Summary
Age-associated thymic involution results in diminished T cell output and function in aged individuals. However, molecular mediators contributing to the decline in thymic function during early thymic involution remain largely unknown. Here we present transcriptional profiling of purified thymic stromal subsets from mice 1, 3, and 6 months of age, spanning early thymic involution. The data implicate novel biological functions for a subset of thymic epithelial cells. The predominant transcriptional signature of early thymic involution is decreased expression of cell cycle associated genes and E2F3 transcriptional targets in thymic epithelial subsets. Also, expression of pro-inflammatory genes increases with age in thymic dendritic cells. Many genes previously implicated in late involution are already deregulated by 3 to 6 months of age. We provide these thymic stromal datasets, along with thymocyte datasets, in a readily searchable web-based platform, as a resource for investigations into thymocyte: stromal interactions and mechanisms of thymic involution.
Introduction
The thymus is spatially organized into cortical and medullary regions, containing heterogeneous stromal cells, including thymic epithelial cells (TECs), dendritic cells, macrophages, fibroblasts, and endothelial cells (Love and Bhandoola, 2011). As thymocytes mature, they migrate through distinct thymic microenvironments, where they undergo bi-directional crosstalk with local stromal cells, essential for the generation of a diverse, and self-tolerant T cell pool (Petrie and Zúñiga-Pflücker, 2007). Signals provided by developing thymocytes are also required for differentiation and maintenance of thymic stromal cells (Nitta et al., 2011). While some molecular signals responsible for this bidirectional signaling have been characterized, many remain to be identified.
Thymocyte:stromal cell crosstalk first occurs in the cortex where thymocyte progenitors encounter cortical TECs (cTECs) that express NOTCH1 ligands, SCF, and IL-7, which are essential for thymocyte survival, proliferation and commitment to the T cell lineage (Anderson and Takahama, 2012; Petrie and Zúñiga-Pflücker, 2007). In addition, cTECs display self-peptide:MHC complexes that promote positive selection of self-MHC restricted thymocytes, and apoptosis of autoreactive cells (McCaughtry et al., 2008). Reciprocally, unidentified signals from early thymocyte progenitors are critical for cTEC differentiation (Klug et al., 1998).
Following positive selection, thymocytes migrate into the medulla where they interact with medullary thymic epithelial cells (mTECs). mTECs can be subdivided into mTEClo and mTEChi subsets, based on differential expression of CD80 and MHC class II. The mTEChi subset expresses the chromatin modulator AIRE, which promotes expression of tissue-restricted antigens (TRAs), genes otherwise expressed in a limited number of differentiated tissues, such as the pancreas or retina (Anderson et al., 2002; Klein et al., 2011). When medullary thymocytes engage TRAs on mTECs, they undergo apoptosis or differentiate into regulatory T cells, thus establishing central tolerance to peripheral self-antigens. Conversely, mTEClo cells must engage thymocytes entering the medulla, via tumor necrosis factor superfamily members, to drive differentiation to the mTEChi stage (Nitta et al., 2011). Thus, bi-directional signaling in the medulla between TECs and maturing thymocytes is critical for thymocyte tolerance and medullary stromal organization.
Thymic dendritic cells also play a critical role in central tolerance. Conventional thymic dendritic cells can be subdivided into Sirpα −CD8+CD11b− (DC) and Sirpα+CD8−CD11b+ (DCS) subsets (Li et al., 2009). Thymic dendritic cells can acquire TRAs from mTECs to mediate deletion of autoreactive thymocytes (Klein et al., 2011). In addition, DCS traffic peripheral antigens into the thymus to mediate negative selection or induction of regulatory T cells (Bonasio et al., 2006; Proietto et al., 2008). Thymic dendritic cells require chemotactic signals from mTECs to accumulate in the medulla and function properly (Lei et al., 2011), underscoring the complex interplay between thymocytes and various stromal subsets required to ensure production of a self-tolerant T cell repertoire.
The thymus involutes in an age-dependent manner, resulting in diminished TEC cellularity and turn-over (Gray et al., 2006), disrupted thymic architecture, decreased thymic output, and reduced T cell function (Chinn et al., 2012; Haynes and Maue, 2009; Nikolich-Žugich et al., 2012). Both hematopoietic age-related dysfunction and degeneration of the thymic stromal compartment likely contribute to thymic involution (Berent-Maoz et al., 2012; Chinn et al., 2012). While reduced levels of the transcription factor Foxn1 contribute to TEC atrophy (Chen et al., 2009), and genetic manipulation of cell-cycle regulators can maintain thymic mass in aged mice (Garfin et al., 2013; Robles et al., 1996), specific molecular pathways driving degeneration of the thymic stroma early in the process of involution remain to be discerned. Furthermore, while manipulation of sex steroids or growth factor levels in aged individuals can transiently increase thymic size (Min et al., 2007; Sutherland et al., 2005), the resultant thymi are not functionally equivalent to young thymi, and may be incapable of maintaining central tolerance (Griffith et al., 2012). Thus, identifying molecular pathways that regulate stromal changes associated with involution remains an important goal.
Here we present global transcriptional profiling of purified thymic stromal subsets from mice at 1, 3, and 6 months of age, enabling the community to query stromal subset-specific gene expression before and during early thymic involution. Griffith et al. previously reported transcriptional profiles of non-sorted thymic stroma from the cortex, medulla and cortico-medullary junction, enabling evaluation of regionalized gene expression in young and aged thymi (Griffith et al., 2012; 2009). The current resource provides complementary information about gene expression in specific stromal cell types during early thymic involution. Our data reveal predicted, as well as unexpected expression of genes in different thymic stromal subsets that suggest novel stromal cell functions. Interestingly, decreased expression of cell-cycle genes and downregulation of E2F3 activity are major hallmarks of early thymic involution in the TEC compartment. Upregulation of proinflammatory genes in thymic dendritic cell subsets also occurs over this time course. Analysis of genes previously implicated in late thymic involution reveals some of these factors may contribute to early involution, while others are likely sequelae of the aging process. To facilitate use of this resource, we provide our thymic stromal gene expression data, along with our previous thymocyte transcriptional profiling data, as an in silico model in the readily searchable web-based platform Gene Expression Commons (GExC) (Seita et al., 2012) (https://gexc.stanford.edu/model/475). Exploration of our datasets on this user-friendly interface will enhance elucidation of the molecular pathways mediating thymocyte-stromal cell crosstalk and age-associated thymic stromal changes.
Results
Transcriptional profiling of thymic stromal subsets at 1, 3, and 6 months of age
We FACS purified thymocyte stromal subsets from mice over the course of early thymic involution to identify subtype specific and age-regulated transcriptional profiles. Thymic cTEC, mTEClo, mTEChi, DC, DCS, and fibroblast populations were FACS purified from C57BL/6J male mice at 1, 3 and 6 months of age as follows: cTEC (Epcam+CD11c−UEA1−MHCII+Ly51+), mTEClo(Epcam+CD11c−UEA1+MHCIIloCD80lo), mTEChi (Epcam+CD11c−UEA1+MHCIIhiCD80hi), DC (MHCII+Epcam−CD11c+Sirpα− CD80+), DCS (MHCII+Epcam−CD11c+Sirpα+CD80+), and fibroblasts (MHCII−CD45− Ter119−CD31−) (Figure 1A). During the sort setup, we ensured that all Epcam+ TECs were included in the initial MHCII+ gate (Figure S1A). Purified mRNA from biological duplicates of each subset at each age was analyzed on Affymetrix Mouse 430 2.0 microarrays. Resultant data were uploaded to the GExC platform for normalization (Seita et al., 2012), and resultant normalized signal intensity values were used for all subsequent bioinformatics analyses herein.
