Summary
Müller glia (MG) in the zebrafish retina respond to retinal injury by generating multipotent progenitors for retinal repair. Here we show that Insulin, Igf-1 and FGF signaling components are necessary for retina regeneration. Interestingly, these factors synergize with each other and with HB-EGF and cytokines to stimulate MG to generate multipotent progenitors in the uninjured retina. These factors act by stimulating a core set of signaling cascades (Mapk/Erk, PI3K, β-catenin and pStat3) that are also shared with retinal injury and exhibit a remarkable amount of crosstalk. Our studies suggest that MG are both the producers and responders of factors that stimulate MG reprogramming and proliferation following retinal injury. The identification of a core set of regeneration-associated signaling pathways required for MG reprogramming not only furthers our understanding of retina regeneration in fish, but also suggests new targets for enhancing regeneration in mammals.
Introduction
Vision is one of our most precious senses. Many blinding eye diseases result from degenerating retinal neurons. Therefore, identifying strategies for regenerating these lost neurons may help restore lost sight. Unfortunately mammals are unable to regenerate a damaged retina. In contrast, teleost fish, like zebrafish exhibit a remarkable regenerative ability that can restore sight to a damaged retina (Lindsey and Powers, 2007; Mensinger and Powers, 1999; Sherpa et al., 2008). Understanding the mechanisms by which zebrafish can regenerate a damaged retina may suggest strategies for stimulating retina regeneration in mammals.
Key to successful retina regeneration are Müller glia (MG), the major glial cell-type in the retina (Bernardos et al., 2007; Fausett and Goldman, 2006; Fimbel et al., 2007). MG are the only cell to span all retinal layers and also extend processes into these layers. These anatomical features facilitate its ability to monitor and communicate with neighboring cells (Bringmann et al., 2009; Reichenbach and Bringmann, 2013). Normally, MG help maintain retinal architecture and homeostasis (Bringmann et al., 2009; Reichenbach and Bringmann, 2013); however, in teleost fish, like zebrafish, MG respond to retinal injury by undergoing a reprogramming event where they acquire properties of a stem cell that produces a proliferating population of multipotent retinal progenitors that regenerate lost neurons (Fausett and Goldman, 2006; Fausett et al., 2008; Kassen et al., 2007; Nagashima et al., 2013; Powell et al., 2013; Qin et al., 2009; Ramachandran et al., 2010a; Ramachandran, 2010; Ramachandran et al., 2012).
The mechanisms driving MG reprogramming are poorly understood. It is interesting that MG elicit a regenerative response regardless if injury affects only photoreceptors, inner retinal neurons or all retinal cell types (Fausett and Goldman, 2006; Fimbel et al., 2007; Montgomery et al., 2010; Vihtelic and Hyde, 2000). Furthermore, in the absence of retinal injury MG can be forced to reprogram by growth factors, like HB-EGF (Wan, 2012) and cytokines (see accompanying manuscript by Zhao et al.) (Kassen et al., 2009). The diversity of injured cell types and secreted factors that stimulate MG reprogramming is intriguing and suggests multiple mechanisms may drive MG reprogramming and retina regeneration.
Previous studies identified a regulatory role for Mapk/Erk in controlling MG reprogramming in response to retinal injury or HB-EGF-treatment of the uninjured retina (Wan, 2012) and Wnt/Gsk3β/β-catenin has been implicated in regulating injury-dependent MG proliferation (Meyers et al., 2012; Ramachandran et al., 2011). The role for FGFR signaling is controversial with one report indicating it stimulates injury-dependent MG proliferation (Hochmann et al., 2012) and another indicating it has little effect (Qin et al., 2011). Finally, in the accompanying paper by Zhao et al., we report an important role for Jak/Stat3 signaling in controlling MG reprogramming and proliferation. Whether additional signaling systems contribute to MG reprogramming and retina regeneration are not known, nor is it known if these signaling cascades reflect the type of stimulus used to induce a MG response.
We were intrigued by reports that MG in the postnatal chick retina could be induced to proliferate in response to intravitreal injection of insulin/FGF2 or IGF-1/FGF2 (Fischer et al., 2002; Fischer and Reh, 2002; Ritchey et al., 2012). Although these treatments stimulate MG proliferation in the chick retina, rarely do these cells survive and regenerate neurons. Here we report that Insulin, IGF-1 and FGF signaling components are necessary for regeneration in the injured zebrafish retina. We show that these factors crosstalk and synergize with each other and with HB-EGF and cytokines to stimulate MG reprogramming and progenitor formation in the uninjured retina. Finally, we found that Mapk and PI3K signaling converge on β-catenin and pStat3 signaling to stimulate MG reprogramming in response to growth factors, cytokines and retinal injury. These MG responses in fish may distinguish them from birds and mammals and thus underlie their unique ability to reprogram and generate progenitors for retinal repair.
Results
Insulin signaling stimulates MG reprogramming and proliferation in the injured and uninjured zebrafish retina
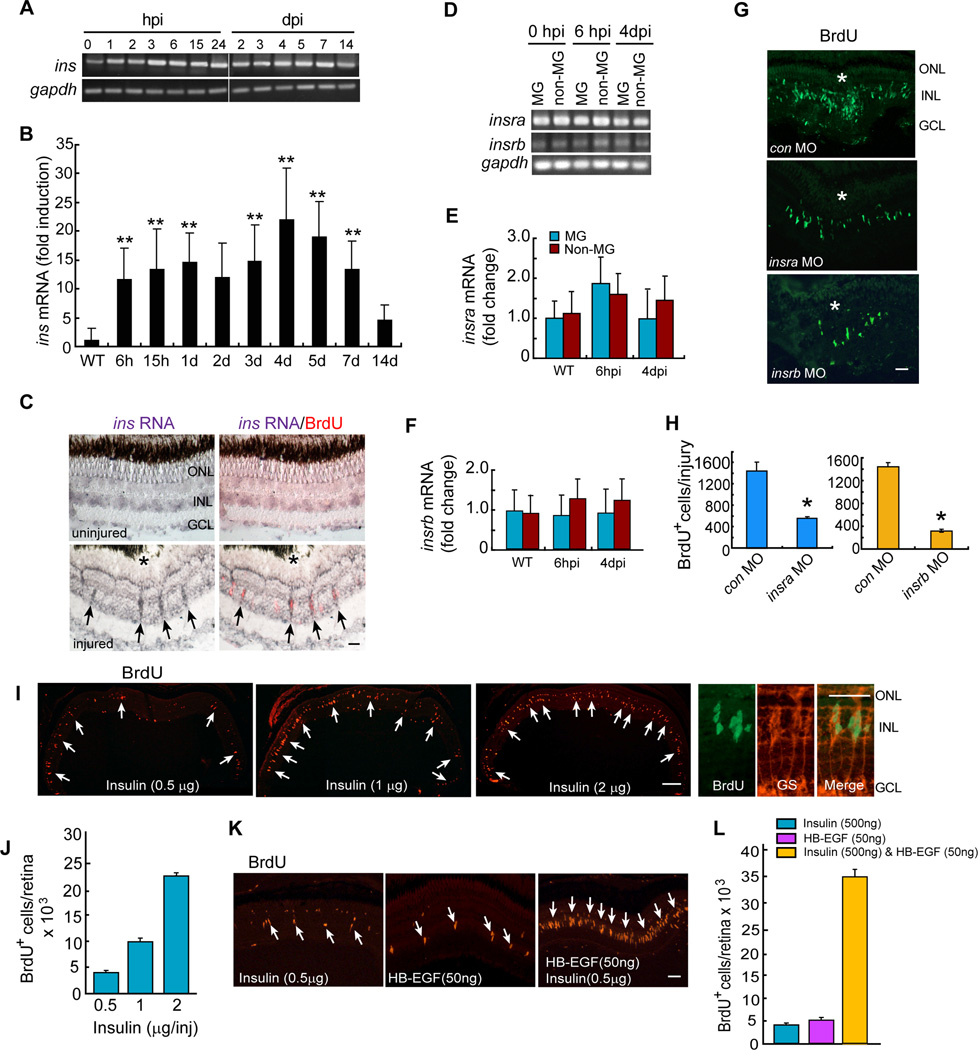
MG proliferation is critical for successful retina regeneration. Intravitreal injection of Insulin into the chick eye has little effect on MG; however, when combined with FGF2, MG proliferation was stimulated (Fischer et al., 2002; Fischer and Reh, 2002). However, this MG response is limited in that it does not result in retina regeneration. To determine if Insulin signaling contributed to retina regeneration we investigated if Insulin signaling components were expressed in injury-responsive MG and regulated during retina regeneration. We previously demonstrated that following a needle poke injury, MG at the injury site proliferate and generate multipotent progenitors for retinal repair (Fausett and Goldman, 2006). Furthermore, those studies showed that essentially all the proliferating cells in the inner nuclear layer (INL) were MG-derived progenitors (Fausett and Goldman, 2006). Interestingly, we found that ins (insulin) mRNA was induced in BrdU+ MG-derived progenitors following retinal injury (Figures 1A–1C). Furthermore, we found constitutive expression of Insulin receptor encoding mRNAs, insra and insrb, in GFP+ MG and GFP-neurons that were purified by FACS using retinas from gfap:gfp fish (uninjured and 6 hours post injury [hpi]) and 1016 tuba1a:gfp fish (4 days post injury [dpi]) (Figures 1D–1F). Importantly, Insra or Insrb knockdown with lissamine-tagged morpholino-modified antisense oligonucleotides (MOs) suppressed the formation of BrdU+ MG-derived progenitors in the injured retina (Figures 1G and 1H).
