Abstract
With eukaryotic genome replication, incomplete telomere synthesis results in chromosome shortening and eventual compromise of genome stability. Telomerase counteracts this terminal sequence loss by synthesizing telomeric repeats through repeated cycles of reverse transcription of its internal RNA template. Using human telomerase domain-complementation assays for telomerase reverse transcriptase protein (TERT) and RNA in combination with the first direct footprinting assay for telomerase association with bound DNA, we resolve mechanisms by which TERT domains and RNA motifs direct repeat synthesis. Surprisingly, we find that product-template hybrid is sensed in a length- and sequence-dependent manner to set the template 5′ boundary. We demonstrate that the TERT N-terminal (TEN) domain determines active-site use of the atypically short primer-template hybrid necessary for telomeric-repeat synthesis. Also against expectation, we show that the remainder of TERT (the TERT ring) supports functional recognition and physical protection of single-stranded DNA adjacent to the template hybrid. These findings establish unprecedented polymerase recognition specificities for DNA-RNA hybrid and single-stranded DNA and suggest a new perspective on the mechanisms of telomerase specialization for telomeric-repeat synthesis.
Keywords: primer-template hybrid, reverse transcriptase, single-stranded DNA, telomerase RNP, telomeric repeats
Introduction
The characteristic telomeric DNA repeats that cap eukaryotic linear chromosome ends are synthesized de novo by telomerase (Blackburn et al, 2006). Telomerase functions as a reverse transcriptase (RT), with its active site contained in the protein subunit TERT and the template for DNA synthesis contained within the TERT-associated telomerase RNA (TER). Telomerase is a promising target for therapies to treat aging-related diseases and cancer. Human gene mutations that reduce telomerase function cause aplastic anemia, pulmonary fibrosis, the bone marrow failure syndrome dyskeratosis congenita, and other tissue renewal failures (Armanios & Blackburn, 2012). Conversely, aberrant telomerase activation contributes to the progression and maintenance of tumor cells (Shay & Wright, 2011). Despite clear relevance for disease therapies, no small-molecule activators or inhibitors of human telomerase have been found that effectively modulate telomerase function in clinical context. Efforts to manipulate telomerase function are crippled by limited insight regarding the structural basis for telomerase specializations that distinguish its activity from better-characterized mobile-element RT enzymes.
Distinct from other RTs, telomerase product DNA is single-stranded rather than a more thermodynamically stable DNA-RNA duplex (Collins, 2011). After primer elongation across the internal TER template to the template 5′ boundary, DNA product is released entirely from template base pairing. Dissociation of the product-template duplex is generally considered rate-limiting for enzyme activity in vitro (Podlevsky & Chen, 2012). Remarkably, most telomerase enzymes can realign the released product 3′ end with the template 3′ end, forming a limited length of template hybrid that can re-engage within the active site. Product DNA retention during this template repositioning gives rise to telomerase repeat addition processivity (RAP). The RAP quantified for purified telomerase enzymes in vitro is likely to be regulated by coordination with other telomere replication factors in vivo, but the general physiological significance of a telomerase mechanism for RAP is supported by measurements of short telomere extension in yeast (Chang et al, 2007) and telomere length maintenance in human cells (Zhao et al, 2011). Consistent with these observations of RAP in biological context, a chemical inhibitor of human telomerase RAP induces telomere shortening (Pascolo et al, 2002), some disease-associated human TERT mutations primarily affect RAP (Robart & Collins, 2010; Alder et al, 2011), and engineered yeast and human TERTs with decreased RAP do not support telomere maintenance (Wyatt et al, 2010). Telomerase RAP implies atypical polymerase specificities of primer recognition that can discriminate a varying length of primer-template hybrid and also retain single-stranded DNA without base pairing to the template.
The telomerase protein-nucleic acid interactions that confer specialized template and primer use derive at least in part from TERT-specific domains not shared with other RTs (Blackburn & Collins, 2011; Podlevsky & Chen, 2012). TERT has four regions considered to be separate domains: the telomerase-essential N-terminal (TEN) domain, TERT RNA-binding domain (TRBD), RT domain, and C-terminal extension (CTE). While vertebrate TERTs share an unusually long linker between the TEN domain and TRBD, TERTs from some species lack this linker and/or a discernable TEN domain. Unique features of the telomerase catalytic cycle depend on TERT-specific protein motifs and motifs within TER. The vertebrate TER secondary structure established by phylogenetic comparison and supported by nuclease and chemical probing, mutagenesis, and high-resolution structure (Chen et al, 2000; Podlevsky et al, 2008; Zhang et al, 2011; Egan & Collins, 2012) includes the conserved template-adjacent pseudoknot (PK) and physically distant conserved regions 4 and 5 (CR4/5; Fig 1A), which together are necessary and sufficient for activity reconstitution in vitro. CR4/5 provides the major affinity of vertebrate TER-TERT interaction and stimulates catalytic activity (Mitchell & Collins, 2000). The telomerase PK has less clear function but known structure, with base triples that create a stably folded triple-helix (Theimer et al, 2005). Sequence changes in the extended PK of human TER (hTR), including disease-linked mutations, result in altered catalytic activity (Ly et al, 2003; Chen & Greider, 2005). Resolution of how the PK motif contributes to enzyme function has been complicated by an inability of mutagenesis-based interrogation to distinguish direct PK loss-of-function from indirect consequences of PK misfolding on positioning of the adjacent TER template.
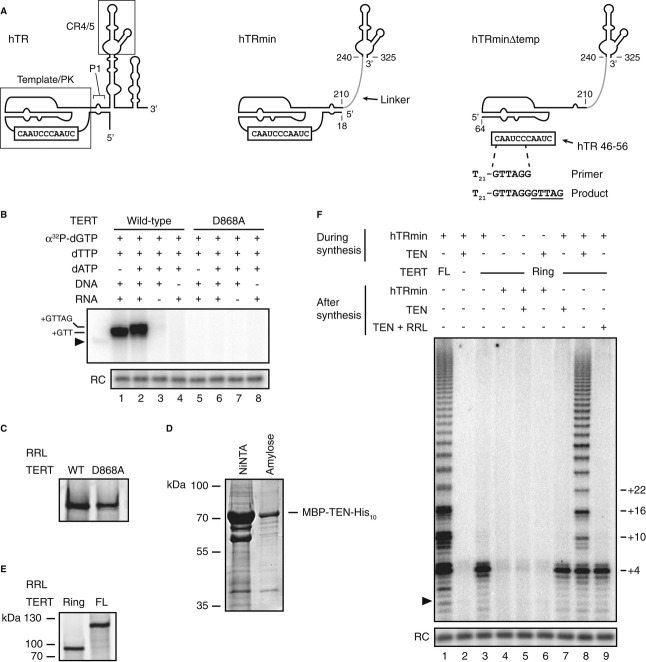
Figure 1. Human telomerase reconstituted with hTRmin supports activity on trans-templates and by trans-complementation of the TEN domain.
- Secondary structures of hTR, hTRmin, and hTRmin△temp with a template oligonucleotide and aligned DNA primer. Nucleotide addition (underlined) extends the primer to form product.
- Activity of wild-type or D868A TERT RNP reconstituted in RRL with hTRmin△temp and assayed with or without hTR 46–56 template, T21-GTTAGG primer, and the indicated nucleotide substrates. In this and subsequent assay panels, an end-labeled DNA recovery control (RC) was added before product precipitation and unextended primer was 5′ end-labeled and run as a size marker (▸).
- SDS–PAGE analysis of TERT expression in RRL by labeling with 35S-methionine.