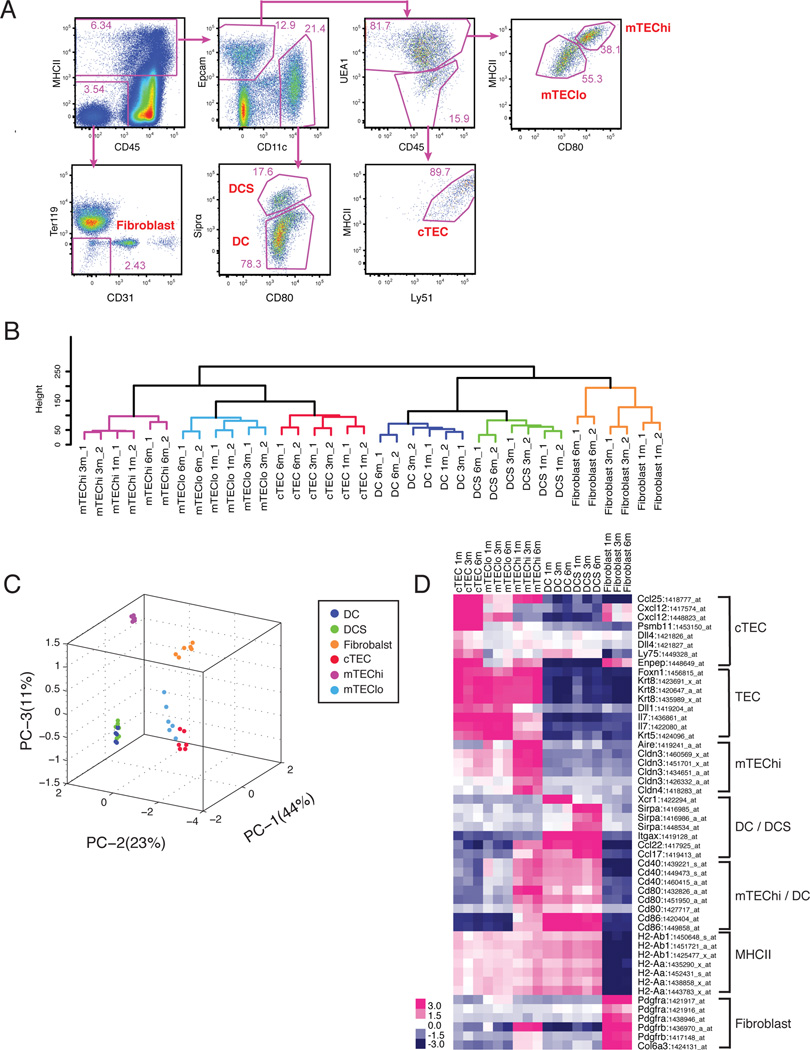
Figure 1. Expression profiling of thymic stromal subsets.
(A) Strategy for sorting thymic stromal subsets (gated on live events; frequencies within gates displayed). (B) Hierarchical clustering and (C) PCA were carried out on the top 30% of genes most variable in expression. Each symbol in the PCA represents a single array. (D) Expression of known thymic stromal subset-specific genes is presented as a heat map, using the average of biological duplicates for each subset. Probesets with a dynamic range ≥ 3 (in GExC) were displayed if multiple probesets were present for one gene.
To assess consistency between duplicate datasets and to globally compare transcriptional profiles between stromal subsets, we performed unsupervised hierarchical clustering and principal components analysis (PCA) on the 30% of genes most variable in expression across subsets (Figure 1B and 1C). Duplicate datasets clustered closely together, indicating reliability of the data. Stromal subtypes clustered together regardless of age (Figure 1B), demonstrating that each stromal subset maintains its transcriptional identity during thymic involution. Stromal subsets at 6 months of age were further from their 1 and 3 month counterparts, indicative of age-related transcriptional changes in the data. Interestingly, age-associated changes were implicated not only in TEC subsets, which have been suggested to be drivers of involution, but also in dendritic cells and fibroblasts. Overall, PCA was consistent with hierarchical clustering (Figure 1C), and showed that mTEChi were distal to other TEC subsets, likely due to expression of diverse TRAs. As expected, dendritic cell subtypes clustered together, while fibroblasts were distinct from other stromal subsets.
To validate our data, we queried genes previously reported to be differentially expressed in distinct stromal subsets (Figure 1D). TECs expressed cytokeratins and the transcription factor Foxn1, confirming their identity (Blackburn et al., 1996; Klug et al., 1998). As expected, cTEC and mTEClo expressed high levels of Il7 (Repass et al., 2009; Ribeiro et al., 2013), while Ccl25, Cxcl12, Dll4, Enpep/Ly51, Ly75/CD205, and Psmb11/β5t were predominantly expressed by cTECs (Murata et al., 2007; Plotkin et al., 2003; Wurbel et al., 2000). Aire, Cldn3, and Cldn4 were expressed specifically in mTEChi cells, as expected (Hamazaki et al., 2007). MHCII genes were expressed by dendritic cells and TECs, consistent with their roles in antigen presentation, while dendritic cells and mTEChi expressed high levels of Cd80 and Cd86. ItgaX (CD11c) was expressed uniquely by dendritic cells, with Sirpα and Xcr1 (Lei et al., 2011) expressed uniquely in the DCS and DC subsets, respectively. Fibroblasts expressed Pdgfrα and Pdgfrβ, and the extracellular matrix (ECM) component Col6a3, as anticipated for mesenchymal cells (Foster et al., 2008). To further validate our data, we sorted an additional biological replicate of thymic stromal subsets from mice 1 month of age, and confirmed subset-specific expression of 25 genes by qRT-PCR analysis on the Fluidigm platform (Figure S1B). Together, these analyses validate the datasets, and demonstrate that thymic stromal subset identity is maintained during aging.
Differentially expressed genes suggest distinct functions of thymic stromal subsets
We identified differentially expressed genes (DEGs) in each stromal subset as those genes uniquely up- or down-regulated by >2 fold relative to all other stromal subsets of the same age, with an adjusted p-value <0.01 (Table S1). As expected from PCA analysis, mTEChi and fibroblast subsets had the most unique DEGs at each age (Figure 2A). We next investigated common functional characteristics of unique DEGs in each subset. The top Gene Ontology (GO) term hit for DEGs from 1-month old fibroblasts was ‘cell adhesion’, with ‘extracellular matrix’ appearing as well, suggesting that although fibroblasts were sorted based on the absence of markers in other subsets, the population is greatly enriched in mesenchymal cells. A heat map of genes in the GO term ‘cell adhesion’ category, reveals ECM components and integrins that are likely relevant substrates for adhesion and migration in the thymus (Figure 2B).