Figure 1. Insulin and HB-EGF are sufficient to stimulate MG proliferation in the uninjured retina.
(A, B) RT-PCR and qPCR analysis of injury-dependent ins gene expression. Error bars are SD; **P<0.01; n=5. (C) In situ hybridization (purple) and BrdU immunofluorescence (red) shows injury-dependent ins gene induction in proliferating MG-derived progenitors at 4 dpi. Arrows point to BrdU+ cells expressing ins RNA. Scale bar is 50 µm. (D) RT-PCR analysis of insra and insrb gene expression in FACS purified MG and non-MG (E, F) qPCR analysis of insra (E) and insrb (F) gene expression in MG and non-MG Error bars are SD; n=4. (G, H) MO-mediated Insra and Insrb knockdown suppresses the generation of MG-derived progenitors at 4 dpi. Scale bar (G) is 50 µm. Asterisk (G) marks the injury site. Error bars (H) are SD; *P<0.05; n=3. (I, J) Insulin stimulates MG proliferation in the uninjured retina. Arrows (I) point to BrdU+ MG. Scale bar is 150 µm. The 3 right-hand panels show BrdU+ progenitors express the MG marker glutamine synthetase (GS). Error bars (J) are SD; n=3. (K, L) HB-EGF and Insulin synergize with each other to stimulate MG proliferation in the uninjured retina. Arrows point to BrdU+ MG. Scale bar is 50 µm. Error bars are SD; n=3. See also Figure S1.
To investigate if Insulin was sufficient to stimulate MG proliferation, we injected 0.5–2 µg of human recombinant Insulin or vehicle intravitreally through the front of the eye for 3 consecutive days and labeled proliferating cells with a pulse of BrdU two days later (Figure S1A). In the accompanying paper by Xiao et al., we validated our intravitreal injection method using HB-EGF to stimulate MG proliferation. A similar series of validations was done for Insulin (data not shown). We suspected that human Insulin would be active in zebrafish because its mature form exhibits extensive amino acid identity (~77%) with that of zebrafish and it was previously shown to activate zebrafish Insulin receptors (Papasani et al., 2006; Toyoshima et al., 2008). Indeed, BrdU immunofluorescence showed a dose-dependent increase in MG proliferation (Figures 1I and 1J). This effect was not a result of Insulin-induced cell death, since Insulin (2 µg) did not stimulate apoptosis (Figure S1B). Insulin did not stimulate microglia proliferation (Figure S1C).
We used a BrdU lineage tracing strategy to test if Insulin-induced MG-derived progenitors were multipotent. For these experiments Insulin was intravitreally injected into the eye, proliferating cells were labeled with BrdU and 10 days later fish were sacrificed for immunofluorescence analysis of BrdU+ cells that co-label with retinal cell type-specific antibodies (Figures S1D–S1F). Although Insulin stimulated the generation of Zpr1+ photoreceptors, HuC/D+ amacrine cells in the INL, Pkc+ bipolar cells and HuC/D+ ganglion cells in the ganglion cell layer (GCL) (Figures S1E and S1F), quantification revealed a preference for generating photoreceptors (Figure S1F). Together, the above studies indicate that Insulin signaling participates in injury-induced retina regeneration and that treating an uninjured retina with Insulin is sufficient to stimulate MG reprogramming and the formation of multipotent progenitors.
Insulin synergizes with HB-EGF to stimulate progenitor formation in the uninjured retina
We previously reported that Hbegfa regulates progenitor formation in the injured zebrafish retina and that recombinant HB-EGF stimulates MG to generate progenitors in the uninjured retina (Wan, 2012). Since Insulin signaling is also necessary for retina regeneration, we investigated if Insulin and HB-EGF collaborate to stimulate progenitor formation. For this analysis, we used concentrations of Insulin (500 ng) and HB-EGF (50 ng) that only stimulated a small amount of MG proliferation in the uninjured retina (Figures 1I, 1J, and S1G–S1I). Even at high concentrations that stimulate robust MG proliferation, neither Insulin nor HB-EGF stimulates microglia proliferation (Figures S1C and S1L); suggesting their effect is specific to MG. Interestingly, when low concentrations of Insulin and HB-EGF were combined, they revealed a remarkable synergy in their ability to stimulate MG proliferation (Figures 1K, 1L, S1J and S1K).
IGF-1 synergizes with FGF2 to stimulate progenitor formation
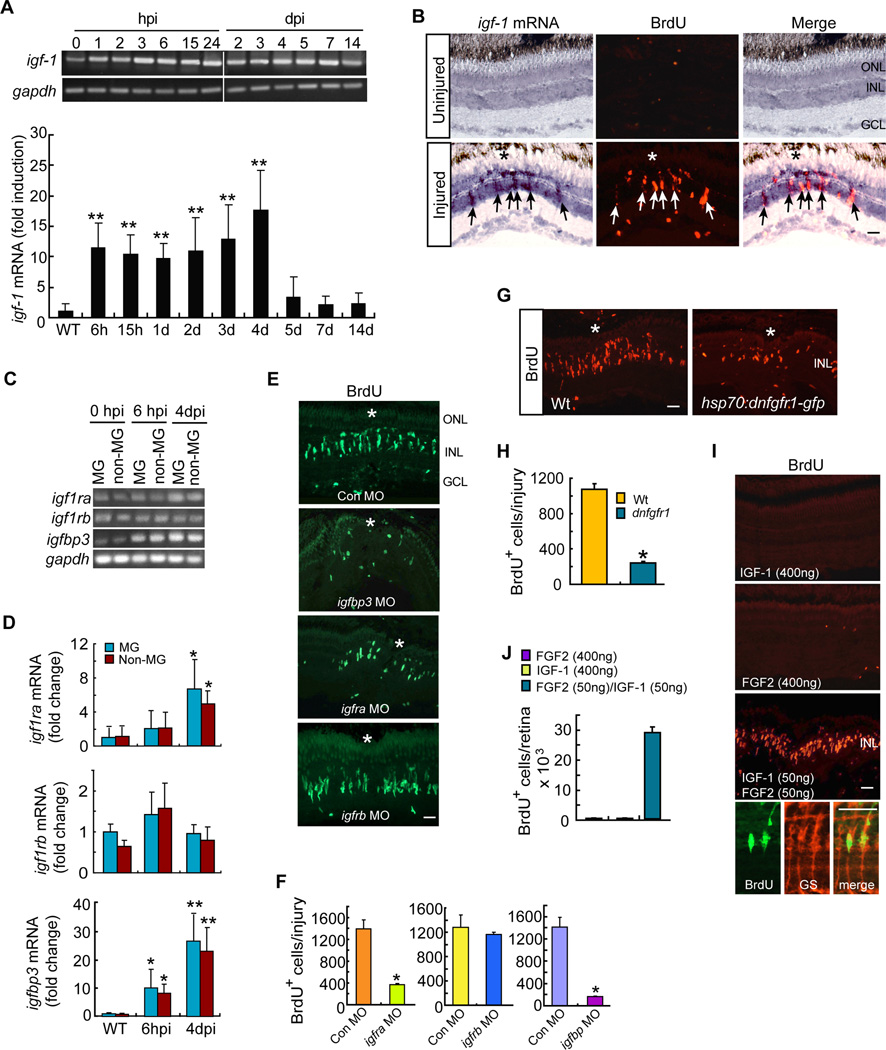
Igf-1 (Insulin-like growth factor 1) is structurally homologous to Insulin and although it can bind the Insulin receptor, its affinity is highest for the Igf-1 receptor (Igf1r) (Wood et al., 2005). Zebrafish harbor 2 igf1r genes, igf1ra and igf1rb that often play overlapping roles (Schlueter et al., 2006). Furthermore, Igf binding proteins stabilize bound Igfs and modulate their activity (Wood et al., 2005). Igf binding protein 3 (Igfbp3) is the most highly expressed Igfbp and binds most abundantly to Igf-1(Wood et al., 2005). To determine if these Insulin-like signaling components were expressed in MG and responsive to retinal injury, we assayed their mRNAs in the uninjured and injured retina. We observed an injury-dependent increase in igf-1 mRNA in BrdU+ MG-derived progenitors (Figures 2A and 2B). Furthermore, IGF-1 receptor mRNAs (igfra and igfrb) were expressed in GFP+ MG and GFP− neurons purified by FACS using retinas from gfap:gfp fish (uninjured and 6 hpi) and 1016 tuba1a:gfp fish (4 dpi) (Figure 2C). igfra mRNA increased at 4 dpi and igfbp3 mRNA increased at 6 hpi in both MG and neurons (Figure 2D). Importantly, MO-mediated knockdown of Igfbp3 or Igfra suppressed progenitor formation, while knockdown of Igf1rb had no effect (Figures 2E and 2F). These data indicate that Igf signaling components are regulated following retinal injury and that Igf signaling regulates the formation of MG-derived progenitors in the injured retina.