- Coomassie-stained SDS–PAGE gel of the bacterially expressed TEN domain after initial partial purification on NiNTA resin and further purification on amylose resin.
- SDS–PAGE analysis of TERT ring or full-length TERT expression in RRL by labeling with 35S-methionine.
- Activity of full-length TERT RNP or TERT ring RNP reconstituted in RRL with hTRmin and assayed with (TTAGGG)3 primer. Purified bacterially expressed TEN domain or TEN domain pre-incubated in RRL for 3.5 h (RRL + TEN) and hTRmin were added before or after TERT ring synthesis as indicated.
Telomerase primer binding and extension can be greatly stimulated by a single-stranded region of primer 5′ of the primer-template hybrid (Morin, 1991; Collins & Greider, 1993; Lee & Blackburn, 1993). This enhancement is proposed to reflect the presence of a telomerase “anchor site”, which based on activity and primer dissociation assays for the human enzyme would engage a length of about 2 telomeric repeats of single-stranded DNA (Morin, 1991; Wallweber et al, 2003). An anchor-site groove has been proposed in the atomic resolution structure of the ciliate Tetrahymena thermophila TERT TEN domain (Jacobs et al, 2006). In support of anchor-site function for the TEN domain, some TEN domain sequence substitutions alter primer Km or RAP (Moriarty et al, 2004; Romi et al, 2007; Zaug et al, 2008). Also, single-stranded DNA cross-links to the protein linker between the TEN domain and TRBD (Romi et al, 2007) or potentially the TEN domain itself (Lue, 2005; Jacobs et al, 2006). The human TERT TRBD, RT, and CTE domains fold together to form a ring encircling the active site (Gillis et al, 2008). RNP assembled in cells with this structural core of TERT (here described as the TERT ring) does not stably bind primer and supports the synthesis of only a single telomeric repeat, but if it is coexpressed with a physically separate TEN domain, the enzyme regains stable primer binding and RAP (Robart & Collins, 2011). These and other findings underlie the widely accepted model that the TEN domain mediates single-stranded DNA binding (Wyatt et al, 2010; Lewis & Wuttke, 2012; Podlevsky & Chen, 2012; Nandakumar & Cech, 2013). However, most assays of purified yeast, ciliate, and human TERT TEN domains show weak if any specific interaction with single-stranded DNA (Xia et al, 2000; O'Connor et al, 2005; Wyatt et al, 2007; Finger & Bryan, 2008; Sealey et al, 2010; Bairley et al, 2011), and truncation of the TEN domain from full-length TERT does not eliminate DNA binding (Wyatt et al, 2007; Finger & Bryan, 2008). Also, sequence substitutions of mapped and putative DNA contact sites in the TEN domain have surprisingly little impact, and sequence substitutions that reduce RAP can result from slower synthesis rather than a compromised stability of product binding (Romi et al, 2007; Eckert & Collins, 2012). Therefore, the structural specializations that underlie telomerase properties of DNA handling remain poorly understood.
Here, we develop and combine TERT and hTR functional complementation assays and a physical nuclease protection assay to establish how telomerase recognizes primer-template duplex and single-stranded DNA. By requiring the active site to elongate a primer-template duplex structurally unconnected from the rest of the RNP, we could distinguish direct from indirect roles of TERT domains and hTR motifs. We find that the telomerase active site inherently limits product synthesis at the template 5′ boundary even without template constraint by flanking RNA motifs. We show that active-site use of a short primer-template hybrid requires the TEN domain, while the TERT ring RNP functionally and physically interacts with single-stranded primer DNA. Our findings provide integrated support for a new structural understanding of telomerase nucleic acid handling, including telomerase specialization for repeat synthesis by TEN domain-dependent recognition of the short initial primer-template hybrid and active-site discrimination of the sequence and length of product-template hybrid. These insights facilitate future development of small-molecule telomerase inhibition therapies.
Results
TERT active-site use of a physically autonomous primer-template hybrid
To gain new insights about primer and template handling by human telomerase, we sought to develop approaches that are not dependent on sequence substitution. We built upon the original trans-template reconstitution for T. thermophila TERT and TER (Miller & Collins, 2002) and a recently reported human telomerase trans-template reconstitution used to infer template translocation by active-site release of product-template hybrid (Qi et al, 2012). Telomerase RNPs with 3xFLAG-tagged TERT were assembled by transient transfection of human cells or using rabbit reticulocyte lysate (RRL) expression conditions that stabilize TERT by coassembly with hTR during protein synthesis (see Materials and Methods). RRL reconstitutions used hTRmin, which connects the PK and CR4/5 with a short linker (Fig 1A). Enzyme reconstituted with hTRmin was comparable in processivity to enzyme reconstituted with full-length hTR or the two-fragment combination of separated template/PK and CR4/5 fragments (Supplementary Fig S1). Activity reconstitution was better with hTRmin than with the full-length hTR prone to H/ACA domain misfolding (Supplementary Fig S1), but unlike the two-fragment system, hTRmin allows the high-affinity CR4/5-TERT interaction to assemble even a truncated template/PK sequence into RNP.
To assay telomerase active-site use of a template physically autonomous from the PK, we reconstituted a template-less version of full-length hTR RNP (hTR△temp) in vivo using telomerase-negative U2OS cells (Supplementary Fig S2) and a template-less version of hTRmin (hTRmin△temp) in vitro, then supplied TERT RNPs with a trans-template RNA oligonucleotide (hTR 46-56; Fig 1A). Both hTR△temp and hTRmin△temp reconstituted robust single-repeat synthesis activity appropriately dependent on the presence of template-cognate dNTPs (Fig 1B and Supplementary Fig S2B, lanes 1–2). As additional controls, assays lacking either RNA template or DNA primer did not give radiolabeled product (Fig 1B and Supplementary Fig S2B, lanes 3–4). To eliminate the possibility of contaminating co-purified polymerase activities, we assayed RNPs assembled by TERT with the RT motif C substitution D868A, which disrupts catalytic metal ion coordination (Weinrich et al, 1997). As expected, D868A TERT did not support trans-template product synthesis (Fig 1B and Supplementary Fig S2B, lanes 5–8) despite RRL expression comparable to wild-type TERT (Fig 1C). Notably, these reconstitutions used the native hTR template sequence instead of a previously assayed heterologous-sequence template (Qi et al, 2012). The results above firmly establish that a human telomerase RNP containing only hTR PK and CR4/5 can accurately position a physically autonomous primer-template hybrid in the active site.
For these and other assays described below, we used primer with a single-stranded region of poly-thymidine (T), typically in the 27-nt primer T21-GTTAGG (Fig 1A, right). The use of a 5′ T-tract obliges base pairing of the primer 3′ end with the template, rather than internal regions, and it precludes primer multimerization by guanosine quadruplex formation. Previous studies establish that in the single-stranded region of a primer, non-telomeric sequence increases primer extension comparably to or better than telomeric-repeat sequence (Morin, 1991; Lee & Blackburn, 1993). Also, 5′ non-telomeric and telomeric-repeat sequence extensions both dramatically stabilize primer DNA physical association with human telomerase (Wallweber et al, 2003). A ˜12-nt length of single-stranded DNA was sufficient for maximum stimulation of human telomerase catalytic activity or primer binding (Morin, 1991; Wallweber et al, 2003). To validate the use of a non-telomeric single-stranded primer 5′ region in our assays, we confirmed that a 5′ T-tract enhanced primer use comparably to telomeric-repeat sequence with both TERT assembled with hTRmin in RRL and TERT assembled with full-length hTR as the cellular holoenzyme (Supplementary Fig S3). We also recapitulated major findings below by testing telomeric-repeat as well as non-telomeric 5′ extensions from a primer-template hybrid.