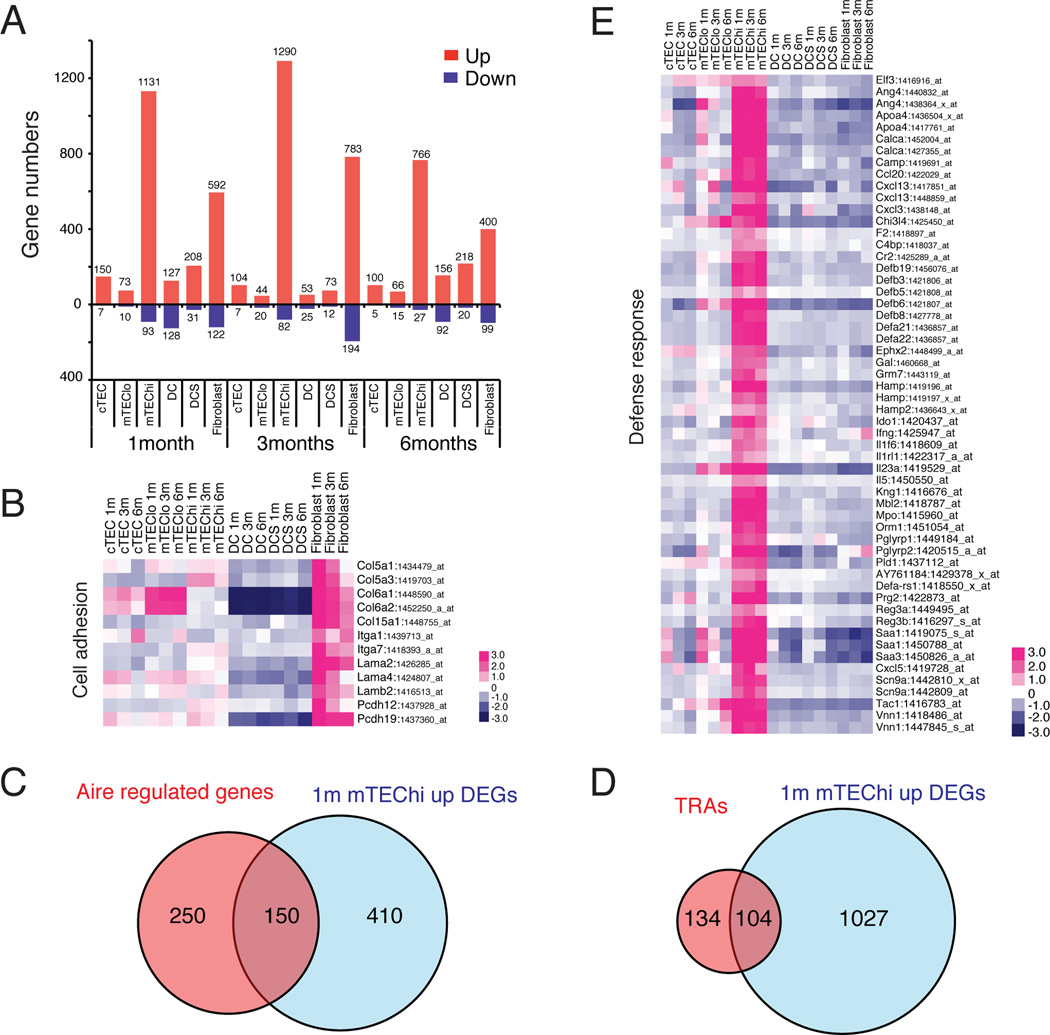
Figure 2. Mesenchymal signature of fibroblasts and defense response function of mTEChi revealed by DEGs.
(A) The number of stromal subset-specific DEGs, identified by pair-wise comparisons between stromal subsets of the same age, is displayed. (B) The heat map displays the relative expression of select up-regulated DEGs from 1-month fibroblasts that overlap with the “cell adhesion” GO term (the heat map of all overlapping genes is in Figure S2A). (C) Overlap between Aire-regulated genes (Derbinski, 2005) and 1 month mTEChi up-regulated DEGs, shared between microarray platforms. (D) Overlap between TRAs (Griffith et al., 2012) and 1 month mTEChi up-regulated DEGs. (E) A heat map displaying the relative expression of up-regulated mTEChi DEGs that overlap with the ‘defense response’ GO term hit.
The large number of DEGs and dissimilarity in overall transcriptional relatedness of mTEChi relative to other stromal subsets (Figures 1C and 2A) may have been due to expression of diverse TRAs (Derbinski et al., 2001). To address this possibility, we identified Aire-regulated genes by analyzing published expression data from Aire deficient versus wild-type mTEChi cells (Derbinski, 2005). After excluding genes not shared between microarray platforms, we compared the list of Aire-regulated genes to mTEChi up-regulated DEGs. Of the 560 mTEChi up-regulated DEGs, only 150 (37%) were Aire-regulated (Figure 2C), suggesting many mTEChi DEGs are Aire-independent. Furthermore, comparison against a list of TRAs identified on the basis of limited tissue expression (Griffith et al., 2012), revealed overlap with only 9% of mTEChi DEGs (Figure 2D). Thus, many mTEChi DEGs may have a biological function, rather than serving as TRAs. Interestingly, the top GO hit term for 1 month mTEChi DEGs was ‘defense response’ (p-value: 8×10−7), which included chemokines (Ccl20, Cxcl13, Cxcl3), defensins (Defb19, Defb3, Defb5, Defb6, Defb8, Defa21, Defa22), and cytokines (Il1f6, Il23a, Il5) (Figure 2E). Only 15 of 55 defense response genes were identified as TRAs (not shown). Thus, mTEChi cells may promote defense responses in the thymus. Consistent with a unique biological state of the mTEChi subset, AIRE has been shown to regulate differentiation of mTEChi cells (Yano et al., 2008).
Because of the transcriptional relatedness of cTEC to mTEClo and DC to DCS (Figure 1B–C), few DEGs were identified in these subsets (Figure 2A). Although these DEGs are likely important for the unique biology of these subsets (Figure S2B and TableS1), the number of DEGs was too low to proceed with downstream bioinformatic analyses. Therefore, we identified additional DEGs in cTEC after omitting mTEClo, and in DC after omitting DCS from the comparisons, and vice versa (Figure S3). GO analyses of these DEGs confirmed epithelial characteristics of cTEC and mTEClo subsets (Figure 3A). The “epithelium development” and “cell adhesion” terms contained previously known as well as unreported genes in cTEC and mTEClo subsets (Figures 3B and S4), including several genes involved in Wnt signaling, such as Wnt4 and the receptors Fzd2 and Fzd3 (consistent with Bredenkamp et al., 2014; Griffith et al., 2012). Fgfr2 likely regulates TEC cellularity, as its ligand KGF causes transient thymic regeneration in aged mice (Min et al., 2007). Cldn8 was expressed in cTECs, while Cldn9 and Cldn12 were expressed in cTEC and mTEClo subsets, demonstrating TEC subset specificity of claudin family members, consistent with expression of Cldn3 and Cldn4 in mTEChi cells (Table S1) (Hamazaki et al., 2007). The top hit from GO analyses of DC and DCS DEGs was ‘immune response’ (Figure 3A), including Toll like receptors, cytokines, and chemokines (Figures 3C and S5), consistent with the central role for dendritic cells in eliciting and modulating immune responses.