Figure 2. IGF-1 and FGF2 synergize with each other to stimulate MG proliferation in the uninjured retina.
(A) RT-PCR and qPCR analysis of injury-dependent induction of igf-1 mRNA. Error bars are SD; **P<0.001; n=6. (B) In situ hybridization shows injury-dependent induction of igf-1 RNA in BrdU+ progenitors (arrows) at 4 dpi. Scale bar is 50 µm. Asterisk marks the injury site. (C, D) RT-PCR (C) and qPCR (D) analysis of igf1ra, igf1rb and igfbp3 gene expression in FACS purified MG and non-MG at different times post retinal injury. Error bars (D) are SD; *P <0.05, **P<0.01; n=4. (E) BrdU immunofluorescence shows effect of morpholino (MO)-mediated knockdown of Igfbp3, Igfra or Igfbp on progenitor formation in the injured retina. Scale bar is 50 µm. Asterisk marks the injury site. (F) Quantification of BrdU+ cells in (E). Error bars are SD; *P <0.05; n=3. (G) BrdU immunofluorescence shows that conditional overexpression of dnFgfr1 in hsp70:dnfgfr1-egfp fish suppresses the generation of BrdU+ progenitors at 4 dpi. Asterisk marks the injury site. Scale bar is 50 µm. (H) Quantification of BrdU+ cells in (G). Error bars are SD; *P <0.05 n=3. (I) BrdU immunofluorescence shows IGF-1 and FGF2 synergize to stimulate MG proliferation in the uninjured retina. Scale bar is 50 µm. Bottom 3 panels show BrdU+ cells are also GS+ MG. (J) Quantification of BrdU+ cells in I. Error bars are SD; n=3. See also Figure S2.
We next investigated if IGF-1 was sufficient to stimulate MG proliferation in the uninjured retina. Therefore, we intravitreally injected recombinant human IGF-1 (400 ng) through the front of the eye for 3 days as described above for HB-EGF and Insulin. We suspected human IGF-1 would act via zebrafish receptors because the mature peptide shares ~81% identity with that of zebrafish (Zou et al., 2009), including key residues important for receptor binding (Gauguin et al., 2008). Furthermore, human and zebrafish IGF-IR share over 80% similarity and key residues involved in ligand binding are preserved (Ayaso et al., 2002; Renteria et al., 2008). Surprisingly, intravitreal injection of IGF-1 did not stimulate MG proliferation in the uninjured retina (Figure 2I, top panel). This result was reminiscent of that found in the post natal chick retina where Igf-1 collaborates with Fgf signaling to stimulate MG proliferation (Ritchey et al., 2012).
Fgfs and their receptors are expressed in the INL of the zebrafish retina (Hochmann et al., 2012); however, the effects of Fgf signaling on MG proliferation are controversial (Hochmann et al., 2012; Qin et al., 2011). Therefore, we investigated Fgf signaling in our mechanical injury model. For this analysis we used hsp70:dnfgfr1-egfp transgenic fish that harbor the hsp70 promoter driving expression of a dominant/negative Fgfr1-GFP fusion protein (dnFgfr1) (Lee et al., 2005). We found that conditional expression of dnFgfr1 suppressed MG proliferation in the injured retina (Figures 2G and 2H), which is consistent with that found when photoreceptors are damaged (Hochmann et al., 2012).
We next investigated if recombinant human FGF2 (400 ng) would stimulate MG proliferation in the uninjured retina. Human and zebrafish FGF2 exhibit ~75% identity and human FGF2 activates the zebrafish Fgf2 receptor (Nicoli et al., 2009). Like IGF-1, intravitreal injection of FGF2 had little effect on MG proliferation in the uninjured retina (Figure 2I, middle panel). However, FGF2 and IGF-1 in combination acted in a synergistic fashion to stimulate MG proliferation (Figures 2I, 2J, S2A, S2B). FGF2/IGF-1 did not stimulate cell death (Figure S2C) or microglia proliferation (Figure S2D).
Using a BrdU lineage tracing strategy, we found that the MG-derived progenitors in IGF-1/FGF2-treated retinas were multipotent (Figures S2E–S2G). Quantification of the generated cell types showed a preference for photoreceptors (Figure S2G), similar to Insulin-treated retinas (Figure S1E). Taken together, our data suggest that retinal injury stimulates MG to release of a variety of growth factors that alone may be ineffective, but together synergize to stimulate MG reprogramming and the production of multipotent progenitors.
Signaling cascades controlling progenitor formation
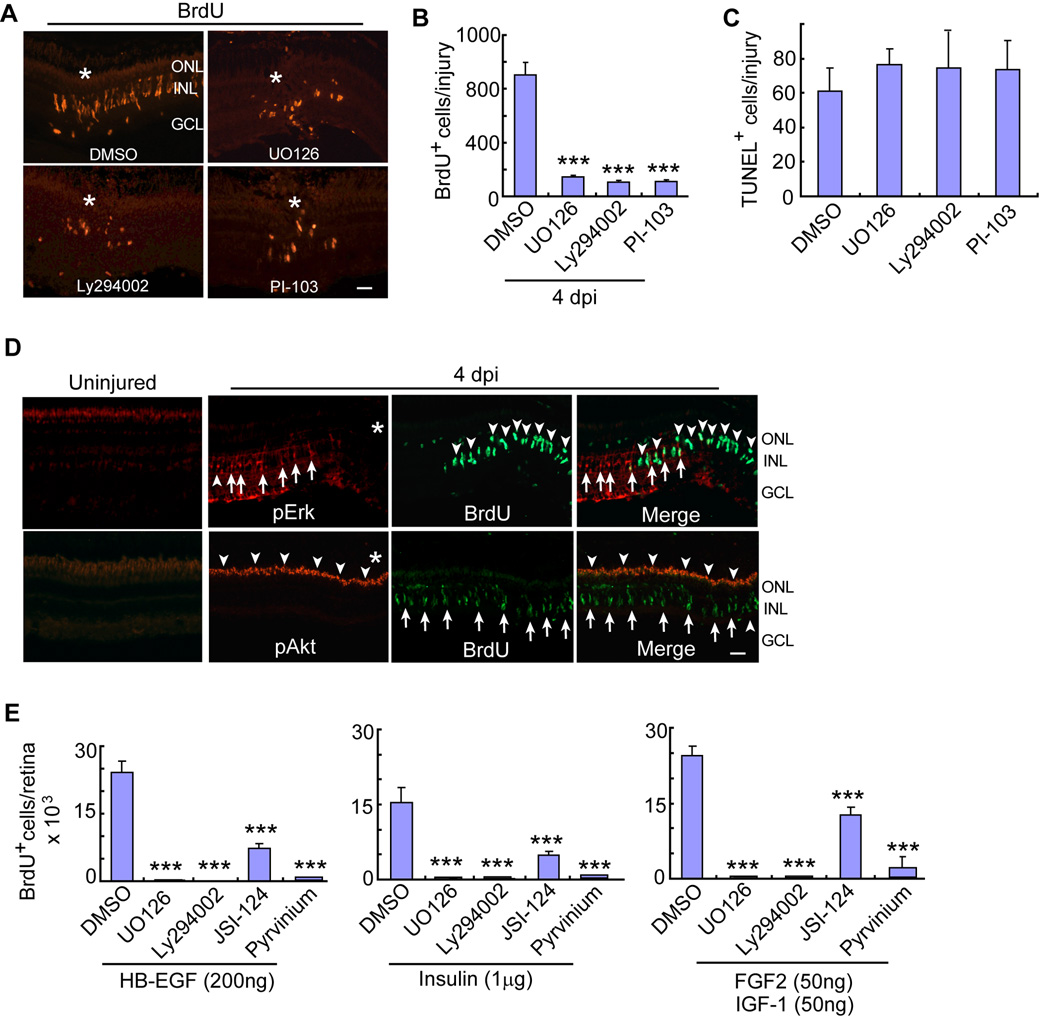
Previous studies (Kassen et al., 2009; Meyers et al., 2012; Nelson et al., 2012; Ramachandran et al., 2011; Wan, 2012), along with the accompanying paper by Zhao et al., indicate that Mapk/Erk, Gsk3 β/β-catenin and Jak/Stat3 signaling regulate MG-dependent progenitor formation in the injured retina. Our finding that Insulin and Igf-1 signaling components are also necessary for progenitor formation (Figures 1 and 2) suggested that PI3K/Akt signaling may also contribute to this process. Indeed, like Mapk inhibition with UO126, injury-dependent formation of BrdU+ MG-derived progenitors was suppressed by exposing injured retinas to PI3K/Akt signaling inhibitors, Ly294002 and PI-103 (Figures 3 A, 3B and S2H). These inhibitors had no significant effect on cell death (Figures 3C and S2I), MG morphology or expression of the MG marker, glutamine synthetase (GS) (Figure S2J).