Human TERT TEN domain function requires co-folding with the TERT ring and hTR
Sequence substitutions of the TEN domain can alter features of template use (Jurczyluk et al, 2010; Wyatt et al, 2010; Bairley et al, 2011; Eckert & Collins, 2012). To investigate how the TEN domain affects active-site handling of primer and template without the caveats of sequence substitution, we built from the TEN domain trans-complementation developed for human telomerase holoenzyme reconstitution in vivo (Robart & Collins, 2011). We bacterially expressed and purified the human TERT TEN domain (TERT amino acids 1-325) in N-terminal fusion to maltose-binding protein (MBP) and C-terminal fusion to a 10-histidine tag (Fig 1D). The human TERT ring (amino acids 326-1132) was expressed in RRL under conditions parallel to full-length TERT (Fig 1E). Paralleling reconstitutions in vivo (Robart & Collins, 2011), the TERT ring RNP reconstituted in vitro supported single-repeat synthesis (Fig 1F, lane 3) and could be complemented by a physically separate TEN domain to attain the RAP of full-length TERT RNP (lanes 8 and 1, respectively).
Curiously, trans-complementation by the purified TEN domain was robust enough to detect by primer extension only if the TEN domain and hTRmin were added to the TERT ring RRL expression reaction before TERT ring synthesis (Fig 1F, lanes 2–8). This requirement could not be bypassed by incubating the TEN domain in RRL in parallel with separate TERT ring synthesis (Fig 1F, lane 9), suggesting that co-folding of the TEN domain, hTR, and TERT ring creates the active conformation of the TEN domain. Co-folding was stable rather than transient, as judged by the enrichment of high-RAP activity upon repurification of the TEN domain after RNP assembly (Supplementary Fig S1C). Removal of the MBP tag from the TEN domain greatly inhibited TEN domain trans-complementation (R.A. Wu & K. Collins, unpublished observations). We note that a large fraction of the isolated TEN domain may have inactive conformation, based on the large stoichiometric excess over TERT ring that is required for activity reconstitution. Thus, studies with an isolated recombinant human TEN domain may not fully recapitulate its important functional properties.
Template-flanking motifs and the TEN domain stimulate use of a primer-template hybrid
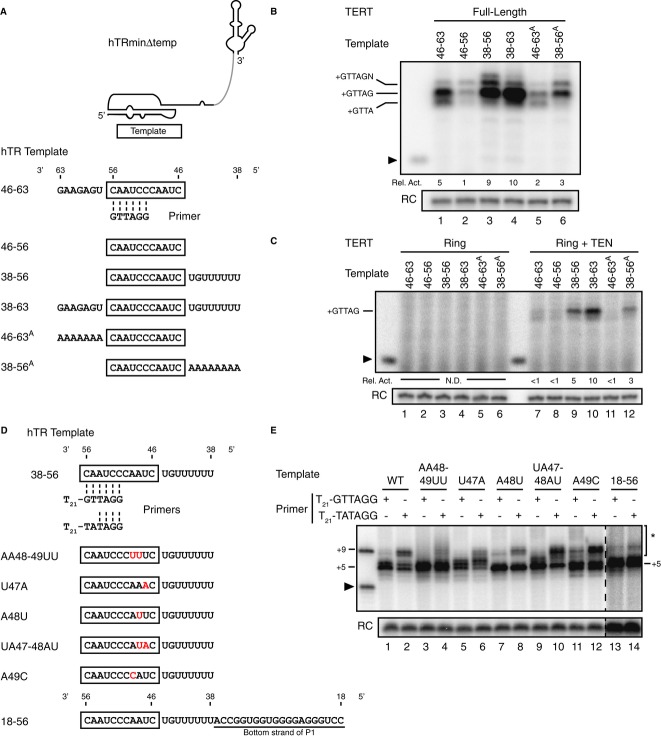
It is surprisingly unknown how template-flanking single-stranded RNA regions influence template use in vertebrate TERs. A T. thermophila TER sequence motif 3′ of the template strongly stimulates trans-template use only in cis-linkage to the template (Miller & Collins, 2002). Also, a T. thermophila TER sequence motif 5′ of the template enforces the cis-linked template 5′ boundary by high-affinity TERT interaction (Lai et al, 2002). Recent mutagenesis and single-molecule FRET studies suggest that these template-flanking RNA elements may define the intervening region as template by storing the strain from template stretching and compression (Berman et al, 2011). However, the 5′ and 3′ template-flanking regions of vertebrate TERs do not have sequence conservation that parallels ciliate TERs or even conservation of a template-flanking stem such as mediates template boundary definition in yeast TERs (Tzfati et al, 2000; Lai et al, 2002; Qi et al, 2013). To investigate how cis-linkage to flanking sequences influences human telomerase template use, we assayed hTRmin△temp RNP for activity with template oligonucleotides extended by the native context of hTR 5′ and/or 3′ template-flanking sequences (Fig 2A).
Figure 2. Template-flanking RNA sequence and the TEN domain stimulate active-site elongation of an autonomous primer-template hybrid.
- Schematic of trans-template RNA oligonucleotides containing template-flanking single-stranded regions with aligned DNA primer GTTAGG.
- Activity from full-length TERT RNP reconstituted in RRL with hTRmin△temp and assayed with the indicated templates and GTTAGG primer. The 6-nt primer was used to accentuate product size differences. For relative activity (Rel. Act.), product DNA intensities were first subtracted for background signal, then normalized to activity from the hTR 38-63 template.
- Activity from TERT ring RNP or TERT ring RNP with trans-complementing TEN domain reconstituted in RRL with hTRmin△temp and assayed with the indicated templates and GTTAGG primer. Product DNA intensities were first subtracted for background signal, then normalized to activity from the hTR 38–63 template. N.D. is not determined.
- Schematic of sequence-substituted template RNA oligonucleotides containing 5′ template-flanking sequences and aligned primers T21-GTTAGG and T21-TATAGG.
- Activity of full-length TERT RNP reconstituted in RRL with hTRminΔtemp and assayed with various templates, T21-GTTAGG or T21-TATAGG primer, and dATP, dCTP, dTTP and radiolabeled dGTP. Lanes 13–14 are shown at a higher exposure. Products of template 5′ boundary bypass are indicated by an asterisk.
Repeat synthesis activity was increased fivefold to tenfold by the presence of a 3′-flanking (hTR 46–63) or 5′-flanking (hTR 38-56) hTR template region (Fig 2B, lanes 1–3) or both (hTR 38–63, lane 4). This effect was highly reproducible in independent triplicate assays. We next tested whether stimulation of template use by hTR template-flanking regions was sequence-dependent, as is the case for T. thermophila TER (Miller & Collins, 2002). Substitution of the wild-type hTR 5′ or 3′ template-flanking sequences for an equal length of poly-adenosine weakened but did not abolish their stimulatory effects (Fig 2B, compare lane 1 to 5 and lane 3 to 6). The sequence of the template 3′-flanking region also affected the fidelity of template copying: The presence of the wild-type sequence reduced non-templated nucleotide addition after complete repeat synthesis, while the non-native poly-adenosine tract reduced complete template copying (Fig 2B, compare product lengths in lanes 1, 2, and 5). Curiously, the presence of template-flanking 5′ sequence did not provide additional template: Synthesis still predominantly halted at the correct template 5′ boundary (Fig 2B, compare product lengths in lanes 2, 3, and 6).