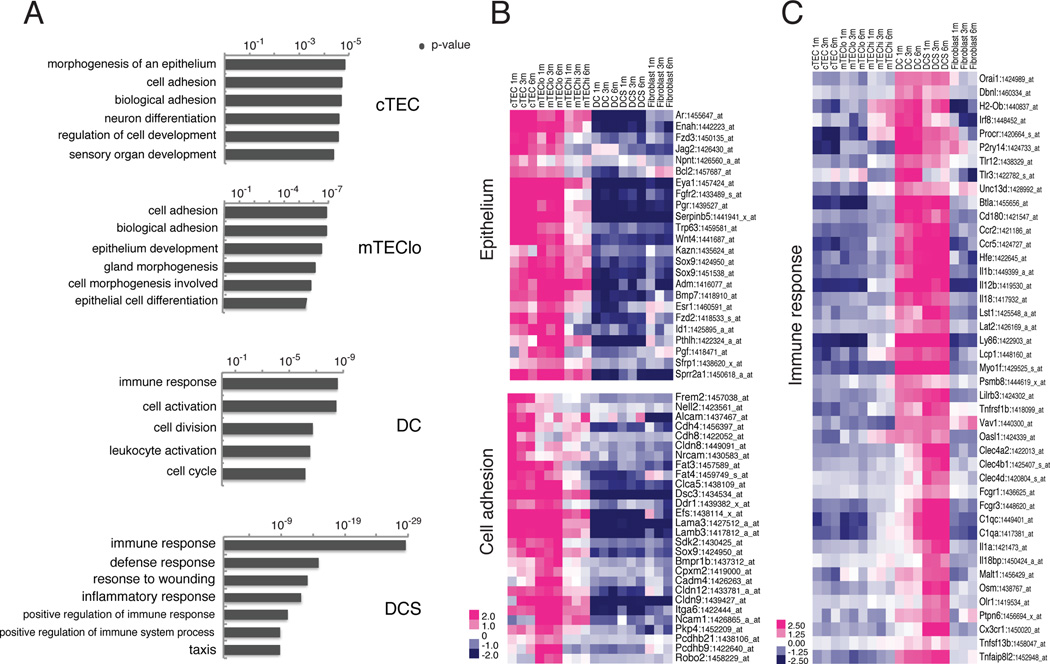
Figure 3. Epithelial identity of cTEC/mTEClo and immune response function of DC/DCS revealed by DEGs.
(A) GO term analyses of DEGs from cTEC/ mTEClo confirm the epithelial identity of these subsets. GO term analysis of DEGs identified in DC/ DCS reveal immune response and inflammatory signatures. See Figure S3. (B) The heat map displays the relative expression of select up-regulated DEGs in cTEC/ mTEClo that overlap with the ‘morphogenesis of an epithelium’ and ‘epithelium development’ (top) and ‘cell adhesion” (bottom) GO term hits (the heat map of all overlapping genes is in Figure S4). (C) The heat map displays the relative expression of select up-regulated DEGs in DC/ DCS that overlap with the ‘immune response’ GO term hit (the heat map of all overlapping genes is in Figure S5).
Thymic involution is associated with down-regulation of cell cycle genes in the mTEClo subset and decreased activity of E2F3 in cTEC and mTEClo cells
Thymocyte and thymic stromal cellularity are greatest in mice around 1 month of age and subsequently decline as the thymus involutes (Gray et al., 2006). To evaluate transcriptional changes associated with early thymic involution, we compared gene expression in stromal subsets from mice at 1, 3, and 6 months of age. We identified aging-associated DEGs as those genes modulated by ≥ 2 fold, with an adjusted p-value ≤ 0.01 in pairwise comparisons of each subset at the three ages. Using these criteria, no aging-associated DEGs were identified in fibroblasts or cTECs, with only 1 DEG in mTEChi (Igha). In contrast, a significant number of DEGs were identified in DC, DCS, and mTEClo subsets (Figure 4A, Table S2). Over 90% of the DEGs in DC and DCS were up-regulated with age, whereas DEGs in mTEClo were both up- and down-regulated with similar frequencies. We conducted K-means clustering on DEGs from each subset to identify groups of genes up- or down-regulated as aging progresses from 1 through 6 months of age. Cluster 3 from mTEClo contained genes progressively and significantly down-regulated with age (Figure 4B–C). We performed GO term analyses on each cluster to identify biological functions associated with age-related changes, and found significant hits only for mTEClo cluster 3. Genes in this cluster were associated with the cell cycle (Figure 4D), implicating a progressive decrease in expression of cell cycle regulators, such as Aurkb, Cdc20, Cdc6, and Ccnb1, in mTEClo during involution (Figure 4E).
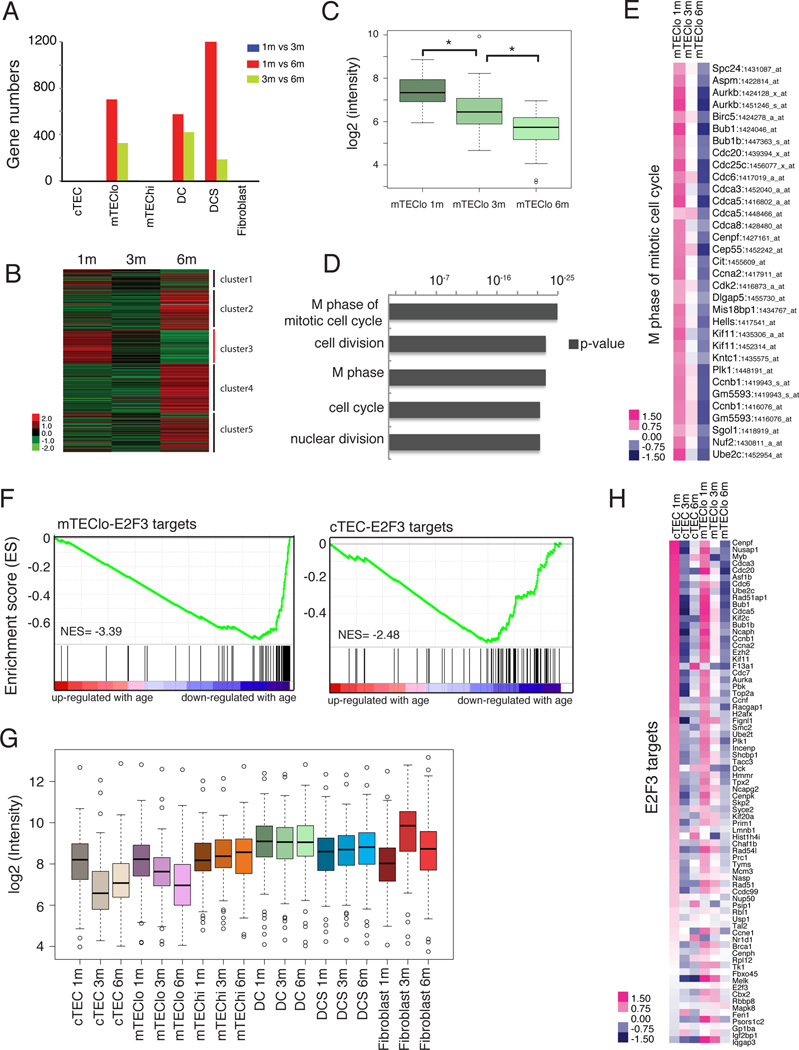
Figure 4. Cell cycle genes and E2F3 target genes are significantly down-regulated in mTEClo and cTEC subsets in early thymic involution.