Figure 3. Mapk/Erk, PI3K/Akt, β-catenin and Jak/Stat signaling regulate injury and growth factor-stimulated MG proliferation.
(A, B) Mapk/Erk inhibitor (UO126) or PI3K inhibitors (Ly294002 or PI-103) reduce the number of BrdU+ progenitors at 4 dpi. Asterisks identify the injury site. Scale bar is 50 µm. Error bars are SD; ***P<0.001; n=6. (C) TUNEL assay shows Mapk and PI3K inhibitors have no effect on cell death. (D) pErk and pAkt immunofluorescence in uninjured and injured (4 dpi) retinas. Asterisks identify the injury site. In the pErk panels, arrows point to pErk+ MG and arrowheads point to BrdU+ cells. In pAkt panels, arrowheads point to pAkt+ cells, while arrows point to BrdU+ cells. Scale bar is 50 µm. (E) In the growth factor stimulated, but uninjured retina, BrdU+ progenitor formation is suppressed by inhibiting Mapk (UO126), PI3K (Ly294002), Jak/Stat (JSI-124) or β-catenin (pyrvinium) signaling. Error bars are SD; ***P<0.001; n=5. See also Figures S2–S4.
In the injured retina, β-catenin (Meyers et al., 2012; Ramachandran et al., 2011) and pStat3 (see accompanying paper by Zhao et al.) are restricted to MG-derived progenitors. To investigate if Mapk/Erk and PI3K/Akt signaling are also restricted to MG-derived progenitors, we investigated pErk and pAkt expression in the uninjured and injured retina using immunofluorescence. Interestingly, pErk and pAkt increased in MG and photoreceptor outer segments, respectively, throughout the retina within 1 hpi (Figure S3 A and S3B). Surprisingly, at 2–4 dpi when BrdU+ MG-derived progenitors can be identified, we observed little co-labeling with either pErk or pAkt antibodies (Figures 3C and S3A). pErk and pAkt expression was dependent on Mapk and PI3K signaling since inhibition of these pathways with UO126 and Ly294002 reduced their expression (Figures S3C).
Because MG proliferation requires activation of Mapk, PI3K, Stat3 and β-catenin signaling pathways in the injured retina, we wondered if these same pathways were required for MG proliferation in the growth factor-treated uninjured retina. For these experiments HB-EGF, Insulin or IGF-1/FGF2 was intravitreally injected, with and without inhibitors of the above signaling cascades, and progenitor formation was quantified. Remarkably, regardless of how we stimulate MG proliferation, this proliferation was suppressed by inhibiting Mapk (UO126), PI3K (Ly294002), Jak (JSI-124) or β-catenin (pyrvinium) signaling (Figures 3E and S4).
Similar to that observed in the injured retina (Figure 3D), we found pErk accumulation in quiescent MG and pAkt in photoreceptor outer segments of the growth factor-treated retina (Figures S3D and S3E). This expression was even evident in retinas treated with FGF2 or IGF-1, which do not stimulate MG proliferation (Figures S3D and S3E). Furthermore, pAkt was also observed to accumulate in putative amacrine cells lining the inner portion of the INL (Figure S3E), which was not observed in the injured retina. Importantly, pAkt accumulation in these cells was dependent on PI3K signaling since it was suppressed by the PI3K inhibitor Ly294002 (Figure S3F). Although our data suggests that pErk and pAkt may indirectly impact the proliferation of MG-derived progenitors in the zebrafish retina, we cannot rule out the possibility that MG-derived progenitors retain increased pErk and pAkt that remain below the limits of detection. Together, these data suggest that activation of Mapk and PI3K signaling is necessary, but not sufficient to drive MG proliferation (see FGF2 and IGF-1 panels in Figures S3D and S3E).
Growth factors converge on β-catenin and pStat3 signaling to stimulate MG proliferation
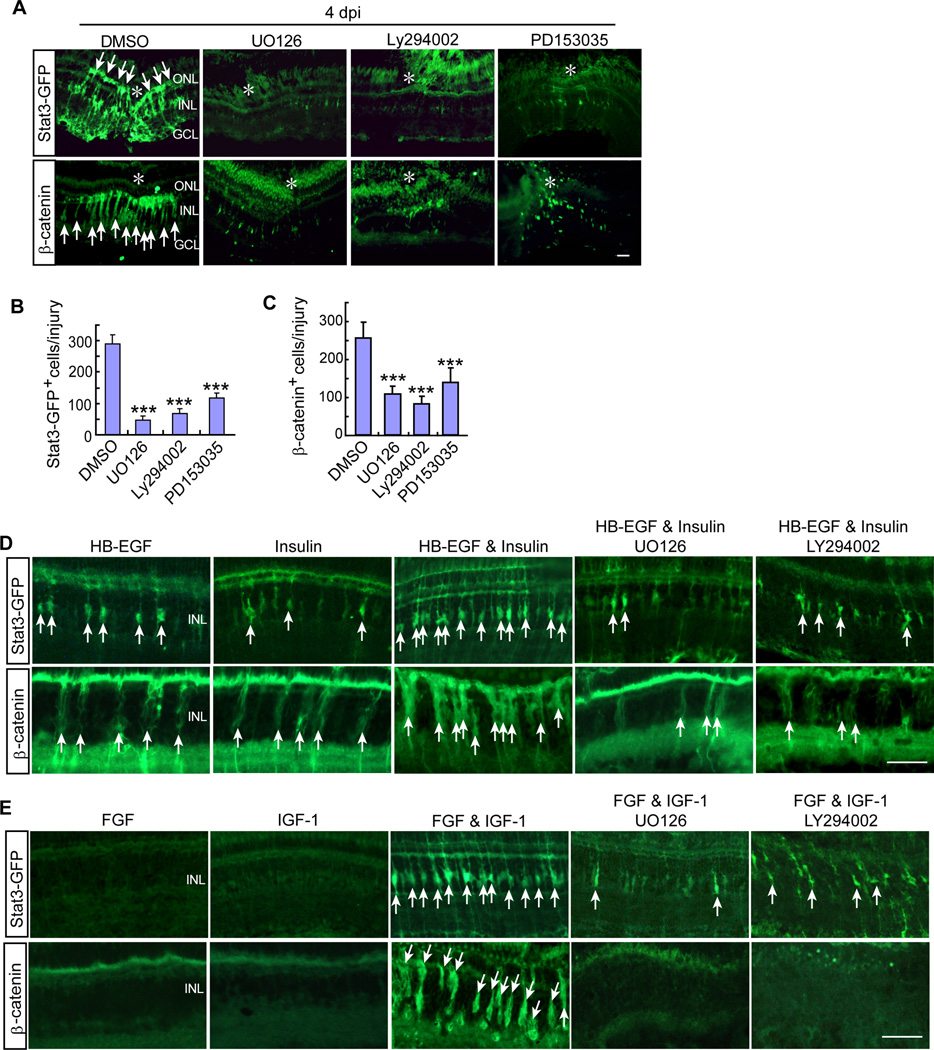
We next investigated if Mapk and PI3K signaling could impact MG-specific β-catenin and pStat3 signaling in the injured retina. We used β-catenin immunofluorescence to detect stabilized β-catenin in reprogrammed MG (Ramachandran et al., 2011) and our gfap:stat3-GFP transgenic fish (see accompanying paper by Zhao et al.) to report pStat3 expression. Interestingly, suppression of Mapk (UO126), PI3K (Ly294002) or Egf receptor (PD153035) signaling dramatically reduced β-catenin and pStat3 levels in the injured retina (Figures 4A–4C) and suggests a hierarchical relationship.
Figure 4. Injury and growth factor-dependent activation of β-catenin and Stat3 signaling in MG-derived progenitors requires Mapk/Erk and PI3K/Akt signaling.