We also compared TERT ring RNP activity on minimal versus extended templates. The hTRmin△temp TERT ring RNP had negligible activity on any trans-template, even if template-flanking 5′ and 3′ regions were both present (Fig 2C, lanes 1–6). However, the same TERT ring RNP complemented with a separate TEN domain could use a trans-template (Fig 2C, lanes 7–12). The 3′ template-flanking sequence had less stimulatory effect on the RNP with trans-complementing TEN domain than full-length TERT (compare Fig 2C, lanes 7 and 8 to Fig 2B, lanes 1 and 2), detected reproducibly across independent assays in triplicate. This finding suggests that the hTR 3′ template-flanking region is recognized by a coordination of the physically linked TEN domain and TERT ring. Overall, these results indicate that the TEN domain greatly stimulates active-site engagement of a primer-template hybrid even when this hybrid has only RNA template sequence (hTR 46-63) and a fully base-paired 6-nt DNA (Fig 2A).
Template 5′ boundary definition inherent to the product-template hybrid
The limited change in product length with 5′-extended trans-template RNAs was unexpected, because it implied that template 5′ boundary definition could be inherent to the product-template hybrid rather than steric constraints provided by template-flanking RNA structure or RNA-protein interaction (Tzfati et al, 2000; Lai et al, 2002; Berman et al, 2011; Blackburn & Collins, 2011). To test this hypothesis, we performed activity assays with trans-template RNAs harboring sequence substitutions in 5′ template positions 47–49 (Fig 2D). We tested each template paired to either primer with a fully template-complementary 3′ end (T21-GTTAGG) or primer with a template 3′-end mismatch (T21-TATAGG) that would reduce the length of primer-template hybrid to four base pairs (as illustrated in Fig 2D).
A shorter initial hybrid length promoted longer product synthesis (Fig 2E, lanes 1–2). Also, given the same primer-template base pairing, some template substitutions also led to longer product synthesis (Fig 2E, compare lanes 1 and 9). Combinations of shorter primer-template hybrid length and template sequence substitution decreased the fidelity of synthesis halt at the template 5′ boundary to a variable extent, suggesting that product-template hybrid length and sequence each contribute to discrimination of the appropriate template boundary. Extension of the 5′ template-flanking region to include the bottom strand of P1 appeared to reduce but not eliminate template 5′ boundary bypass induced by shorter initial primer-template hybrid length (Fig 2E, compare lanes 1 and 2 to lanes 13 and 14). Taken together, our findings support the conclusion that for human telomerase, the precision of repeat synthesis derives in part from a high specificity of discrimination for the length and sequence of product-template hybrid. Discrimination inherent to features of the product-template hybrid can explain the absence of template-flanking sequence conservation among vertebrate TERs and uncovers an active-site recognition of product DNA-RNA hybrid that is unprecedented in other polymerases.
A PK stem is required for productive coupling of the TEN domain and TERT ring and for TERT ring use of primer-template hybrid
The activity inhibition observed in previous PK mutagenesis studies could arise from cis-linked template mis-positioning in the global hTR fold. To discriminate this indirect loss-of-function from a direct PK role such as primer alignment or TEN domain positioning (Qiao & Cech, 2008; Robart & Collins, 2011), we combined the trans-template assay with TEN domain trans-complementation. The entire PK or only the 5′ strand of the P2a.1/P2a stem extension was removed from hTRmin△temp (△PK or 91-end; Fig 3A). For comparison, we introduced substitutions that disrupt PK base triples (illustrated in Fig 3B) formed either in the P2b loop (U100C) or in the loop between P2a.1 and P3 (A172U), both of which are critical for enzyme activity (Theimer et al, 2005).
Figure 3. The hTR PK has distinct structural requirements for activity of the TERT ring RNP with or without trans-complementing TEN domain.
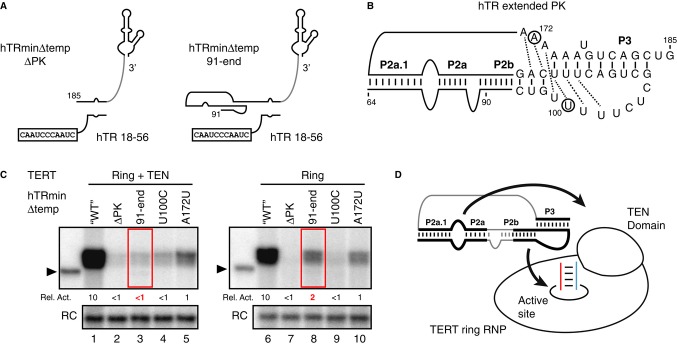
- Secondary structures of hTRmin with truncation of the entire PK or the P2a.1/P2a bottom strand (hTRmin△temp 91-end) and the trans-template RNA hTR 18–56.
- Sequence and secondary structure within the PK. Dotted lines indicate base triple pairing. Positions altered by mutagenesis are circled.
- Activity from TERT ring RNP with or without trans-complementing TEN domain reconstituted in RRL and assayed with template hTR 18–56 and T21-GTTAGG primer. Product DNA intensities were first subtracted for background signal, then normalized to activity from unsubstituted hTRmin temp (indicated as WT).
- The P2a.1/P2a extended stem influences functional coupling of the TEN domain and the TERT ring RNP, and the entire PK increases engagement of primer-template hybrid by the TERT ring RNP.
Without the PK, the TERT ring RNP complemented with a TEN domain showed greatly reduced but still detectable trans-template copying activity (Fig 3C, lane 2), which was reproducible in more than 3 independent assays. Removal of the P2a.1/P2a bottom strand decreased activity similarly to removal of the entire PK (Fig 3C, lane 3). Disruption of the conserved P2b loop triple-helix also strongly decreased activity, more so than disrupting a base triple formed by the P2a.1/P3 loop (Fig 3C, lanes 4 and 5). In comparison, for the TERT ring RNP alone, removing the PK decreased activity to a nearly undetectable level, as did disrupting the P2b loop triple-helix (Fig 3C, lanes 7 and 9). On the other hand, truncating the bottom strand of P2a.1/P2a had less impact on activity, comparable to disrupting a base triple of the P2a.1/P3 loop (Fig 3C, lanes 8 and 10). Thus, the presence of P2a.1/P2a was more critical for activity in the presence versus absence of TEN domain (Fig 3C, compare lanes 1 and 3 to lanes 6 and 8). Overall, these results suggest that the vertebrate-extended P2a.1/P2a influences coupling of the TEN domain and TERT ring and support important direct function of the PK in active-site engagement of primer-template hybrid (Fig 3D).
TERT RNP without TEN domain functionally senses single-stranded primer DNA
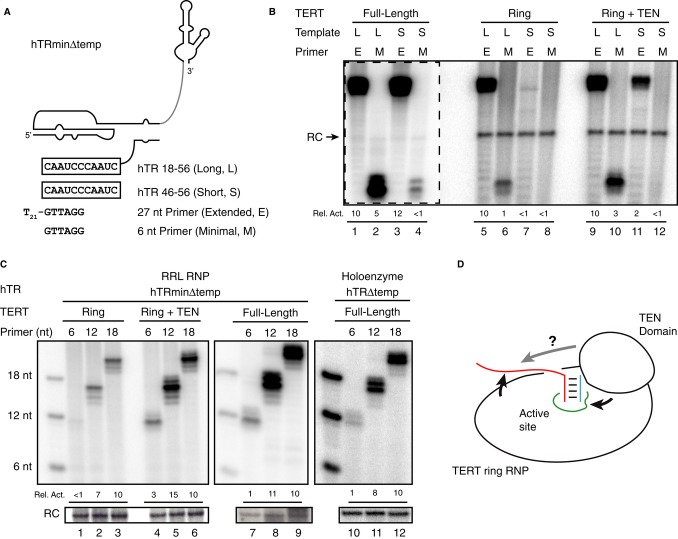
Current models of telomerase structure and catalytic cycle mechanism predict the TEN domain to be required for activity stimulation by single-stranded DNA 5′ of the primer-template hybrid. To test this expectation, we compared the activity of RRL-assembled RNPs containing full-length TERT, TERT ring or TERT ring and trans-complementing TEN domain using primers with or without a 5′ single-stranded extension from the primer-template hybrid (Fig 4A, E and M primers). We used each primer in combination with the shortest, 11-nt trans-template lacking any non-template sequence or a longer, 5′-extended trans-template RNA (Fig 4A, S and L templates).
