast cells were incubated with 2 mg/ml chloramphenicol for 3 h prior to mitochondrial isolation (Mick et al, 2010).
Synthesis of radiolabeled proteins and import into isolated mitochondria
Full-length open reading frames of INA22 (YIR024C) and ATP3, as well as ATP synthase subunits ATP14, ATP16 and ATP19 with three additional C-terminal methionine residues were cloned into the pTNT™ vector (Promega). Open reading frames of full-length INA17 (AIM43), RIP1, ATP4, ATP5 and ATP21 as well as DNA encoding the Ina171–129 and Ina171–157 fragments for pull-down assays were amplified using a forward primer containing the SP6 polymerase binding site and reverse primer with three additional codons for extra methionine residues prior to the stop codon. All constructs were in vitro transcribed using mMESSAGE mMACHINE® SP6 Kit (Life Technologies). The obtained RNA was used for in vitro translation by Flexi® Rabbit Reticulocyte Lysate System (Promega) in the presence of [35S]methionine. Radiolabeled proteins were imported into isolated mitochondria according to published procedures (Wiedemann et al, 2006) and analyzed by SDS-PAGE or BN-PAGE. Gels were dried and exposed on phosphorimager screens (GE Healthcare) for detection of radioactive signal by digital autoradiography.
In organello labeling
Labeling of mitochondrial translation products with [35S] methionine was performed essentially as described previously (Westermann et al, 2001). In brief, mitochondria were incubated in translation buffer (900 mM sorbitol, 225 mM potassium chloride, 22.5 mM potassium phosphate, 30 mM Tris/HCl pH 7.5, 4.5 mg/ml BSA, 6 mM ATP, 0.75 mM GTP, 9 mM α-ketoglutarate, 10 mM phosphoenolpyruvate, 0.15 mM methionine-free amino acid mix, 7.5 μg/ml cycloheximide, 19 mM magnesium sulfate, 38 μg/ml pyruvate kinase) in the presence of 20 μM [35S] methionine (10 mCi/ml) at 30°C.
Protein localization assays
For membrane association analysis, mitochondria were treated with sodium carbonate buffers, pH 10.8 and 11.5, or lyzed with 0.1% Triton X-100. Soluble fractions were separated from membranes by centrifugation for 45 min at 100,000 g, 4°C. The samples were precipitated with trichloroacetic acid (TCA) and analyzed by SDS-PAGE and immunoblotting. For proteinase protection assays, mitochondria were suspended in isoosmotic SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS, pH 7.2), hypotonic EM buffer (1 mM EDTA, 10 mM MOPS, pH 7.2) for 20 min on ice or sonicated in the presence of 0.1% Triton X-100 and subsequently treated with indicated amounts of proteinase K (PK) for 10 min on ice. The samples were TCA-precipitated and analyzed by SDS-PAGE and immunoblotting.
Enzyme activity measurements
Activity of respiratory chain complexes III and IV in isolated mitochondria was determined by assessing cytochrome c reduction and oxidation, respectively. Complex III activity was determined in 10 mM potassium phosphate buffer, supplemented with 0.5 mM NADH and 10 mM KCN. Mitochondria were suspended at 0.1 mg/ml and the reaction was started by addition of non-reduced cytochrome c to a final concentration of 1.5 mg/ml. The rate of absorbance increase at 550 nm, corresponding to cytochrome c reduction, in wild-type mitochondria was set to 100% activity. For complex IV activity determination, mitochondria were suspended in 10 mM potassium phosphate buffer, pH 7.4, at a concentration 0.25 mg of mitochondrial protein per ml, and reduced cytochrome c was added to a final concentration of 1.5 mg/ml. The activity was determined as the rate of absorbance change at 550 nm.