(A) Stat3-GFP and β-catenin immunofluorescence shows that inhibition of Mapk (UO126), PI3K (Ly294002) or the Egf receptor (PD153035) suppresses injury-dependent Stat3-GFP and β-catenin accumulation in gfap:stat3-gfp transgenic fish. Asterisks mark the injury site and arrows point to Stat3-GFP+ and β-catenin+ progenitors. Scale bar is 50 µm. (B and C) Quantification of data shown in (A). Error bars are SD; *P<0.05; ***P<0.001; n=4. (D, E) Stat3-GFP and β-catenin immunofluorescence shows that intravitreally injected HB-EGF (50ng) and Insulin (0.5 µg) (D) or FGF2 and IGF-1 (individually at 400ng or together at 50ng each) (E) act in a synergistic fashion to stimulate Stat3-GFP and β-catenin accumulation in the uninjured retina of gfap:stat3-gfp transgenic fish and their action is suppressed by inhibition of Mapk (UO126) or PI3K (Ly294002) signaling. Arrows point to Stat3-GFP+ and β-catenin+ progenitors. Scale bar is 50 µm.
To determine if growth factors acted on these same signaling cascades to stimulate MG proliferation in the uninjured retina, we assayed β-catenin and pStat3 expression with and without Mapk and PI3K inhibition in the growth-factor treated retina. HB-EGF or Insulin concentrations that only stimulated a small amount of MG proliferation (Figures 1J and 1K), had a correspondingly small effect on the number of β-catenin and pStat3-positive cells detected in the INL (Figure 4D). FGF2 or IGF-1, that were ineffective in stimulating MG proliferation in the uninjured retina (Figures 2I and 2J) were also ineffective in stimulating β-catenin and pStat3 expression (Figure 4E). However, like their synergistic effect on MG proliferation (Figures 1K, 1L, 2I and 2J), HB-EGF and Insulin or IGF-1 and FGF2 together, dramatically stimulated β-catenin and pStat3 expression (Figures 4D and 4E). Furthermore, these effects on β-catenin and pStat3 expression were suppressed by Mapk (UO126) and PI3K (Ly294002) inhibition (Figures 4D and 4E), suggesting a similar hierarchial relationship among these signaling cascades as noted in the injured retina.
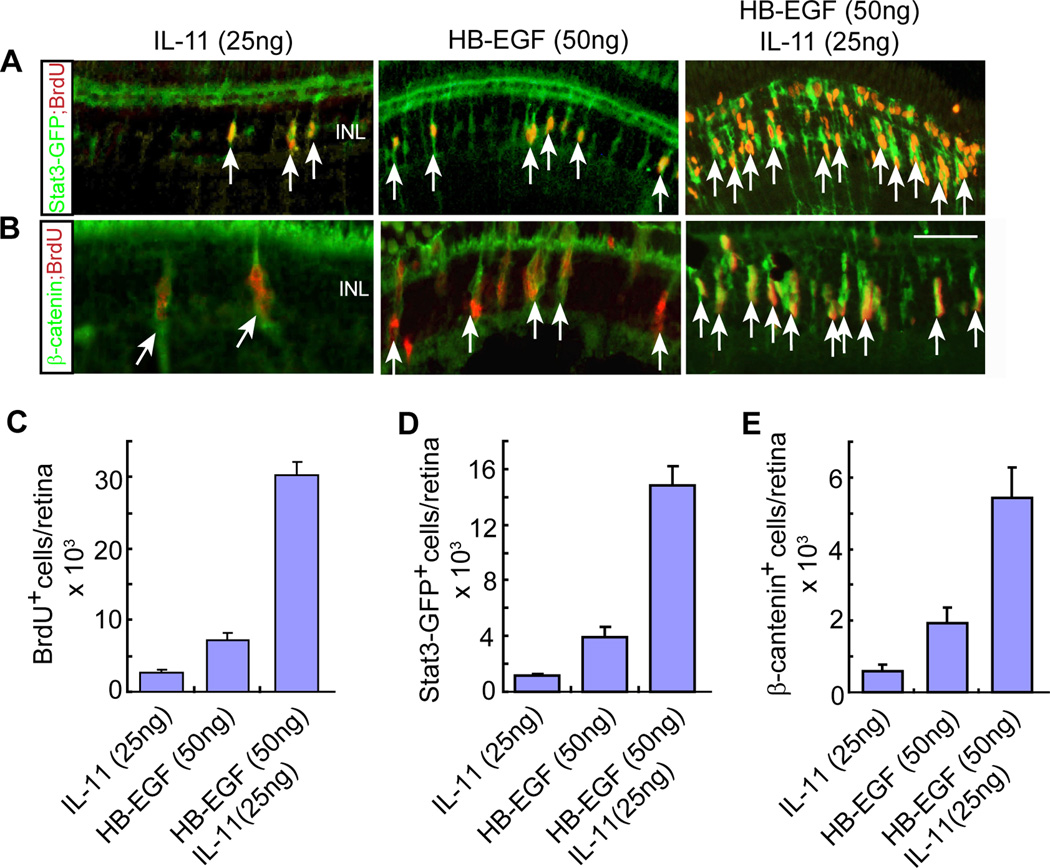
HB-EGF synergizes with IL-11 to stimulate progenitor formation
In the accompanying manuscript by Zhao et al., we report that cytokines, like Leptin and IL-11, are necessary and sufficient to stimulate MG reprogramming and progenitor formation in the injured and uninjured retina. Our in situ hybridization data suggests that these cytokines are released along with growth factors from MG at the injury site. This raised the possibility that growth factors might not only synergize with other growth factors, but also with cytokines to stimulate MG proliferation. To test this idea, wild type and gfap:stat3-GFP transgenic fish received intravitreal injections of HB-EGF and IL-11, either individually or together, at concentrations that alone would only cause a small amount of MG proliferation (Figure 5A–5C). Interestingly, HB-EGF and IL-11 acted in a synergistic fashion to stimulate the formation of BrdU+ MG-derived progenitors and this was highly correlated with β-catenin and pStat3 expression (Figure 5A–5E). The green immunofluorescence detected in the ONL appears to be non-specific since it is not detected when we used a secondary antibody coupled to a red Fluor (Figure S5). Together, our data suggests that 1) retinal injury results in the release of a variety of growth factors and cytokines by MG at the injury site; 2) these factors synergize with each other and act in an autocrine/paracrine manner to stimulate MG reprogramming and proliferation; and 3) this proliferation is mediated by activation of β-catenin and pStat3 signaling pathways in injury-responsive MG.
Figure 5. HB-EGF and IL-11 synergize with each other to stimulate MG proliferation and activation of the Stat3 and β-catenin signaling components in the uninjured retina.
(A, B) HB-EGF and IL-11 synergize with each other to stimulate the generation of BrdU+ progenitors and the accumulation of Stat3 (A) and β-catenin (B) signaling components in the uninjured retina of gfap:stat3-gfp transgenic fish. Arrows point to double-labeled cells. Scale bar is 50 µm. The green immunofluorescence signal in the photoreceptor layer is non-specific since it does not show up when using a secondary antibody coupled to a red Fluor (Figure S7). (C–E) Quantification of BrdU+ (C), Stat3-GFP+ (D) and β-catenin+ (E) progenitors in retinas intravitreally injected with HB-EGF, IL-11 or HB-EGF/IL-11. Error bars are SD; n=3. See also Figure S5.
Extensive crosstalk underlies the effectiveness of factors that stimulate MG proliferation
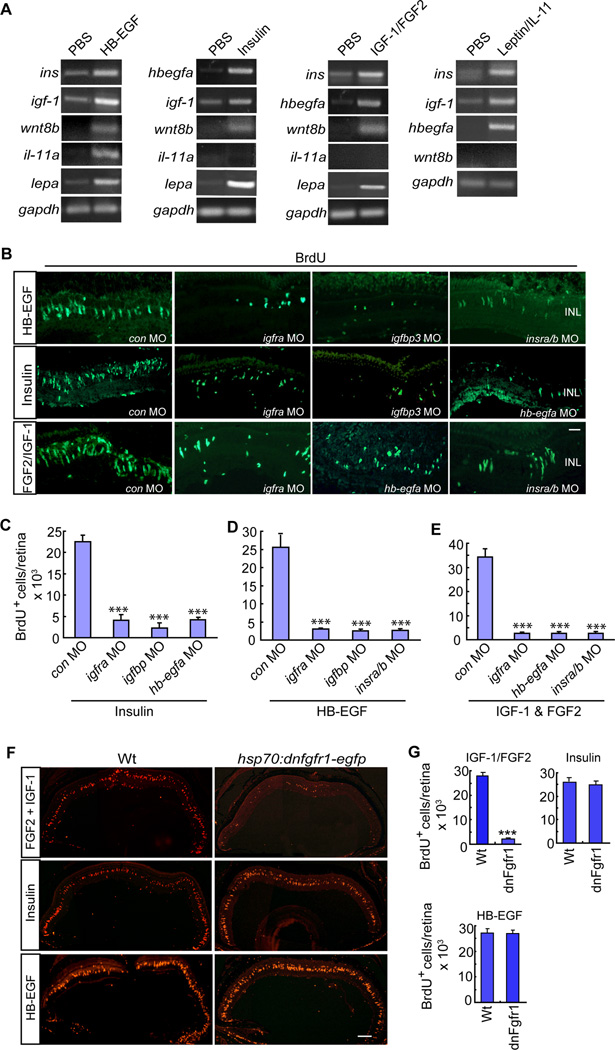
Our studies have revealed a remarkable number of secreted factors that stimulate MG proliferation by converging on Erk, PI3K, β-catenin and pStat3 signaling. The effectiveness of these factors in activating multiple signaling cascades may lie in their ability to stimulate each other’s expression. To test this idea we intravitreally injected HB-EGF, Insulin, IGF-1/FGF2 or IL-11/Leptin into the eyes of fish and assayed the expression of mRNAs encoding a variety of secreted factors that participate in retina regeneration (Figure 6A). Interestingly, HB-EGF-treated eyes exhibited increased expression of ins, igf-1, il-11a,wnt8b and lepa mRNAs; Insulin-treated eyes exhibited increased expression of hbegfa, igf-1, wnt8b and lepa mRNAs; IGF-1/FGF2-treated eyes exhibited increased expression of ins, hbegfa, wnt8b and lepa mRNAs; and IL-11/Leptin-treated eyes exhibited increased expression of ins, igf-1, and hbegfa mRNAs (Figure 6A). We note that although wnt4a mRNA has been reported to be induced by retinal injury (Ramachandran et al., 2011), we could not detect its induction by the growth factors or cytokines tested here (data not shown). Interestingly, in situ hybridization assays showed that the growth factor and cytokine-dependent gene inductions were predominantly in proliferating MG (Figure S6).