Figure 4. The TEN domain stimulates elongation of an entirely template-paired primer.
- Secondary structure of hTRmin△temp with the longest template RNA hTR 18–56 (L) or the shortest template RNA hTR 46–56 (S) aligned with the 5′-extended primer T21-GTTAGG (E) or the minimal template-paired primer GTTAGG (M).
- Activity of full-length TERT RNP, TERT ring RNP, or TERT ring RNP with trans-complementing TEN domain reconstituted in RRL with hTRmin△temp and assayed with the indicated S or L template and M or E primer. All product intensities were first subtracted for background signal, then normalized to activity from the L template and E primer within each set of assays.
- Activity of TERT ring RNP, TERT ring RNP with trans-complementing TEN domain or full-length TERT RNP reconstituted in RRL with hTRmin△temp, or full-length TERT RNP reconstituted in U2OS cells with hTR△temp, each assayed with hTR 18–56 template and 6-nt (GTTAGG), 12-nt (T6-GTTAGG), or 18-nt (T12-GTTAGG) primer. Product intensities were first subtracted for background signal, then normalized to activity on the 18-nt primer within each set of assays. Unextended 5′ end-labeled primers are shown as size markers.
- The single-stranded region of DNA (DNA shown in red) stimulates activity of the TERT ring RNP, and the TEN domain stimulates activity through the primer-template hybrid (template shown in blue). Not resolved is whether the TEN domain also senses single-stranded DNA (gray arrow).
The full-length TERT RNP used the long or short template paired with either the minimal or extended primer (Fig 4B, lanes 1–4). An activity stimulation of more than tenfold required only extended primer or the longer template, with no additional activity increase from their combination (Fig 4B, compare lanes 2–4). The TERT ring RNP with trans-complementing TEN domain also showed activity stimulation by the presence of a single-stranded region of RNA or DNA, with an additional boost from their combination (Fig 4B, lanes 9–12). The TERT ring RNP alone was more dependent on the combination of single-stranded RNA and primer regions for robust activity (Fig 4B, lanes 5–8). Comparing enzymes with and without the TEN domain, the TEN domain stimulated activity not only with the extended primer but also with the minimal primer, which lacks a single-stranded DNA region for putative TEN domain interaction (Fig 4B, compare lanes 6 and 10). Even more surprisingly, the TERT ring RNP was stimulated by the presence of single-stranded DNA (Fig 4B, compare lane 6 to 5 and lane 8 to 7). This stimulation was highly reproducible across multiple independent assays. These results suggest that the TEN domain stimulates active-site use of a primer-template hybrid even without single-stranded DNA, and TERT ring RNP senses a single-stranded DNA extension from the primer-template hybrid.
We next explored the length dependence of TERT ring RNP activity stimulation by single-stranded DNA. Primers of 6, 12, or 18 nt (GTTAGG, T6-GTTAGG, T12-GTTAGG) progressively stimulated RRL-assembled TERT ring RNP activity on the hTR 18-56 trans-template (Fig 4C, lanes 1–3). This stimulation was comparable to or greater than single-stranded DNA stimulation of trans-template copying by the RRL-assembled hTRmin△temp full-length TERT RNP or TERT ring RNP with a trans-complementing TEN domain (Fig 4C, lanes 4–9) or the hTR△temp holoenzyme reconstituted in vivo (lanes 10–12). We extended these findings to the TERT ring RNP assembled in vivo on full-length hTR, which also showed stimulation by single-stranded DNA comparable to or greater than that of full-length TERT RNP assembled in vivo on full-length hTR, each assayed with primers containing a 5′ T-tract or telomeric-repeat extension from the primer-template hybrid (Supplementary Fig S4). These results provide strong evidence that the TERT ring RNP can functionally sense single-stranded DNA (Fig 4D).
TERT RNP without TEN domain physically protects single-stranded DNA
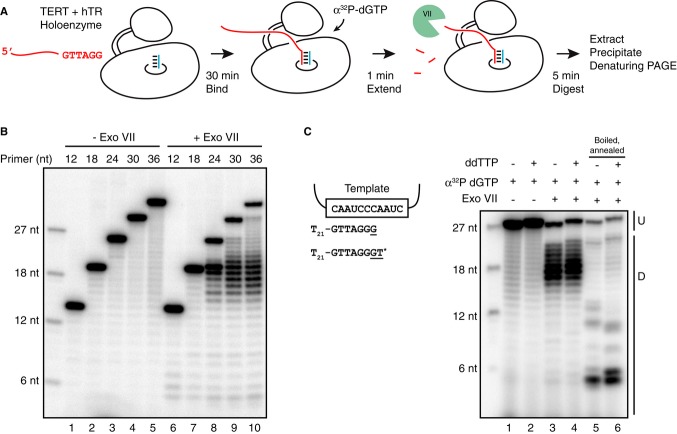
To parallel the functional assays above, we sought to test telomerase physical association with DNA. Because there is no previously reported benchmark for such an assay, we used holoenzyme with full-length hTR to develop a nuclease protection assay specific for active enzyme complexes (Fig 5A). First, the telomerase active site was used to label a productively bound DNA primer by the addition of a single radiolabeled dGTP (Fig 5B, lanes 1–5). After labeling, bound product was trimmed of accessible single-stranded DNA by brief digestion with exonuclease VII (ExoVII). The total length of protected product was then assessed by denaturing PAGE, with end-labeled DNA oligonucleotide markers and some undigested product DNA retained for size comparison (Fig 5B, lanes 6–10). Because the end-labeled oligonucleotides share the 5′ monophosphate of ExoVII products (Chase & Richardson, 1974), they are accurate migration standards. The full-length TERT holoenzyme protected product lengths of ˜18 nt, including both the product-template duplex and single-stranded region. Importantly, beyond 18 nt, the length range of DNA protection was not dependent on initial primer length (Fig 5B, lanes 8–10) and was consistent over a range of nuclease reaction times and amounts (R.A. Wu & K. Collins, unpublished observations). RRL-reconstituted full-length TERT hTRmin RNP also protected an ˜18-nt length of DNA (Supplementary Fig S5), indicating that holoenzyme proteins other than TERT do not associate stably with single-stranded product. The initial and protected product length both increased by 1 nt if dGTP and dideoxythymidine (ddTTP) were added in primer extension (Fig 5C, lanes 1–4), and a correspondingly 1-nt longer length of product-template duplex could be detected if product DNA was boiled and annealed with excess template RNA before ExoVII digestion (lanes 5–6).
Figure 5. A limited length of telomerase product DNA is protected from ExoVII digestion.
- Schematic of the ExoVII protection assay. Bound DNA is labeled by the addition of radiolabeled dGTP for 1 min, followed by treatment with ExoVII for 5 min. The protected, labeled DNA is extracted and resolved by denaturing PAGE.