Activity of the F1Fo-ATP synthase was determined as ATP hydrolysis rate, measured by inorganic phosphate release (Chen et al, 1956; Ackerman & Tzagoloff, 2007). Mitochondria (0.2 mg/ml) were suspended in assay buffer (50 mM Tris-sulfate, pH 8.5, 4 mM MgSO4) with or without 50 μM oligomycin. The reaction was started by addition of 20 mM ATP and stopped after 10, 20 or 30 min at 37°C by adding 10% TCA. Reference samples were treated in the same manner, except that the TCA was added prior to ATP. After a clarifying spin (5 min, 20,000 g), the amount of inorganic phosphate in the supernatant was determined by an ammonium molybdate colorimetric assay. For this, an equal volume of ammonium molybdate reagent (2% ascorbic acid, 0.5% ammonium molybdate, 1.2 N sulfuric acid) was added, the samples were incubated at 37°C for 2 h, and the absorbance at 820 nm was monitored. The amount of released phosphate was determined based on calibration curve with known phosphate concentrations.
In-gel activity staining
After separation of protein complexes by BN-PAGE, in-gel activity staining for F1Fo-ATP synthase was performed according to established protocols (Wittig & Schägger, 2005). In brief, a gel stripe was pre-incubated in 35 mM Tris, 220 mM glycine, pH 8.3, for 3 h, then transferred to F1Fo-ATP synthase buffer (35 mM Tris, 220 mM glycine, pH 8.3, 8 mM ATP, 14 mM MgSO4, 0.2% Pb(NO3)2) and incubated at room temperature.
IgG affinity chromatography and co-immunoprecipitation
Protein complexes were purified from the strains bearing Protein A tags on Ina22 and Ina17 by means of IgG affinity chromatography. For this, mitochondria were solubilized in solubilization buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 1% digitonin, 1 mM PMSF) for 20 min on ice, cleared by centrifugation, and IgG sepharose beads were added at a ratio of 5 μl beads per 1 mg of solubilized mitochondria. After 2 h of binding at 4°C, the beads were washed 10 times with 10 bed volumes of washing buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.6% digitonin), and the bound proteins were eluted by SDS gel loading buffer or (in case of stable isotope-labeled samples for mass-spectrometry analysis) cleaved overnight at 4°C with 0.4 mg/ml TEV protease. The eluates were analyzed by SDS-PAGE with subsequent Western blotting or mass spectrometry. Immunoprecipitation of Coa3 was performed according to the same procedure, using rabbit Coa3 antiserum coupled to Protein A sepharose (GE Healthcare) using dimethyl pimelimidate crosslinking.
Mass spectrometry and data analysis
After separation by SDS-PAGE, protein bands were analyzed by in-gel digestion followed by LC-MALDI-TOF/TOF mass spectrometry as described (Schulz et al, 2011). In brief, after reduction with DTT and carbamidomethylation with iodoacetamide, protein bands were in-gel digested with trypsin and the peptides separated and analyzed using LC-MALDI-TOF/TOF mass spectrometry. The data was used to identify proteins in the NCBInr protein database using the search program Mascot (matrix science, London).
For stable isotope labeling experiments, the ARG4 gene was replaced by a kanMX4 resistance cassette in wild-type YPH499 and Ina22ProtA strains (see Supplementary Table S1). The cells were grown on minimal medium containing stable isotope-labeled L-arginine (U-13C6, 99%; U-15N4, 99%) and L-lysine (U-13C6, 99%; U-15N2, 99%; Cambridge Isotope Laboratories). Mitochondrial isolation and complex purification using IgG beads were performed as described above. Following affinity purification, proteins of differentially SILAC-labeled Ina22 complexes were precipitated using acetone and resuspended in 8 M urea/50 mM NH4HCO3. Cysteine residues were reduced and subsequently alkylated using 5 mM TCEP for 30 min at 37°C and 50 mM iodoacetamide for 30 min at room temperature in the dark, respectively. Urea concentration was adjusted to 2 M with 50 mM NH4HCO3 and proteins tryptically digested overnight at 37°C. Peptides were dried in vacuo and reconstituted in 0.1% (v/v) TCA. LC/MS analyses of three independent replicates were performed using an UltiMate 3000 RSLCnano HPLC system (Thermo Scientific, Dreieich, Germany) and an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany). Peptides were separated on a 50 cm × 75 μm C18 reversed-phase nano LC column (Acclaim PepMap RSLC column; 2 μm particle size; 100 Å pore size; Thermo Scientific) using a binary solvent system consisting of 0.1% (v/v) formic acid (solvent A) and 50% (v/v) methanol/30% (v/v) acetonitrile in 0.1% formic acid (solvent B). After washing the LC column with 15% solvent B for 15 min, peptides were eluted applying a 135-min gradient ranging from 15 to 72% solvent B followed by an increase to 100% B within 15 min. The flow rate was 250 nl/min. MS survey scans (m/z 370–1,700) were acquired in the orbitrap at a resolution of 60,000 (at m/z 400) with automatic gain control (AGC) set to 5 × 105 ions and a maximum fill time of 500 ms. Simultaneously, up to five of the most intense multiply charged precursor ions were further fragmented by collision-induced dissociation in the linear ion trap at a normalized collision energy of 35%, an activation q of 0.25, an activation time of 30 ms, an AGC of 104 ions, and a maximum fill time of 100 ms. The dynamic exclusion time for previously fragmented precursor ions was 45 s.