Figure 6. Crosstalk between HB-EGF, Insulin, IGF-1/FGF and IL-11/Leptin signaling components contribute to MG proliferation in the uninjured retina.
(A) RT-PCR shows the effect of intravitreal injection of HB-EGF, Insulin, IGF-1/FGF2 or Leptin/IL-11 on each other’s expression and also the regeneration-associated factor, wnt8b. (B) BrdU immunofluorescence shows that MO-mediated knockdown of specific HB-EGF, Insulin and IGF-1 signaling components suppress each other’s ability to stimulate MG proliferation in the uninjured retina. Scale bar is 50 µm. (C–E) Quantification of data presented in (B). Error bars are SD; ***P<0.001; n=4. (F) Conditional overexpression of dnFgfr1 in hsp70:dnfgfr1-egfp fish inhibits FGF2/IGF-1-dependent MG proliferation, but not Insulin or HB-EGF-dependent MG proliferation in the uninjured retina. Scale bar is 150 µm. (G) Quantification of data presented in (F). Error bars are SD. ***P<0.001; n=3. See also Figure S6.
These data suggest that growth factor or cytokine injection into the uninjured retina stimulates the expression of additional growth factors and cytokines and this may contribute to their effectiveness in inducing MG proliferation. To test this idea, we knocked down specific growth factor signaling components and determined if this knockdown affected MG proliferation elicited by different growth factors. For this analysis, we injected a particular growth factor and knockdown MO through the back of the eye. This procedure causes a focal retinal injury at the injection site where MG will respond by proliferating; however, the growth factor stimulates MG proliferation throughout the retina. Thus, by assaying proliferation throughout the retina we can determine if the MO affected MG proliferation induced by the growth factor. Consistent with the crosstalk observed at the gene expression level (Figure 6A), this study revealed that MG proliferation in HB-EGF-treated eyes required Insulin and Igf signaling components (Figures 6B and 6C); MG proliferation in Insulin-treated eyes required Igf and Hbegfa signaling components (Figures 6B and 6D) and MG proliferation in IGF-1/FGF2-treated eyes required Insulin, Igf and Hbegfa signaling components (Figures 6B and 6E).
We also tested if FGF receptor signaling was necessary for growth factor-dependent MG proliferation in the uninjured retina. For these studies we used hsp70:dnfgfr1-egfp transgenic fish to inhibit FGF receptor signaling and found that conditional overexpression of dnFgfr1only blocked MG proliferation in retinas treated with IGF-1/FGF2, but not in retinas treated with Insulin or HB-EGF (Figures 6F and 6G). This result suggests that regeneration-associated signaling pathways acting downstream of the Fgf receptor are sufficiently stimulated in HB-EGF and Insulin-treated retinas, but not in IGF-1-treated retinas and hence explains why IGF-1 requires FGF2 to stimulate MG proliferation. Together, these studies indicate a remarkable amount of crosstalk between growth factors, cytokines and their signaling cascades and suggest that this crosstalk contributes to the effectiveness of these factors ability to stimulate MG proliferation.
Discussion
Zebrafish MG are able to respond to retinal injury by reprogramming their genome so they acquire retinal stem cell characteristics that allow them to proliferate and regenerate all retinal cell types. The nature of the injury signals, their derivation and their mechanism of action are poorly understood. Our studies provide important new information on these processes. In this report we show that Insulin, Igf-1 and Fgf signaling components are required for MG proliferation in response to retinal injury and that these factors can stimulate MG proliferation in the uninjured retina. We found that these factors are expressed by injury-responsive MG, suggesting an autocrine/paracrine mechanism of action. Furthermore, we revealed a remarkable synergy among growth factors and cytokines that allows them to stimulate MG proliferation at low concentrations. Importantly, we found that Mapk, PI3K, β-catenin and pStat signaling must be activated in order for MG to reprogram in response to retinal injury or as a result of growth factor or cytokine stimulation in the uninjured retina. Finally, our studies revealed an extensive crosstalk among the growth factors and cytokine signaling systems that is necessary for MG reprogramming and proliferation.
Although zebrafish have been known for some time to have the capacity to regenerate a damaged retina, the endogenous factors mediating injury-dependent MG reprogramming and proliferation remain poorly characterized. The studies report here, along with that of Zhao et al. in the accompanying paper, suggest a remarkable variety of secreted factors contribute to MG reprogramming and proliferation in the injured retina. Interestingly, all these factors are specifically induced in MG that are reprogramming for retinal repair; they are not detected in quiescent MG. Furthermore, recombinant versions of these factors, individually or in combination, are sufficient to stimulate MG proliferation in the uninjured retina and exhibit a remarkable synergy in their action. This local induction and synergistic action ensures that only MG at the injury site will mount a regenerative response. We speculate that factors released from injured cells, like TNFα (Nelson et al., 2013), stimulate growth factor and cytokine induction in MG at the injury site where they act in an autocrine and/or paracrine fashion to drive MG reprogramming and proliferation. Interestingly, Insulin, IGF-1 and FGF-2 stimulate MG proliferation in the postnatal chick retina (Fischer et al., 2002; Ritchey et al., 2012). However, these cells rarely survive and make new neurons; perhaps reflecting differences in growth factor-regulated signaling cascades between fish and birds.
The finding that growth factors and cytokines can stimulate each other’s expression in injury-responsive MG allows for signal amplification and versatility in response. It is interesting that MG mount a similar response to a variety of injuries regardless if these injuries are restricted to specific cell types or more widespread (Fausett and Goldman, 2006; Fimbel et al., 2007; Montgomery et al., 2010; Vihtelic and Hyde, 2000). Whether these different injury paradigms act in a similar fashion to elicit a MG response is not known; however, our data suggest that as long as a certain threshold of growth factor and/or cytokine induction is achieved that is reflected in by increased pStat3 and β-catenin expression in injury-responsive MG, a regenerative response will ensue.
Our data suggest that once MG are activated in response to injury by expressing growth factor and cytokine-like genes, these gene products then act in an autocrine/paracrine manner to drive MG reprogramming and proliferation. This suggests that a common set of signaling cascades activated by these growth factors and cytokines may underlie MG reprogramming and proliferation. Indeed, we found that Mapk/Erk, PI3K, β-catenin and Jak/Stat signaling pathways must be activated for MG reprogramming and proliferation in the injured retina or the uninjured retina. However, only β-catenin and pStat3 were detected in MG-derived progenitors, while pErk and pAkt appear to be restricted to other cell types. Of course we cannot rule out the possibility that increases in pErk and pAkt in reprogrammed MG remained below the limits of detection. Interestingly, pErk is localized to proliferating MG in the injured chick retina (Fischer et al., 2009) and pAkt is found in Xenopus and rodent photoreceptor outer segments and correlated with cell survival (Dilly and Rajala, 2008; Ivanovic et al., 2011; Jomary et al., 2006; Li et al., 2008; Rajala et al., 2002). It is intriguing that pErk appears confined to quiescent MG in the injured zebrafish retina, yet localizes to proliferating MG in the injured chick retina. Whether this contributes to the noted differences in progenitor survival or multipotency between these species is not known. Importantly, our data suggests a hierarchical relationship amongst regeneration-associated signaling cascades since Mapk and PI3K signaling is necessary for β-catenin and pStat3 accumulation in injury-responsive MG and their subsequent proliferation. Thus, all conditions known to stimulate MG reprogramming and the generation of multipotent progenitors converge on β-catenin and pStat3 signaling.