- ExoVII protection assay using 12-, 18-, 24-, 30-, and 36-nt primers (Tn-GTTAGG with n = 6, 12, 18, 24, 30) with full-length TERT RNP reconstituted in 293T cells with full-length hTR. Unextended 5′ end-labeled 6-, 12-, 18-, and 27-nt oligonucleotides are shown as size markers.
- ExoVII protection assay using full-length TERT RNP reconstituted in 293T cells with full-length hTR and primer T21-GTTAGG extended with radiolabeled dGTP with or without ddTTP (T*). Product DNA was left enzyme-bound or boiled. Boiled samples were supplemented with 2.5 μM hTR 46–56 to re-form the template hybrid. In this and subsequent figures, undigested telomerase product DNA (U) and ExoVII-digested telomerase products (D) are indicated.
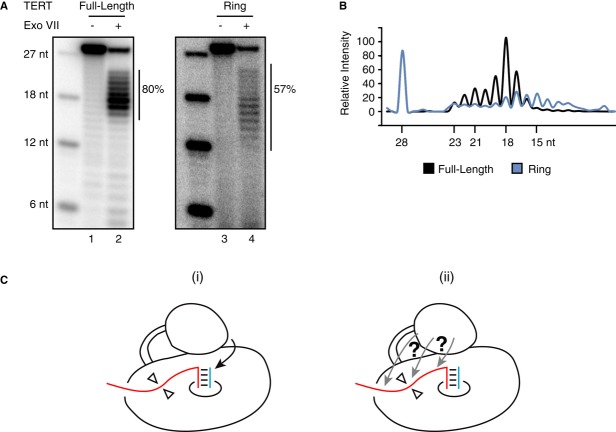
We next tested physical protection of product DNA by the TERT ring RNP. Remarkably, the TERT ring RNP reconstituted in vivo with full-length hTR protected a product length roughly comparable to the full-length TERT holoenzyme reconstituted in parallel (Fig 6A). Product protection by the TERT ring RNP appeared less quantitatively robust than its protection by full-length TERT RNP, based on the lower summed product intensity normalized to the amount of initial product before nuclease digestion (Fig 6A, percentage of initial product converted to protected product is indicated). This suggests a higher rate of product dissociation or a more dynamic enzyme-DNA interaction. In addition, the products protected by the TERT ring RNP appeared more evenly spread over a length distribution than the products protected by full-length TERT RNP (Fig 6A), with a size range quantified as 14–18 nt (Fig 6B). Together with the activity assays described above, the physical nuclease protection results suggest the surprising conclusion that single-stranded DNA threads from the product-template hybrid along a surface of the TERT ring RNP (Fig 6C, open triangles indicate an anchor site not necessarily localized to one surface of the TERT ring RNP; see Discussion). The TEN domain enhances placement of the initial primer-template hybrid in the active site (Fig 6C, left), with or without a direct contribution to the surface and/or stability of single-stranded DNA interaction (Fig 6C, right).
Figure 6. Exonuclease VII protection of product DNA resolves roles of the TERT ring RNP and TEN domain in DNA handling.
- ExoVII protection of elongated T21-GTTAGG using full-length TERT RNP or TERT ring RNP reconstituted in 293T cells with full-length hTR. Unextended 5′ end-labeled 6-, 12-, 18-, and 27-nt oligonucleotides are shown as size markers. The percentage of cleaved but protected product DNA is quantified relative to the total product amount before ExoVII digestion from a separate reaction shown in the adjacent lane.
- Protected product profiles for full-length TERT RNP and TERT ring RNP in the gel lanes from (A).
- Telomerase DNA handling includes TERT ring RNP interaction (⊳◃) with single-stranded DNA and TEN domain stabilization of primer-template hybrid in the active site (i). Unresolved is whether the TEN domain has any contribution to binding of single-stranded DNA, using an interaction site integrated with or separable from DNA interaction by the TERT ring RNP (ii).
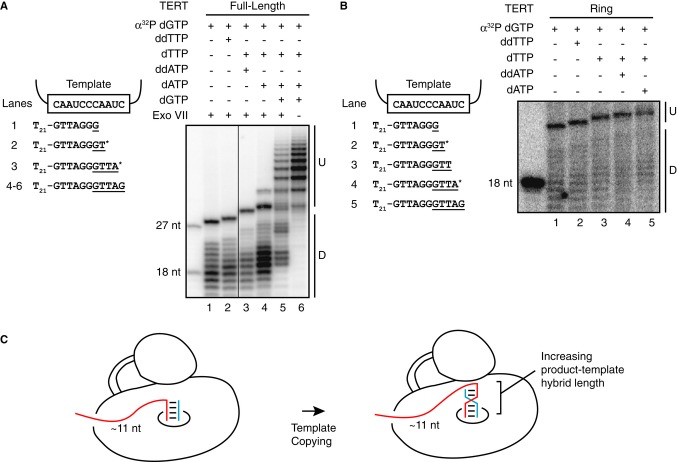
To explore how product DNA is protected during repeat synthesis, we performed the nuclease protection assay on products with 3′ ends extended to successive positions of the template using combinations of deoxy- and dideoxynucleotides (ddNTPs). In general, the protected products of full-length TERT holoenzyme increased in length with synthesis across the template (Fig 7A, lanes 1–4; Supplementary Fig S6). Roughly similar lengths of protection were observed when the primer contained 5′ telomeric sequence rather than a T-tract extension (Supplementary Fig S7), with the caveat that the 5′ edge of protection of telomeric-repeat product DNA is determined in part by sequence-differential ExoVII cleavage (Chase & Richardson, 1974). Paralleling the results for full-length TERT holoenzyme, protected products of the TERT ring RNP increased in length as synthesis proceeded across the template (Fig 7B). This comparison suggests that the TEN domain did not alter the register of telomerase interaction with protected product DNA during repeat synthesis.
Figure 7. Changes in ExoVII protection of product DNA during a telomerase catalytic cycle.
A, B At left, schematic of the extent of DNA primer elongation prior to ExoVII treatment. Nucleotides added by primer extension are underlined, with asterisks denoting ddNTPs. At right, ExoVII footprinting with full-length TERT RNP (A) or TERT ring RNP (B) reconstituted in 293T cells with full-length hTR. Processive repeat synthesis by full-length TERT RNP was supported by adding unlabeled dGTP (dGTP) and incubating for 10 min before ExoVII digestion. Unextended 5′ end-labeled 18- and/or 27-nt oligonucleotides are shown as size markers.
C Single-stranded DNA register does not change during active-site progression across the template, consistent with the model of a relatively constant length of single-stranded DNA (˜11 nt) and an increasing length of product-template hybrid.
Curiously, the register of the 5′ end of protected product DNA changed little with active-site progression across the template: Product elongation by 4 nt was accompanied by a 3-nt increase in protected product length (Fig 7A, lanes 1–4; Supplementary Fig S6). This finding suggests that during repeat synthesis, single-stranded DNA could largely retain its position relative to the active site while the product-template hybrid increases in length (Fig 7C). If repeat synthesis was allowed to proceed with RAP before ExoVII digestion using 10-min rather than the standard 1-min interval of primer extension (Fig 5A), protected product lengths paralleled those from single-repeat synthesis (Fig 7A, lanes 4–6). This result suggests that product DNA threads out of the RNP rather than forming a large loop with retained binding of the original single-stranded primer 5′ region. The minority population of longer protected DNA observed upon ExoVII digestion of high-RAP products (Fig 7A, lane 5) could reflect ExoVII-resistant guanosine quadruplex structures in enzyme-bound or released product or product elongation after ExoVII cleavage. Overall, these physical and functional assays support a new perspective on telomerase mechanism, including unique polymerase specificities of nucleic acid interaction that discriminate different states of primer-template hybrid as well as single-stranded DNA (Fig 8).