(A) The number of up- or down-regulated DEGs identified from pairwise comparisons of stromal subsets at different ages is shown. (B) The heat map displays K-means clustering of all age-associated DEGs identified in the mTEClo subset. (C) The boxplot displays expression levels of mTEClo DEGs in cluster 3 from B, which are progressively down-regulated with age (p-value < 2.2 × 10−16). (D) The top 5 GO term hits from cluster 3 of mTEClo DEGs reveal down-regulation of cell-cycle associated genes during aging. (E) The heat map displays the relative expression of down-regulated age-associated DEGs in mTEClo that overlap with the ‘M phase of mitotic cell cycle’ GO term. (F) GSEA reveals expression of E2F3 target genes is down-regulated with age in mTEClo and cTEC subsets. (cTEC NOM p-value < 10−3 NES=−2.48; mTEClo NOM p-value < 10−3, NES=− 3.39) (G) The boxplot shows expression levels of E2F3 target genes in stromal subsets at 1, 3, and 6 months of age. Decreased expression of E2F3 target genes is apparent in aging cTEC and mTEClo subsets. (H) Relative expression of E2F3 target genes in cTEC and mTEClo subsets at 1, 3, and 6 months of age.
We next utilized gene set enrichment analysis (GSEA) to identify aging-associated gene sets, without imposing an arbitrary fold-change cutoff. Strikingly, E2F3 target genes (Kong et al., 2007) were significantly down-regulated with age in both cTEC and mTEClo subsets (Figure 4F). This decline was gradual in mTEClo cells from 1 to 6 months, but precipitous in cTECs between 1 and 3 months (Figure 4G). E2F3 is a transcription factor critical for normal cellular proliferation (Humbert et al., 2000), and many target genes are regulators of the cell cycle, such as Cdc6, Ccna2, Aurka, and Cdc7 (Figure 4H). Together, these analyses suggest that a decline in E2F3 activity results in decreased cell-cycle progression in cTEC and mTEClo cells, likely contributing to the decline in TEC cellularity early in the process of thymic involution.
DC and DCS have an increasingly pro-inflammatory signature with age
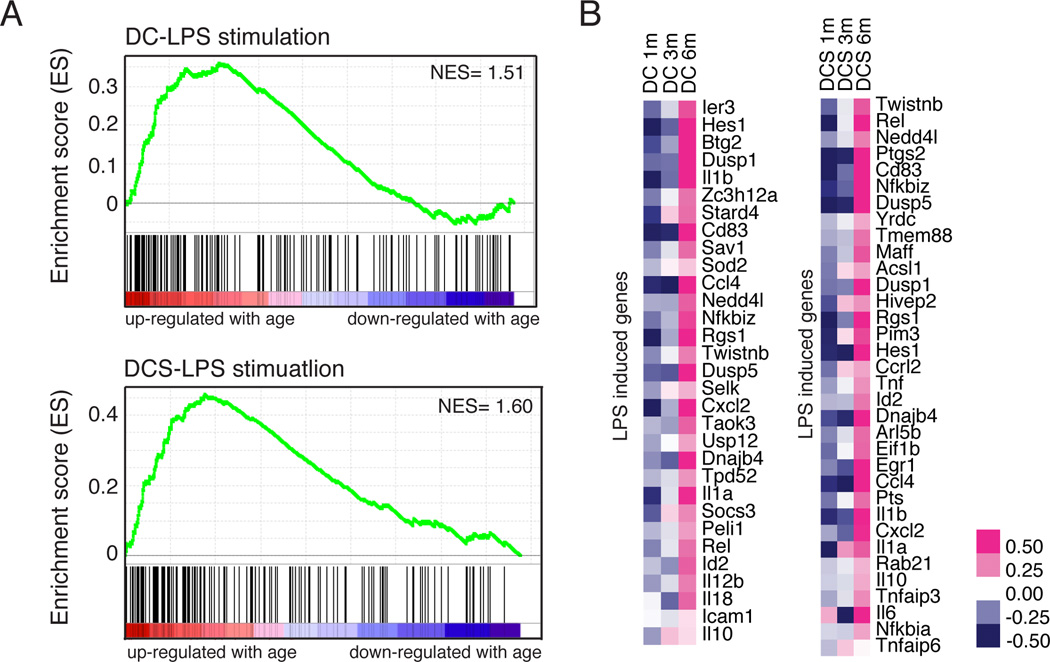
Although GO term analyses did not yield significant hits for aging-associated DEGs in DC and DCS subsets, GSEA revealed an increasingly pro-inflammatory signature with age. Genes induced after LPS simulation in human monocytes were up-regulated with age in thymic DC and DCS (Figure 5A) (Dower et al., 2008). Aging is associated with increased inflammation (Chung et al., 2009) and inflammatory cytokines become elevated in human thymi with age (Sempowski et al., 2000). Both previously described and unreported pro-inflammatory molecules, including Il1a, Il1b, Cxcl2, Il-6, Il12b, Il18, and Tnf were upregulated in thymic dendritic cells at 6 months of age (Figure 5B), suggesting that an increasingly inflammatory environment generated by aging dendritic cells contributes to early thymic involution.
Figure 5. Increased expression of pro-inflammatory genes in aging DC and DCS subsets.
(A) GSEA analysis reveals a positive correlation between LPS stimulated genes and genes up-regulated with age in DC and DCS (NOM p-value=0.054, NES=1.51 for DC, NOM p-value=0.025, NES=1.6 for DCS). (B) The heat map displays the relative expression of some LPS induced genes that are positively correlated with aging in DC and DCS (the heat map of all overlapping genes is in Figure S6).