It is interesting that in mammals, β-catenin has been associated with a very limited amount of MG proliferation while pStat3 has been associated with injury-induced gliosis (Osakada et al., 2007; Peterson et al., 2000; Rhee et al., 2013; Xue et al., 2011). Why pStat3 contributes to gliosis in mammals and regeneration in fish is not known; perhaps these two events share some common elements. Regardless, it seems likely that these different responses to injury in fish and mammals is a result of differences in the responding signaling cascades and differences in the targets they act on.
In conclusion, our studies have revealed a diverse set of conditions (injury, growth factors and cytokines) that activate a common core set of signaling pathways that are necessary for MG reprogramming and retina regeneration. We found that injury-responsive MG secrete a variety of factors that drive their own reprogramming and proliferation which, along with extensive crosstalk among these factors, allows for signal amplification and localization. These aspects of retina regeneration in fish provide a robust system that is able to respond to a variety of insults. The signaling pathways uncovered in this study represent new targets for stimulating MG reprogramming and retina regeneration in mammals.
Experimental Procedures
Animals
Zebrafish were kept at 26–28°C with a 10/14 hr light/dark cycle. Adult fish from 6 to 12 months of age were used for experiments. 1016 tuba1a:gfp fish and Tg(hsp70l:dnfgfr-EGFP) fish were previously described (Fausett and Goldman, 2006; Lee et al., 2005). The gfap:stat3-gfp transgenic fish are described in the accompanying paper by Zhao et al.
Fluorescence-activated cell sorting (FACS)
FACS was performed as previously described (Ramachandran et al., 2010a). Briefly, GFP+ MG were purified from 4 uninjured retinas of gfap:gfp transgenic fish and 25 injured retinas of 1016 tuba1a:gfp transgenic fish whose retinas received 10 lesions by needle poke. Retinas were collected in 0.5 ml L15 medium, treated with hyaluronidase and dissociated in trypsin with frequent trituration. A single-cell suspension was confirmed by microscopy and cells were sorted on a BC Biosciences FACSViDa 3 laser high speed cell sorter.
RNA isolation and PCR
PCR primers are listed in Table S1. Total RNA was isolated using Trizol (Invitrogen). cDNA synthesis and PCR reactions were as previously described (Fausett et al., 2008; Ramachandran et al., 2010a). Real-time qPCR reactions were carried out in triplicate with ABsolute SYBR Green Fluorescein Master Mix (Thermo Scientific) on an iCycler real-time PCR detection system (BioRad). The ΔΔCt method was used to determine relative expression of mRNAs in control and injured retinas and normalized to gapdh mRNA levels.
Retinal injections and BrdU incorporation
Fish were anesthetized in tricaine and the left eye (control) was injected with 1 µl of vehicle (PBS plus 0.1% BSA) and the right eye was injected with one or more of the following recombinant human growth factors: HB-EGF (R&D Systems), FGF-2 (R&D Systems), IGF-1 (R&D Systems), Insulin (Gibco, pH7.6), IL-11 (R&D Systems), Leptin (Amylin Pharmaceutical/BMS). We intravitreally injected 0.5–2 µl of recombinant protein through the front of the eye. This was accomplished by first making a small incision with either a double-edge sapphire blade (World Precision Instruments, Inc.) or a 30 gauge beveled needle attached to a Hamilton syringe. If a sapphire blade was used to make the incision, a Hamilton syringe equipped with a blunt 33 gauge needle was used to deliver molecules behind the lens. If a Hamilton syringe equipped with a 30 gauge beveled needle was used to make an incision, recombinant molecules were delivered through this needle. Similar results were obtained regardless of the method used for intravitreal injection. Estimates of growth factor and cytokine intravitreal concentrations are reported in Table S2. Recombinant proteins were injected once daily for 3 days, and 4 days after the first injection, fish received an intraperitoneal injection of BrdU (20 µl of 20 mM stock) 3 hr prior to sacrifice. Experimenters remained blind to the material injected into the vitreous until after data analysis. For lineage tracing, fish retinas were injured and then fish received in i.p. injection of BrdU at 4 dpi before being sacrificed 10 days later. All studies were repeated at least 3 times.
Retinal lesion and morpholino (MO)-mediated gene knockdowns
Retinas were injured and electroporated with MOs as previously described (Fausett et al., 2008; Ramachandran et al., 2010a). Briefly, fish were anesthetized and the right retina was poked 4 times, once in each quadrant, using a 30 gauge needle inserted through the sclera to the length of the bevel (~5mm). Approximately 0.5 µl of Lissamine-MOs (Gene Tools, LLC) were delivered at the time of injury using the same needle to poke the retina. MO uptake by cells was facilitated by electroporation (Fausett et al., 2008; Ramachandran et al., 2010a). The insra MO is CAAAGTCCGCAGCCGCATTTTGACC and the insrb MO is TGTGTCCAGCCGCATTCTGCCTCGC. The igfbp3, igf1ra and igf1rb MOs were previously characterized and published (Schlueter et al., 2006; Zhong et al., 2011).
Signaling pathway inhibitors
Control fish were treated with DMSO (1:200). Mapk/Erk inhibitor, UO126 (Tocris Bioscience); PI3K/Akt inhibitor, Ly294002 (Cayman Chemical) and PI-103 (Tocris); EGF receptor inhibitor, PD153035; β-catenin inhibitor, pyrvinium (Sigma); and Jak/Stat inhibitor JSI-124 (Tocris) were used in this study. Fish were immersed in fish water containing the inhibitor (10 µM) or injected intravitreally through the front of the eye (1 µl of 1 µM).
Tissue preparation, immunohistochemistry and in situ hybridization
Adult fish were overdosed with tricaine and the eyes were dissected; the lens was removed and the eye cup was fixed in 4% paraformaldehyde. Fixed samples were prepared for immunofluorescence as previously described (Fausett and Goldman, 2006; Ramachandran et al., 2010a; Ramachandran, 2010). For pErk and pAkt immunofluorescence, retinas were fixed in 2% paraformaldehyde for 20 min. Anti-pAkt (Thr308) (Sigma) and anti-pErk (Thr202/Tyr204) antibodies (Cell Signaling) were used at 1:50 dilution. The anti-BrdU, anti-β-catenin and retinal cell type-specific antibodies used in this study were previously described (Fausett and Goldman, 2006; Ramachandran et al., 2010a; Ramachandran, 2010). For anti-BrdU immunofluorescence, sections were treated with 2N HCL at 37°C for 20 min, rinsed with 0.1M sodium borate solution (pH 8.5) for 10 min, and then processed using standard immunohistochemical procedures. In situ hybridization was performed as described previously (Barthel and Raymond, 2000). Digoxigenin-labeled RNA probes were prepared using the DIG RNA labeling kit (Roche Diagnostics).
TUNEL
We used an in situ Cell Death Detection Kit (TMR red; Applied Science) to detect apoptosis cells. Insulin (2 µg) or FGF2 (50 ng)/IGF-1 (50 ng) was delivered intravitreally through the front of the eye for 3 consecutive days and 1 day after the 3rd injection, eyes were collected, sectioned and used in a TUNEL assay according to the manufacturer's protocol. Eyes intravitreally injected of 3µM ouabain were included as a positive control (Fimbel et al., 2007).
Heat shock
The day before retinal injury, WT and Tg(hsp70l:dnfgfr-EGFP) fish were immersed in a water bath at 37.5°C for 2 hours and then returned to system water at 28 °C. Over the next 4 days, heat shock was performed at 37.5 °C for 1h and repeated three times per day.
Cell quantification and statistical analysis
Cell counts were determined by counting BrdU+ or GFP+ cells in retinal sections visualized using fluorescence microscopy. All experiments were performed at least in triplicate and repeated at least twice. Non-parametric Mann-Whitney analysis was used with an n of only 3, ANOVA/post-hoc test was applied for multiple comparisons and a two tailed unpaired Student t test was used for single comparison. Error bars are standard deviation (SD).
Microscopy
Imagines were captured by a Zeiss Axiophot fluorescence microscope or an Olympus FluoView FV1000 confocal microscope.