Figure 8. Multiple specificities of primer-template hybrid recognition provide a new perspective on the telomerase catalytic cycle.
The stop sign indicates product-template hybrid determination of the template 5′ boundary. See text for discussion.
Discussion
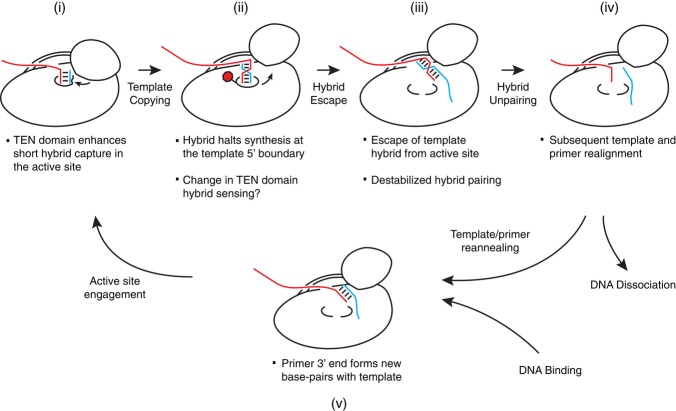
The telomerase catalytic cycle obliges a distinctive, dynamic positioning of different lengths of primer-template hybrid in the active site. Here, we describe new functional and physical assay methods that allowed us to define previously unknown nucleic acid recognition specificities of the TEN domain and TERT ring RNP for primer-template hybrid, template-flanking RNA sequences, and primer single-stranded DNA. Our findings provide integrated support for a model in which the TEN domain functions to enhance catalytic activity and confer RAP by increasing active-site use of primer-template hybrid, rather than by binding a 5′ single-stranded region of primer distant from the active site (Fig 8, state i). This accounts for why even single-repeat primers hybridized entirely with the hTR template are elongated with RAP yet still require the TEN domain for stable binding (Morin, 1991; Robart & Collins, 2011). Upon copying to the template 5′ end, additional synthesis is halted by features inherent to the native length and/or sequence of product-template hybrid (Fig 8, state ii). Curiously, although template-flanking RNA regions improve human telomerase trans-template use, they are not required for template 5′ boundary definition. Active-site release of the product-template hybrid as previously proposed (Qi et al, 2012) would facilitate strand separation (Fig 8, states iii-iv). The TEN domain then promotes active-site capture of the product 3′ end re-annealed at the template 3′ end (Fig 8, state v to i). The TEN domain may have evolved to reduce the minimum required base pairing between primer and template. This would account for why a TEN domain sequence substitution in yeast restricts the registers of primer-template alignment (Bairley et al, 2011) and why TEN domain sequence substitutions in yeast, ciliate, or human enzymes can be compromised for synthesis at only template positions that oblige a particularly short primer-template hybrid (Lue, 2005; Jurczyluk et al, 2010; Eckert & Collins, 2012).
Because the active site must release product-template hybrid to complete a catalytic cycle, the TEN domain may favor stable engagement of the primer-template hybrid only for lengths or sequences that do not include the template 5′ end. We suggest that functional coordination of the TEN domain with the changing primer-template hybrid could be mediated by the template-proximal PK stem, hTR P2a. Because the P2a stem has a particularly extended length in mammalian TERs (Podlevsky et al, 2008), the mammalian TEN domain may have stronger interaction with TER and/or more stable positioning relative to the active site than the TEN domain of other TERTs. Consistent with this hypothesis, the minimal recombinant human telomerase RNP has more robust TEN domain trans-complementation and higher RAP than does the T. thermophila minimal RNP, which requires additional holoenzyme proteins for TEN domain trans-complementation or high RAP (Eckert & Collins, 2012).
Ciliate and yeast telomerases establish the template 5′ boundary relative to a template-flanking RNA structure or protein-RNA interactions (Tzfati et al, 2000; Lai et al, 2002; Berman et al, 2011; Blackburn & Collins, 2011). For human telomerase, disruption of the hTR P1 stem can reduce template boundary fidelity (Chen & Greider, 2003; Moriarty et al, 2005), as does physical discontinuity of the TEN domain and TERT ring in an RNP reconstituted with full-length hTR in vivo (Robart & Collins, 2011). Results here indicate that at least for human telomerase, the product-template hybrid itself also plays a major role. Sequence-based discrimination of the product-template hybrid would account for why substitutions of the human template sequence alter the profile and rate of product synthesis (Drosopoulos et al, 2005). Recognition of the product-template hybrid appears more autonomous of template-flanking sequences in the human enzyme than its ciliate or yeast counterparts. To achieve this recognition, we propose that human telomerase retains most or all of the possible product-template base pairing, rather than a limited length of base-paired duplex as is the precedent from budding yeast (Förstemann & Lingner, 2005). Several lines of evidence support this speculation. First, primer mismatched to a trans-template 3′ end induced template 5′ boundary bypass, suggesting that the template 3′ hybrid influences template 5′ boundary fidelity. In addition, for holoenzyme RNP assembled in vivo, the length of product DNA protected from exonuclease digestion increased with synthesis to the template 5′ boundary. As illustrated in Fig 7C, this is consistent with a constant protection of ˜11 nt of single-stranded DNA and an increasing length of product-template hybrid. We note that previous human telomerase trans-template assays are also fully consistent with accommodation of a 10- or 11-base pair product-template hybrid in the telomerase active site (Qi et al, 2012).
Numerous approaches have attempted to identify physical constituents of the telomerase anchor site. Previous binding and activity assays suggest that human telomerase interacts with a single-stranded primer region of 7–12 nt extending from the primer-template hybrid (Morin, 1989; Wallweber et al, 2003). Using both functional and physical assays, we demonstrate that the TERT ring RNP lacking a TEN domain can account for this inferred length of anchor-site interactions. We propose that anchor-site interactions occur along surfaces that can thread the single-stranded DNA without a specific register of binding other than imposed by distance from the primer-template hybrid. In future studies, it will be of high interest to define whether single-stranded DNA has a unique binding site on the TERT ring RNP or instead is occluded from nuclease digestion without the requirement for specific side-chain contacts. Both the TERT ring and TEN domain have a high isoelectric point, reflecting a predominance of basic side chains that could form an electrostatically favorable path or general surface area for anchor-site associations. Electrostatic interactions without a specific binding cleft would account for telomerase activity on primers with base-paired as well as single-stranded 5′ extensions (Oganesian et al, 2006). We note that telomerase holoenzyme proteins other than TERT can provide additional anchor-site DNA interactions, such as T. thermophila Teb1 (Min & Collins, 2009).
Although active-site stabilization of primer-template hybrid is sufficient to account for TEN domain function and the TERT ring RNP can account for anchor-site interactions, the TEN domain may contribute to the anchor site as well. Activity assays do not provide evidence for TEN domain interaction with single-stranded DNA, but they are not inconsistent with the possibility. For example, activity with a short trans-template and 5′-extended primer is higher in the presence of the TEN domain (Fig 4B, compare lanes 7 and 11), which could reflect TEN domain association with single-stranded DNA in addition to its stabilization of primer-template hybrid in the active site. Physical protection of product DNA differed quantitatively and qualitatively in the presence versus absence of the TEN domain, which again could reflect TEN domain association with single-stranded DNA. With or without a direct single-stranded DNA interaction surface, the general position of the TEN domain relative to the active site (Jiang et al, 2013) suggests that it could constrain product-template hybrid length or escape of the product single-stranded region from the RNP. Future studies can exploit the new reconstitution systems and assays developed in this work for additional insights about the telomerase structures responsible for DNA handling and how they coordinate with other factors required for telomere replication.