Diminished niche activity, declining TEC homeostasis, and a decrease in growth factors are associated with early thymic involution
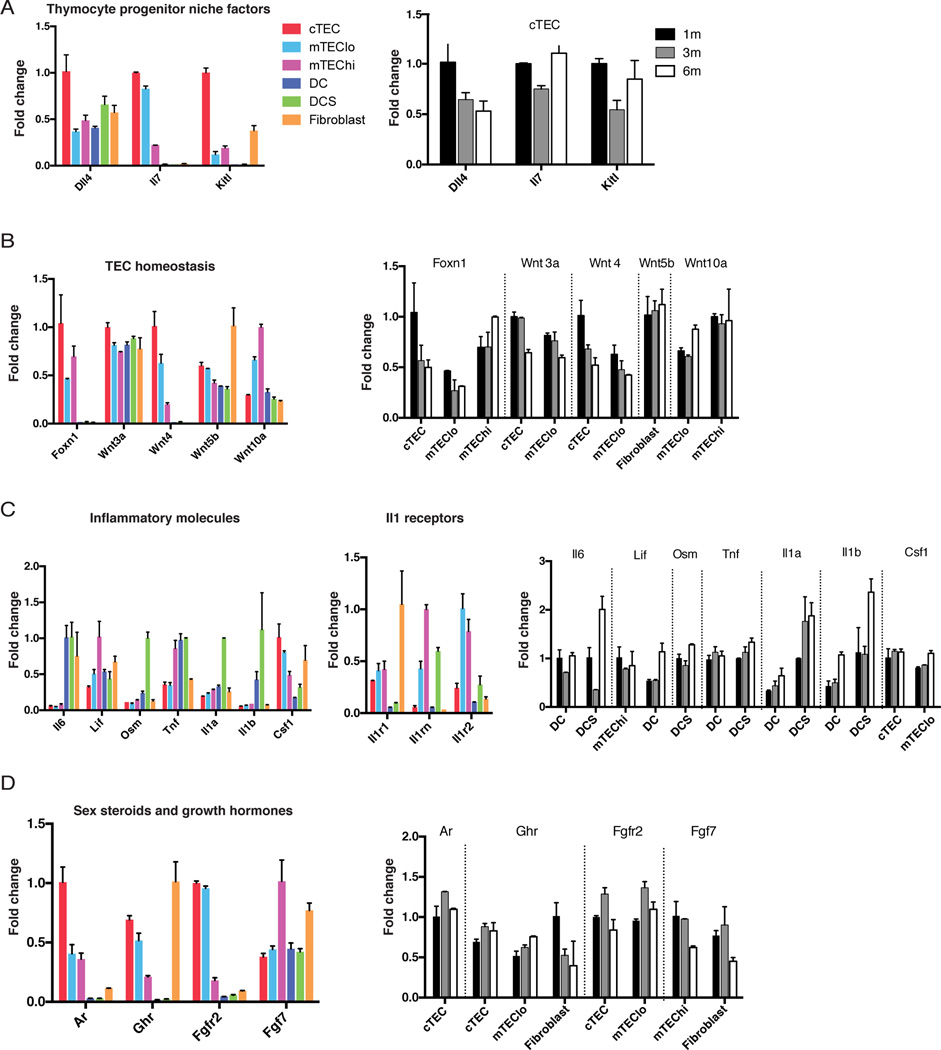
We analyzed pathways and genes previously implicated in late thymic involution to determine if their expression was altered in stromal subsets during the early stages of thymic degeneration. Dll4, Il7, and KitL, which are all required for a functional thymocyte progenitor niche, were expressed at the highest levels by cTECs. Only Dll4 expression diminished over 1 to 6 months (Figure 6A), suggesting a decline in Notch signaling contributes to early thymic involution.
Figure 6. A subset of genes implicated in late thymic involution are deregulated in thymic stroma by 3 to 6 months.
Graphs display the relative expression levels of genes involved in (A) thymocyte progenitor niche activity, (B) TEC homeostasis, (C) inflammation, and (D) sex steroid and growth hormone signaling in the thymus. For all graphs, expression values are normalized to the subset expressing the highest level of each gene at 1 month of age. The color graphs on the left display expression levels of genes of interest for each stromal subset at 1 month. The grayscale graphs on the right display expression levels of the same genes from 1 to 6 months for the indicated thymic stromal subsets. Graphs depict the mean plus or minus range of expression values.
Several studies have implicated decreased Foxn1 expression as a major contributor to thymic involution (Bredenkamp et al., 2014; Chinn et al., 2012). Foxn1 was not identified as an aging-associated DEG because it did not meet the <0.01 p-value criterion. However, Foxn1 expression declined ~2 fold in both cTEC and mTEClo subsets from 1 to 6 months of age (Figure 6B), consistent with previous findings at later stages of involution (Bredenkamp et al., 2014; Ortman et al., 2002). Two recent studies associated late thymic involution with altered expression of Wnt pathway genes (Bredenkamp et al., 2014; Griffith et al., 2012). Consistent with these studies, Wnt3a and Wnt4 were downregulated in early thymic involution, specifically in cTEC and mTEClo cells, while Wnt10a was upregulated in the mTEClo subset (Figure 6B). Consistent with Griffith et al., Wnt5b was slightly upregulated by 6 months; however, this increase occurred in fibroblasts. Taken together, some Wnt family members and Foxn1 are deregulated early in thymic involution, suggesting that TEC homeostasis is impaired, consistent with decreased TEC proliferation and E2F3 activity described above.
Inflammatory cytokines are upregulated in aging human thymi (Sempowski et al., 2000). Similiarly, Lif, Il6, and Osm were upregulated in murine thymic dendritic cell subsets over 1 to 6 months (Figure 6C). Furthermore, Tnf, Il1a and Il1b were upregulated by DCS. Interestingly, the activating receptor Il1r1 was expressed by all TECs and fibroblasts, while the IL1 antagonists Il1rn and Il1r2 were expressed by mTECs, suggesting IL1 likely impinges preferentially on cTECs and fibroblasts. We also note a trend of increased Csf1 expression by mTEClo over 1 to 6 months, which can drive dendritic cell differentiation (Fancke et al., 2008).
Sex steroids and growth factors can modulate thymic size (Lynch et al., 2009). For example, castration transiently increases thymic size and output (Griffith et al., 2012; Sutherland et al., 2005). Androgen receptor, Ar is expressed predominantly on cTEC, but is not upregulated over 1 to 6 months (Figure 6D). Administration of growth hormone and KGF also induce growth of involuted thymi (Lynch et al., 2009). Expression of the growth hormone receptor Ghr was not altered over 1 to 6 months in TECs (Figure 6D); however, it was decreased with age in fibroblasts, which expressed the highest levels of Ghr at 1 month. The expression of Fgfr2/Kgfr on cTEC and mTEClo cells did not decline over 1 to 6 months. However, expression of the ligand, Fgf7/Kgf diminished in both mTEChi cells and fibroblasts between 3 to 6 months (Figure 6D). Taken together, our data suggest that reduced growth factor signaling during early involution could affect both TECs and fibroblasts.