Protein identification and relative quantification was performed using the MaxQuant software suite (version 1.3.0.5; Cox & Mann, 2008; Cox et al, 2011). MS/MS data were searched against the Saccharomyces Genome database (SGD; http://www.yeastgenome.org) applying the MaxQuant default settings (unless stated otherwise) including Arg10 and Lys8 as heavy labels. Protein identification was based on ≥ 1 unique peptide with a minimum length of 7 amino acids and a false discovery rate of < 1% on peptide and protein level. Protein abundance ratios (Ina22ProtA/WT) were calculated based on unique peptides and ≥ 1 ratio count. Ina22ProtA/WT ratios were logarithmized and the mean log10 protein ratio calculated across ≥ 2 replicates was plotted against the P-value determined for each protein in a one-sided t-test.
Pull-down analyses
Soluble intermembrane space domain of Ina22 (Ina22IMS, amino acids 64–216) was cloned in pProEx HTc vector with an N-terminal His6 tag, expressed in E. coli BL21 and purified using HisTrap columns (GE Healthcare). The purified protein was covalently bound to CNBr-activated Sepharose 4B (GE Healthcare), and the beads were stored at 4°C. For pull-down experiments using solubilized mitochondria, mitochondria were solubilized in Triton X-100 buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 mM PMSF), cleared by centrifugation, and the Ina22IMS sepharose beads were added at a ratio of 10 μl beads per 1 mg of solubilized mitochondria. After 2 h of binding at 4°C, the beads were washed five times with five bed volumes of solubilization buffer, and the bound proteins were eluted with SDS gel loading buffer. The eluates were analyzed by SDS-PAGE and Western blotting. For pull-down experiments using in vitro translated proteins, Ina22IMS-coupled sepharose beads were incubated with [35S]-labeled translation products in Triton X-100 buffer for 1 h at 4°C, washed 10 times with 10 bed volumes of the Triton X-100 buffer and eluted with one bed volume of SDS gel loading buffer. Bound proteins were detected by autoradiography.
Fluorescence microscopy
Fluorescence microscopy of yeast cell was performed as previously described (Alkhaja et al, 2012). Yeast cells transformed with pSU9-DHFR-GFP were grown overnight in selective SGG medium at 30°C. Cells were directly used for fluorescence microscopy. Images were collected by using a DeltaVision microscope (Olympus IX71; Applied Precision, Issaquah, WA, USA) and deconvoluted by using Softworx, version 3.5.1 (Great Falls, MT, USA).
Bioinformatic analysis
Mitochondrial targeting signal prediction was performed using the MitoProt Server (Claros & Vincens, 1996). Transmembrane spans of Ina22 and Ina17 were predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Coiled-coil domain in Ina22 and Ina17 were found using the Pfam database (Sonnhammer et al, 1997) and the COILS algorithm (Lupas et al, 1991).
Miscellaneous
Protein complexes were analyzed by Blue native electrophoresis (BN-PAGE) and Clear native electrophoresis (CN-PAGE) according to published procedures (Schägger & von Jagow, 1991; Wittig & Schägger, 2005). SDS gel electrophoresis (SDS-PAGE) and Western blotting were performed by standard protocols; signals were detected using fluorescently labeled secondary antibodies (LI-COR) and a FLA-9000 scanner (Fujifilm) or HRP-coupled secondary antibodies and enhanced chemiluminescence system (GE Healthcare). No image processing, other than cropping, scaling and contrast adjustment using Adobe Photoshop CS3 and Adobe Illustrator CS3, was applied. All quantitative data are presented as mean ± standard error of the mean.