Materials and Methods
Telomerase reconstitution in vitro
The hTR RNAs were synthesized from a pUC119 plasmid linearized with EcoRI using in vitro transcription and purified by denaturing gel electrophoresis. Telomerase was reconstituted using the TNT-T7 coupled reticulocyte lysate system (Promega) with reactions containing 40 ng/μl pCITE 3xFLAG-TERT expression construct and 100 ng/μl purified hTRmin incubated at 30°C for 3.5 h. The MBP-TEN domain-His10 fusion was expressed using pET28a in E. coli BL21 (DE3) RP cells, partially purified on NiNTA resin (Qiagen) with 300 mM imidazole elution, and further purified using amylose resin (New England Biolabs) eluted with 10 mM maltose in buffer containing 50 mM Tris–HCl at pH 7.9, 400 mM KCl, 1 mM MgCl2, 10% glycerol, 5 mM DTT, and 0.1 mM PMSF.
Telomerase holoenzyme reconstitution
Human U2OS or 293T cells were transiently transfected using calcium phosphate with a pcDNA 3xFLAG-TERT expression construct and full-length hTR or hTR△temp construct based on pBSU3hTR500, which contains the U3 small nucleolar RNA promoter and 500 bp of downstream genomic region following the mature hTR 3′ end (Fu & Collins, 2003). After 48 h, cells were lysed by three freeze-thaw cycles in hypotonic lysis buffer (HLB; 20 mM HEPES at pH 8, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, and 0.1 mM PMSF). NaCl was added to 400 mM final concentration, and the whole-cell extract was cleared by centrifugation. The extract was diluted to bring NaCl concentration to 150 mM before immunopurification with FLAG M2 monoclonal antibody resin (Sigma-Aldrich) by end-over-end rotation at room temperature for 2 h. The resin was then washed three times with HLB containing 150 mM NaCl, 0.1% Triton X-100, and 0.2% CHAPS at room temperature.
Activity assays
Activity assays were performed in 20-μl reaction mixtures containing extract-purified RNP immobilized on FLAG resin or 2 μl in vitro reconstituted telomerase RNP. Reaction mixtures contained 50 mM Tris-acetate at pH 8, 3 mM MgCl2, 1 mM EGTA, 1 mM spermidine, 5 mM DTT, 5% glycerol, 0.1 μM α-32P dGTP (3000 Ci/mmol, 10 mCi/ml, Perkin-Elmer) and if included, 250 μM of other dNTPs and/or 500 μM of ddNTPs. Trans-template assays used 20 μM pre-annealed RNA/DNA hybrid, prepared by mixing and heating to 80°C for 5 min followed by slow cooling to 10°C in buffer containing 10 mM Tris–HCl at pH 7.9, 1 mM EDTA, and 200 mM NaCl. Reaction mixtures were incubated at room temperature for 60 min. The products were then extracted, precipitated, and resolved on 12% or 18% polyacrylamide/7 M urea/0.6× Tris borate-EDTA gels.
Nuclease protection assay
Telomerase enzyme was combined with 1 pmol DNA primer in a total volume of 10 μl and incubated at room temperature for 30 min. The primer extension reaction was initiated by the addition, to final concentration, of 50 mM Tris-acetate at pH 8, 3 mM MgCl2, 1 mM EGTA, 1 mM spermidine, 5 mM DTT, 5% glycerol, 0.1 μM α-32P dGTP and, if included, 250 μM of other dNTPs and/or 500 μM of ddNTPs in a final volume of 20 μl. The reaction mixtures were incubated at room temperature for 1 min, followed by the addition of 5 units of exonuclease VII (ExoVII, Affymetrix). The reaction mixtures were incubated at room temperature for 5 min, then stopped by the addition of 80 μl stop buffer (50 mM Tris–HCl at pH 7.9, 20 mM EDTA, and 0.2% SDS). The products were extracted, precipitated, and resolved on 18% polyacrylamide/7 M urea/0.6× Tris borate-EDTA gels. For heat denaturation, reaction mixtures were boiled for 10 min after the initial 1-min elongation, then supplemented with 2.5 μM hTR 46–56 and incubated on ice for 5 min, followed by treatment with 5 units of ExoVII for 5 min. For footprinting during processive synthesis, unlabeled dGTP was added to a final concentration of 5 μM and the reaction mixture was incubated at room temperature for 10 min after the initial 1-min elongation, followed by treatment with 5 units of ExoVII for 5 min. Protected products were quantified using Semi-Automated Footprinting Analysis (SAFA) version 11b (Das et al, 2005).
Acknowledgments
This work was supported by N.I.H. Grants HL079585 and GM054198 to K.C. and an N.S.F. predoctoral fellowship to R.A.W.
Author contributions
R.A.W. performed the experiments. R.A.W. and K.C. designed and analyzed the experiments and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Alder JK, Cogan JD, Brown AF, Anderson CJ, Lawson WE, Lansdorp PM, Phillips JA, 3rd, Loyd JE, Chen JJ, Armanios M. Ancestral mutation in telomerase causes defects in repeat addition processivity and manifests as familial pulmonary fibrosis. PLoS Genet. 2011;7:e1001352. doi: 10.1371/journal.pgen.1001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairley RC, Guillaume G, Vega LR, Friedman KL. A mutation in the catalytic subunit of yeast telomerase alters primer-template alignment while promoting processivity and protein-DNA binding. J Cell Sci. 2011;124:4241–4252. doi: 10.1242/jcs.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–1375. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, Richardson CC. Exonuclease VII of Escherichia coli. Mechanism of action. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. Proc Natl Acad Sci USA. 2005;102:8080–8085. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol. 2011;15:643–648. doi: 10.1016/j.cbpa.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Direnzo R, Prasad VR. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J Biol Chem. 2005;280:32801–32810. doi: 10.1074/jbc.M506319200. [DOI] [PubMed] [Google Scholar]
- Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–12814. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2010;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly H, Blackburn EH, Parslow TG. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol Cell Biol. 2003;23:6849–6856. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci USA. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol Cell Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TJ, Marie-Egyptienne DT, Autexier C. Regulation of 5′ template usage and incorporation of noncognate nucleotides by human telomerase. RNA. 2005;11:1448–1460. doi: 10.1261/rna.2910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Morin GB. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, Garin-Chesa P, Rettig WJ, Damm K, Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat Res. 2012;730:3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li Y, Honda S, Hoffmann S, Marz M, Mosig A, Podlevsky JD, Stadler PF, Selker EU, Chen JJ. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Xie M, Brown AF, Bley CJ, Podlevsky JD, Chen JJ. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 2012;31:150–161. doi: 10.1038/emboj.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem. 2010;285:4375–4386. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc Natl Acad Sci USA. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealey DC, Zheng L, Taboski MA, Cruickshank J, Ikura M, Harrington LA. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010;38:2019–2035. doi: 10.1093/nar/gkp1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Sem Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- Wallweber G, Gryaznov S, Pongracz K, Pruzan R. Interaction of human telomerase with its primer substrate. Biochemistry. 2003;42:589–600. doi: 10.1021/bi026914a. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol Cell Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci USA. 2011;108:20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.