A searchable web-based platform containing thymocyte and thymic stromal cell gene expression data
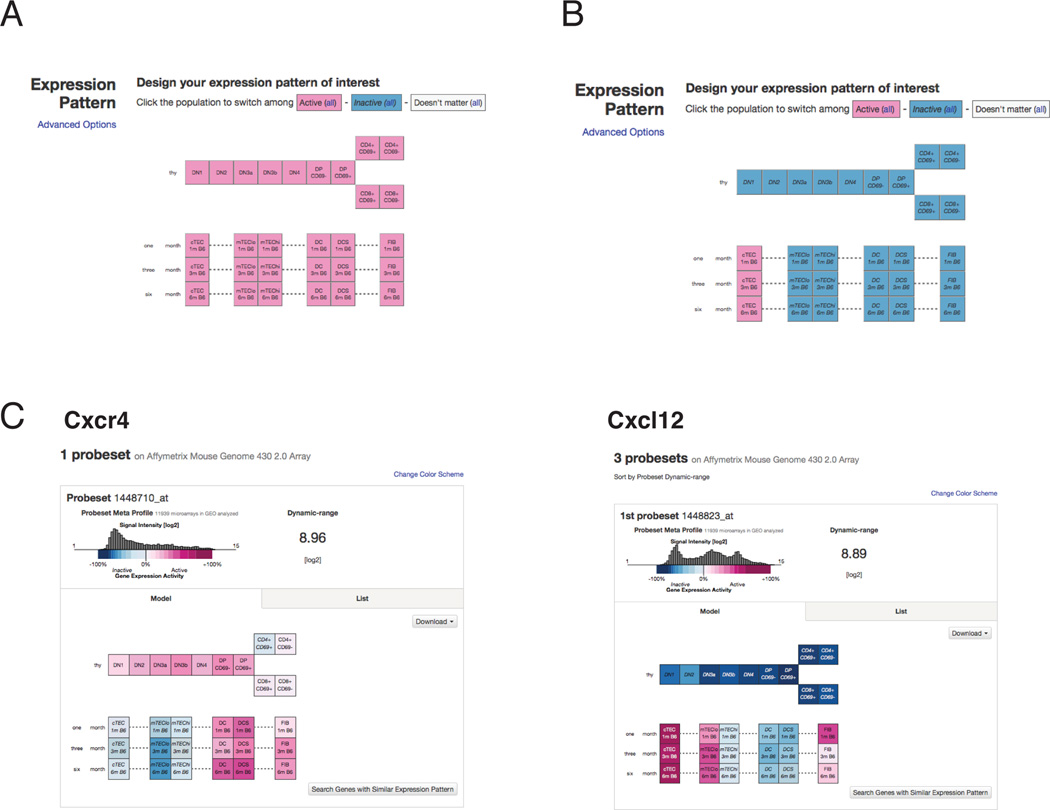
To make our thymic stromal expression data easily accessible, we have uploaded the datasets to the web-based platform Gene Expression Commons (GExC), where we have generated an online model containing both thymic stromal microarray datasets and our previously published thymocyte datasets (Seita et al., 2012) (https://gexc.stanford.edu/model/475). Users can readily query expression of genes of interest (GOI) in thymocyte or thymic stromal subsets (Figure 7A). One of the most powerful features of this platform is that data are normalized against a common reference of ~12,000 diverse datasets; thus, gene expression values can be compared against the full dynamic range of expression values for each probe set (Seita et al., 2012). In addition to normalized signal intensity values, “gene expression activity values” are provided on GExC. After using a step function to determine the threshold of expression for each probe set, each intensity value above and below this cutoff was assigned a percentile rank, reflecting the distribution of values in the common reference, as seen in the histograms (Figure 7C–D). For example, if a 30% “gene expression activity” value is obtained, the subset queried expresses the gene at a level above the first 30% of the common reference datasets over the threshold, and below the remaining 70%. This enables users to determine how robustly a gene is expressed relative to the range of expression values across diverse tissues. A second key advantage is the ability to readily search for expression patterns of interest, such as genes uniquely expressed in cTEC (Figure 7B). Finally, the combination of thymic stromal data with our previously published thymocyte data (Seita et al., 2012), will enable users to search for partner genes, such as receptors and ligands, that may contribute to thymocyte: stromal cell crosstalk. For example, the chemokine receptor Cxcr4 is expressed in cortical double negative (DN) and double positive (DP) thymocytes, as well as in thymic dendritic cells and fibroblasts, while its ligand Cxcl12 is highly expressed by cTECs (Figure 7C). This in silico model should greatly facilitate discovery of molecular mediators of thymocyte: stromal cell crosstalk, thymic stromal function, and altered gene expression in stromal cells during early thymic involution.
Figure 7. GExC platform for exploring expression data from thymocytes and thymic stromal subsets.
(A) The “Complete thymocyte: stromal interaction” model in GExC (https://gexc.stanford.edu/model/475). Eleven thymocyte subsets (at 1 month of age) and six thymic stromal subsets (at 1, 3, or 6 months of age) are represented as boxes, with colors representing the average expression level of data from biological replicates (n=3 and n=2 replicates for thymocyte and stromal subsets, respectively). (B) In this example of the pattern search feature on GExC, high expression in cTECs relative to all other subsets will be queried. (C) The database is useful for querying ligand-receptor pairs involved in thymocyte: stromal cell crosstalk. For example, the chemokine Cxcr4 is expressed by cortical thymocytes, as well as dendritic cells, while the ligand Cxcl12, is expressed at high levels by cTEC. The dynamic range of each probeset is displayed as a histogram, enabling users to ascertain expression levels relative to the range of values in publically available biological space (>12,000 diverse datasets are represented).
Discussion
We present global gene expression data from six thymic stromal subsets over the course of early thymic involution (1, 3, and 6 months of age) on a web-based platform as a resource for the scientific community (https://gexc.stanford.edu/model/475). Unsupervised clustering reveals the datasets are highly consistent (Figure 1), and qRT-PCR analyses on independently sorted samples validate the microarray expression data (Figure S1B). Nonetheless, there are several potential caveats to consider when querying the GExC database. First, thymic fibroblasts were sorted based on the absence of markers on hematopoietic cells, erythrocytes and endothelial cells, enhancing the possibility for contamination with other cell types. Nevertheless, strong transcription of adhesion molecules, ECM components, and platelet-derived growth factor receptors reveal this population is highly enriched in mesenchymal cells. Second, the sorted populations may be heterogeneous. The transcriptional profile of rare TEC progenitors included in cTEC or mTEClo subsets would be entangled in the larger population. As additional TEC subsets are identified, it will be important to compare their transcriptional profiles against the parental TEC population. Third, when viewing gene expression activity values in GExC, it is important to remember that gene expression activity values are scaled relative to the large common reference. This can cause a visual flattening of expression changes; for example, because Foxn1 is uniquely expressed in TECs, the ~2 fold decrease in expression over 1 to 6 months in cTEC and mTEClo cells (Figure 6B) is not as visually striking as the high expression in TECs relative to all other cell types. Also, gene expression activity values can be saturated (seen as 100%), as in the case of Psmb11/b5t in cTEC, obscuring the 20–50-fold increase in expression relative to other TEC subsets. In both cases above, it is important to compare the normalized signal intensity values, provided for download or on the “list” tab of GExC, to assess relative changes in expression with age or between subsets, respectively.
Surprisingly, our analyses revealed a significant association of mTEChi DEGs with the ‘defense response’ GO term. mTEChi is the most distal thymic stromal population by PCA analysis (Figure 1C), and its distinct transcriptional profile could result from expression of diverse TRAs. TRA expression is largely driven by Aire (Anderson et al., 2002), and 38% of genes uniquely expressed in mTEChi overlapped with Aire-regulated genes (Figure 2C). However, not all Aire-regulated genes are TRAs (Derbinski, 2005), and the remaining 62% of mTEChi DEGs may serve a function beyond elimination of autoreactive thymocytes. The GO term category ‘defense response’, includes genes such as defensins and cytokines (Figure 2E), which are critical for early innate immune responses against pathogens. Some of these genes (Defb8, Defb3) are expressed at high levels in mTEChi relative to canonical TRAs (Crp, Gad67) (not shown), which tend to be expressed at low levels (Derbinski et al., 2001; 2008). Therefore, we speculate that these ‘defense response’ genes may serve a functional role in mTEChi biology, perhaps to protect the thymus against infection.