Acknowledgments
We thank A. Chernev, M. Köppelmann, O. Bernhard, K. Lobenwein and B. Knapp for technical support, and Dr. S. Dennerlein for discussion. Antibodies to Cbp3 and Cbp4 were a gift from Dr. M. Ott. Work in the authors' laboratories was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746 (M.v.d.L) & 860 (P.R.), the Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS; M.v.d.L and B.W.), the Ph.D. program “Molecular Biology” – International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB; DFG grant GSC 226/1) at the Georg August University Göttingen, and the Max-Planck-Society (P.R.).
Author contributions
OL, NN, SO, BS, KvdM, MD performed the experiments. OL, NN, SO, BS, MD, BW, MvdL and PR analyzed the data. PR and MvdL led the project. OL, MvdL and PR wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary informationfor this article is available online: http://emboj.embopress.org
References
- Ackerman S, Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci USA. 1990;87:4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman S, Tzagoloff A. Methods to determine the status of mitochondrial ATP synthase assembly. In: Leister D, Herrmann J, editors. Mitochondria. Totowa, NY: Humana Press; 2007. pp. 363–377. (eds) [DOI] [PubMed] [Google Scholar]
- Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, Schliebs W, Riedel D, Urlaub H, Jakobs S, Deckers M. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arselin G, Giraud M-F, Dautant A, Vaillier J, Brethes D, Coulary-Salin B, Schaeffer J, Velours J. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur J Biochem. 2003;270:1875–1884. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Capaldi R, Aggeler R. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem Sci. 2002;27:154–160. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Toribara T, Warner H. Microdetermination of phosphorus. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- Chen Y-C, Taylor EB, Dephoure N, Heo J-M, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meirleir L, Seneca S, Lissens W, De Clercq I, Eyskens F, Gerlo E, Smet J, Van Coster R. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41:120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenish RJ, Prescott M, Roucou X, Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim Biophys Acta. 2000;1458:428–442. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dudkina N, Sunderhaus S, Braun H, Boekema E. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580:3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- Fernández-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim Biophys Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Fillingame RH, Angevine CM, Dmitriev OY. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 2003;555:29–34. doi: 10.1016/s0014-5793(03)01101-3. [DOI] [PubMed] [Google Scholar]
- Fox TD. Mitochondrial protein synthesis, import, and assembly. Genetics. 2012;192:1203–1234. doi: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert M, Schrempp SG, Mehnert CS, Heißwolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, Hunte C, Meyer HE, Haferkamp I, Guiard B, Warscheid B, Pfanner N, van der Laan M. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol. 2012;197:595–604. doi: 10.1083/jcb.201110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gruschke S, Kehrein K, Römpler K, Gröne K, Israel L, Imhof A, Herrmann JM, Ott M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol. 2011;193:1101–1114. doi: 10.1083/jcb.201103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Römpler K, Hildenbeutel M, Kehrein K, Kühl I, Bonnefoy N, Ott M. The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria. J Cell Biol. 2012;199:137–150. doi: 10.1083/jcb.201206040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, Giaever G, Costanzo M, Troyanskaya OG, Caudy AA. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- Kehrein K, Bonnefoy N, Ott M. Mitochondrial protein synthesis: efficiency and accuracy. Antioxid Redox Signal. 2013;19:1928–1939. doi: 10.1089/ars.2012.4896. [DOI] [PubMed] [Google Scholar]
- Kucharczyk R, Zick M, Bietenhader M, Rak M, Couplan E, Blondel M, Caubet S-D, di Rago J-P. Mitochondrial ATP synthase disorders: molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793:186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F1-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- Ludlam A, Brunzelle J, Pribyl T, Xu X, Gatti DL, Ackerman SH. Chaperones of F1-ATPase. J Biol Chem. 2009;284:17138–17146. doi: 10.1074/jbc.M109.002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mayr JA, Havlícková V, Zimmermann F, Magler I, Kaplanová V, Jesina P, Pecinová A, Nusková H, Koch J, Sperl W, Houstek J. Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 epsilon subunit. Hum Mol Genet. 2010;19:3430–3439. doi: 10.1093/hmg/ddq254. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, Kinosita K. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Osman C, Wilmes C, Tatsuta T, Langer T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell. 2007;18:627–635. doi: 10.1091/mbc.E06-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767–775. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Pagliarani A, Nesci S, Ventrella V. Modifiers of the oligomycin sensitivity of the mitochondrial F1Fo-ATPase. Mitochondrion. 2013;13:312–319. doi: 10.1016/j.mito.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brèthes D, di Rago J-P, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pícková A, Potocký M, Houstek J. Assembly factors of F1Fo-ATP synthase across genomes. Proteins. 2005;59:393–402. doi: 10.1002/prot.20452. [DOI] [PubMed] [Google Scholar]
- Prescott M, Bush NC, Nagley P, Devenish RJ. Properties of yeast cells depleted of the OSCP subunit of mitochondrial ATP synthase by regulated expression of the ATP5 gene. Biochem Mol Biol Int. 1994;34:789–799. [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, Zhang J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2012;2:1399–1410. doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Tzagoloff A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci U S A. 2009;106:18509–18514. doi: 10.1073/pnas.0910351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Zeng X, Brière J-J, Tzagoloff A. Assembly of Fo in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1793:108–116. doi: 10.1016/j.bbamcr.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 2011;30:920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Zahedi R. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol. 2011;195:643–656. doi: 10.1083/jcb.201105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi RP, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Fox JL, Winge DR. Biogenesis of the cytochrome bc1 complex and role of assembly factors. Biochim Biophys Acta. 2012;1817:276–286. doi: 10.1016/j.bbabio.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E, Eddy S, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Liu J, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2011;1817:883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel R, Khayat M, Shalev SA, Horovitz Y, Mandel H, Hershkovitz E, Barghuti F, Shaag A, Saada A, Korman SH, Elpeleg O, Yatsiv I. TMEM70 mutations are a common cause of nuclear encoded ATP synthase assembly defect: further delineation of a new syndrome. J Med Genet. 2011;48:177–182. doi: 10.1136/jmg.2010.084608. [DOI] [PubMed] [Google Scholar]
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart R. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraco A, Verrigni D, Rizza T, Meschini MC, Vazquez-Memije ME, Martinelli D, Bianchi M, Piemonte F, Dionisi-Vici C, Santorelli FM, Bertini E, Carrozzo R. TMEM70: a mutational hot spot in nuclear ATP synthase deficiency with a pivotal role in complex V biogenesis. Neurogenetics. 2012;13:375–386. doi: 10.1007/s10048-012-0343-8. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Barrientos A, Neupert W, Herrmann JM. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J Biol Chem. 2004;279:19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- Von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- Vukotic M, Oeljeklaus S, Wiese S, Vögtle FN, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Wagner K, Rehling P, Sanjuán Szklarz LK, Taylor RD, Pfanner N, van der Laan M. Mitochondrial F1Fo-ATP synthase: the small subunits e and g associate with monomeric complexes to trigger dimerization. J Mol Biol. 2009;392:855–861. doi: 10.1016/j.jmb.2009.07.059. [DOI] [PubMed] [Google Scholar]
- Wagner K, Perschil I, Fichter CD, van der Laan M. Stepwise assembly of dimeric F1Fo-ATP synthase in mitochondria involves the small Fo-subunits k and i. Mol Biol Cell. 2010;21:1494–1504. doi: 10.1091/mbc.E09-12-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Wang ZG, Sheluho D, Gatti DL, Ackerman SH. The alpha-subunit of the mitochondrial F1 ATPase interacts directly with the assembly factor Atp12p. EMBO J. 2000;19:1486–1493. doi: 10.1093/emboj/19.7.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Senior AE. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- Westermann B, Herrmann JM, Neupert W. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 2001;65:429–438. doi: 10.1016/s0091-679x(01)65025-8. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Rehling P. Import of precursor proteins into isolated yeast mitochondria. Methods Mol Biol. 2006;313:373–383. doi: 10.1385/1-59259-958-3:373. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- Zeng X, Neupert W, Tzagoloff A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol Biol Cell. 2007;18:617–626. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.