Supplementary Material
Acknowledgements
This research was supported by NEI grant RO1 EY 018132 from the NIH, a Research to Prevent Blindness Innovative Ophthalmic Research Award and a gift from the Marjorie and Maxwell Jospey Foundation. We thank Alon Kahana (University of Michigan) for sharing igfr1a, igfr1b and igfbp3-targeting MOs; ZIRC (Eugene, Oregon) for hsp70:dnfgfr1-egfp fish; Martin Meyers (University of Michigan) and Amylin Pharmaceutical/BMS for Leptin; Peter Hitchcock (University of Michigan) for 4C4 antibody; the University of Michigan Flow Cytometry Core for cell sorting; Randall Karr and Joshua Kirk for fish care and the Goldman laboratory for comments and suggestions during the course of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayaso E, Nolan CM, Byrnes L. Zebrafish insulin-like growth factor-I receptor: molecular cloning and developmental expression. Mol Cell Endocrinol. 2002;191:137–148. doi: 10.1016/s0303-7207(02)00083-7. [DOI] [PubMed] [Google Scholar]
- Barthel LK, Raymond PA. In situ hybridization studies of retinal neurons. Methods Enzymol. 2000;316:579–590. doi: 10.1016/s0076-6879(00)16751-5. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann A, Iandiev I, Pannicke T, Wurm A, Hollborn M, Wiedemann P, Osborne NN, Reichenbach A. Cellular signaling and factors involved in Muller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28:423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Dilly AK, Rajala RV. Insulin growth factor 1 receptor/PI3K/AKT survival pathway in outer segment membranes of rod photoreceptors. Invest Ophthalmol Vis Sci. 2008;49:4765–4773. doi: 10.1167/iovs.08-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, McGuire CR, Dierks BD, Reh TA. Insulin and fibroblast growth factor 2 activate a neurogenic program in Muller glia of the chicken retina. J Neurosci. 2002;22:9387–9398. doi: 10.1523/JNEUROSCI.22-21-09387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Exogenous growth factors stimulate the regeneration of ganglion cells in the chicken retina. Dev Biol. 2002;251:367–379. doi: 10.1006/dbio.2002.0813. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Scott MA, Tuten W. Mitogen-activated protein kinase-signaling stimulates Muller glia to proliferate in acutely damaged chicken retina. Glia. 2009;57:166–181. doi: 10.1002/glia.20743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauguin L, Delaine C, Alvino CL, McNeil KA, Wallace JC, Forbes BE, De Meyts P. Alanine scanning of a putative receptor binding surface of insulin-like growth factor-I. J Biol Chem. 2008;283:20821–20829. doi: 10.1074/jbc.M802620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmann S, Kaslin J, Hans S, Weber A, Machate A, Geffarth M, Funk RH, Brand M. Fgf Signaling is Required for Photoreceptor Maintenance in the Adult Zebrafish Retina. PLoS One. 2012;7:e30365. doi: 10.1371/journal.pone.0030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovic I, Allen DT, Dighe R, Le YZ, Anderson RE, Rajala RV. Phosphoinositide 3-kinase signaling in retinal rod photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:6355–6362. doi: 10.1167/iovs.10-7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomary C, Cullen J, Jones SE. Inactivation of the Akt survival pathway during photoreceptor apoptosis in the retinal degeneration mouse. Invest Ophthalmol Vis Sci. 2006;47:1620–1629. doi: 10.1167/iovs.05-1176. [DOI] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, C TB, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Kassen SC, Thummel R, Campochiaro LA, Harding MJ, Bennett NA, Hyde DR. CNTF induces photoreceptor neuroprotection and Muller glial cell proliferation through two different signaling pathways in the adult zebrafish retina. Exp Eye Res. 2009;88:1051–1064. doi: 10.1016/j.exer.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Li G, Rajala A, Wiechmann AF, Anderson RE, Rajala RV. Activation and membrane binding of retinal protein kinase Balpha/Akt1 is regulated through light-dependent generation of phosphoinositides. J Neurochem. 2008;107:1382–1397. doi: 10.1111/j.1471-4159.2008.05707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey AE, Powers MK. Visual behavior of adult goldfish with regenerating retina. Vis Neurosci. 2007;24:247–255. doi: 10.1017/S0952523806230207. [DOI] [PubMed] [Google Scholar]
- Mensinger AF, Powers MK. Visual function in regenerating teleost retina following cytotoxic lesioning. Vis Neurosci. 1999;16:241–251. doi: 10.1017/s0952523899162059. [DOI] [PubMed] [Google Scholar]
- Meyers JR, Hu L, Moses A, Kaboli K, Papandrea A, Raymond PA. beta-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012;7:30. doi: 10.1186/1749-8104-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, Hyde DR. A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J Comp Neurol. 2010;518:800–814. doi: 10.1002/cne.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O'Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR. Stat3 defines three populations of Muller glia and is required for initiating maximal muller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520:4294–4311. doi: 10.1002/cne.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, De Sena G, Presta M. Fibroblast growth factor 2-induced angiogenesis in zebrafish: the zebrafish yolk membrane (ZFYM) angiogenesis assay. J Cell Mol Med. 2009;13:2061–2068. doi: 10.1111/j.1582-4934.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasani MR, Robison BD, Hardy RW, Hill RA. Early developmental expression of two insulins in zebrafish (Danio rerio) Physiol Genomics. 2006;27:79–85. doi: 10.1152/physiolgenomics.00012.2006. [DOI] [PubMed] [Google Scholar]
- Peterson WM, Wang Q, Tzekova R, Wiegand SJ. Ciliary neurotrophic factor and stress stimuli activate the Jak-STAT pathway in retinal neurons and glia. J Neurosci. 2000;20:4081–4090. doi: 10.1523/JNEUROSCI.20-11-04081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Kidd AR, Thomas JL, 3rd, Poss KD, Hyde DR, Raymond PA, Thummel R. FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp Eye Res. 2011;93:726–734. doi: 10.1016/j.exer.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala RV, McClellan ME, Ash JD, Anderson RE. In vivo regulation of phosphoinositide 3-kinase in retina through light-induced tyrosine phosphorylation of the insulin receptor beta-subunit. J Biol Chem. 2002;277:43319–43326. doi: 10.1074/jbc.M206355200. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010a;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Reifler A, Parent JM, Goldman D. Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J Comp Neurol. 2010;518:4196–4212. doi: 10.1002/cne.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D. Ascl1a/Dkk/{beta}-catenin signaling pathway is necessary and glycogen synthase kinase-3{beta} inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D. Insm1a–mediated gene repression is essential for the formation and differentiation of Muller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14:1013–1023. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, Bringmann A. New functions of Muller cells. Glia. 2013;61:651–678. doi: 10.1002/glia.22477. [DOI] [PubMed] [Google Scholar]
- Renteria ME, Gandhi NS, Vinuesa P, Helmerhorst E, Mancera RL. A comparative structural bioinformatics analysis of the insulin receptor family ectodomain based on phylogenetic information. PLoS One. 2008;3:e3667. doi: 10.1371/journal.pone.0003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KD, Nusinowitz S, Chao K, Yu F, Bok D, Yang XJ. CNTF-mediated protection of photoreceptors requires initial activation of the cytokine receptor gp130 in Muller glial cells. Proc Natl Acad Sci U S A. 2013;110:E4520–E4529. doi: 10.1073/pnas.1303604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey ER, Zelinka CP, Tang J, Liu J, Fischer AJ. The combination of IGF1 and FGF2 and the induction of excessive ocular growth and extreme myopia. Exp Eye Res. 2012;99:1–16. doi: 10.1016/j.exer.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter PJ, Royer T, Farah MH, Laser B, Chan SJ, Steiner DF, Duan C. Gene duplication and functional divergence of the zebrafish insulin-like growth factor 1 receptors. Faseb J. 2006;20:1230–1232. doi: 10.1096/fj.05-3882fje. [DOI] [PubMed] [Google Scholar]
- Sherpa T, Fimbel SM, Mallory DE, Maaswinkel H, Spritzer SD, Sand JA, Li L, Hyde DR, Stenkamp DL. Ganglion cell regeneration following whole-retina destruction in zebrafish. Dev Neurobiol. 2008;68:166–181. doi: 10.1002/dneu.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y, Monson C, Duan C, Wu Y, Gao C, Yakar S, Sadler KC, LeRoith D. The role of insulin receptor signaling in zebrafish embryogenesis. Endocrinology. 2008;149:5996–6005. doi: 10.1210/en.2008-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AW, Duan C, Bern HA. Insulin-like growth factor signaling in fish. Int Rev Cytol. 2005;243:215–285. doi: 10.1016/S0074-7696(05)43004-1. [DOI] [PubMed] [Google Scholar]
- Xue W, Cojocaru RI, Dudley VJ, Brooks M, Swaroop A, Sarthy VP. Ciliary neurotrophic factor induces genes associated with inflammation and gliosis in the retina: a gene profiling study of flow-sorted, Muller cells. PLoS One. 2011;6:e20326. doi: 10.1371/journal.pone.0020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Lu L, Zhou J, Li Y, Liu Y, Clemmons DR, Duan C. IGF binding protein 3 exerts its ligand-independent action by antagonizing BMP in zebrafish embryos. J Cell Sci. 2011;124:1925–1935. doi: 10.1242/jcs.082644. [DOI] [PubMed] [Google Scholar]
- Zou S, Kamei H, Modi Z, Duan C. Zebrafish IGF genes: gene duplication, conservation and divergence, and novel roles in midline and notochord development. PLoS One. 2009;4:e7026. doi: 10.1371/journal.pone.0007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.