Thymic involution results in diminished T cell output, leading to decreased immune function with age (Chinn et al., 2012; Lynch et al., 2009). Both CD4+ and CD8+ T cells generated in aged mice have impaired effector functions (Haynes and Maue, 2009; Nikolich-Žugich et al., 2012). These deficiencies can be attributed in part to the impact of an aged thymic environment, as fully functional T cells were generated when bone marrow precursors from aged mice were transferred into young, but not old recipients (Eaton et al., 2008). Therefore, it is important to understand the cellular and molecular drivers of thymic involution, with the ultimate goal of restoring functional T cells to aged individuals or patients following cytoablative therapies or infections.
Unbiased analyses of aging-associated transcriptional changes during early thymic involution revealed that cell cycle related genes and E2F3 transcriptional targets were significantly down-regulated in the mTEClo and cTEC subsets (Figure 4). This is consistent with the reduced proportion and number of cycling TECs with age (Gray et al., 2006), and the finding that overexpression of cyclinD1 or inactivation of Rb family members prevents thymic involution (Garfin et al., 2013; Robles et al., 1996). Our data add the novel perspective that E2F3 target genes, many of which are required for cell cycle progression, are down-regulated in both cTEC and mTEClo subsets by 6 months of age, suggesting that diminished cell cycle progression in TECs is an early hallmark of thymic involution. A recent study revealed that Foxn1 is an E2F3 target gene (Garfin et al., 2013). Thus, diminished E2F3 activity could directly reduce Foxn1 levels, consistent with the reduction in Foxn1 expression in TECs by 6 months of age (Figure 6B). Foxn1 is a critical regulator of thymic involution, as diminished Foxn1 expression results in premature involution (Chen et al., 2009), while enforced re-expression in aged TECs increases TEC cellularity and function and thymocyte output (Bredenkamp et al., 2014). However, maintenance of Foxn1 expression in TECs was insufficient to prevent thymic involution (Zook et al., 2011), suggesting modulation of other E2F3 targets and pathways contribute to involution. Foxn1 has also been implicated in regulating genes associated with cell cycle, such as Ccnd1 (Bredenkamp et al., 2014); thus, there is likely a complex feedback mechanism driving both diminished cell cycle progression and Foxn1 levels. Expression of e2f3 itself is not significantly altered in cTEC and mTEClo from 1 to 6 months (not shown), suggesting that E2F3 activity is modulated over the course of thymic involution. Identifying initial drivers of reduced E2F3 function will likely reveal critical regulators of thymic involution.
Diminished expression of Wnt pathway genes has also been implicated in age-associated TEC degeneration (Bredenkamp et al., 2014; Griffith et al., 2012). In agreement with these studies, we find that Wnt3a is diminished ~1.5 fold in cTEC and mTEClo at 6 months, while Wnt4 is decreased ~2 fold in cTEC (Figure 6B). As Wnt4 can induce Foxn1 expression (Balciunaite et al., 2002) and Wnt3a is implicated in epithelial proliferation (Liu et al., 2010), diminished expression could contribute to impaired TEC homeostasis. Furthermore, the subtle increase in expression of Wnt5b by fibroblasts, which may drive adipogenesis (van Tienen et al., 2009), suggests thymic mesenchymal cells could promote the age-associated increase in adipose tissue early in involution. We did not find evidence for altered expression of other Wnt family members (not shown), suggesting that deregulation of some Wnts (Bredenkamp et al., 2014; Griffith et al., 2012) may occur late in the involution process.
Reduced potency of the thymocyte progenitor niche may also impair thymopoiesis during involution. IL-7, Dll4, and Kitl, expressed by TECs, are all critical for promoting survival and differentiation of early thymocyte progenitors (Anderson and Takahama, 2012). Of these niche factors, only expression of Dll4 was diminished in cTECs by 6 months of age (Figure 6A), suggesting the thymic niche may have a reduced capacity to stimulate Notch signaling, thus impairing thymopoiesis in early involution. IL-7 levels are reduced in late thymic involution (Ortman et al., 2002); however, the lack of transcriptional downregulation over 6 months suggests that diminished IL-7 may be due to reduced cTEC numbers in the involuted thymus.
Signaling via sex steroids and growth hormones has also been implicated in thymic involution (Lynch et al., 2009). Our data do not reveal altered expression levels of Ar in TECs over 1 to 6 months (Figure 6D) or in estrogen or progesterone receptors (not shown). Taken together with the findings that sex steroid ablation induces only transient and partial regeneration of the thymus (Griffith et al., 2012) and that sex steroids diminish with age, while the thymus continues to involute (Chinn et al., 2012), our data do not suggest that sex steroids play a significant role in early thymic involution. However, it is possible that altered sensitivity to signaling by sex steroid hormones in aging TECs could promote involution. Our data suggest that altered growth factor signaling could contribute to early thymic involution because Ghr and Fgf7/Kgf expression was decreased in fibroblasts by 6 months (Figure 6D). Because both TECs and fibroblasts express Ghr, thymic rebound induced by growth hormone (Chen et al., 2003) may be due to signaling in both stromal subsets. Reduced expression of Fgf7/Kgf by aging fibroblasts may diminish signaling through FGFR2 on cTEC and mTEClo cells, contributing to TEC atrophy. Consistent with this notion, treatment with KGF causes transient thymic regeneration (Min et al., 2007).
Unbiased analyses of our data revealed an increasingly proinflammatory signature of DC and DCS with age (Figure 5). A previous study demonstrated that expression of Osm, Lif, Scf, and Il6 increased in aged human thymi, and administration of these cytokines promoted thymic atrophy in mice (Sempowski et al., 2000). Our analyses add further insight that Osm, Lif and Il6 are upregulated by thymic dendritic cells early in the course of involution, though we found no evidence for increased Scf, either due to species or temporal differences. We also identified additional genes associated with inflammation that are upregulated by thymic dendritic cells by 6 months of age (Figure 5). Il1a and Il1b are strikingly upregulated by thymic dendritic cells, whereas activating, but not inhibitory Il-1 receptors are expressed on cTEC and fibroblasts (Figure 6C). IL-1 administration promotes thymic atrophy (Morrissey et al., 1988), and Nlrp3 deficient mice, which cannot generate active IL-1, preferentially maintain the cTEC compartment with age (Youm et al., 2012). Taken together, these data suggest that IL-1 produced by thymic dendritic cells is likely a potent mediator of early thymic involution, signaling to both cTEC and thymic fibroblasts. Interestingly, Csf1, which can promote dendritic cell differentiation, is upregulated in mTEClo at 6 months (Figure 6C). Thus, a feedback loop may exist in which aging TECs promote differentiation of dendritic cells, whose inflammatory cytokines in turn promote TEC degeneration. Further elucidation of the mechanisms driving DCs to adopt a pro-inflammatory signature and causing TECs to reduce E2F3 activity and cycling will be key to understanding the etiology of age associated thymic involution.
Experimental Procedures
Mice
C57BL/6J mice were housed at the UT Austin animal facility, under conditions approved by the Institutional Animal Care and Use Committee.
FACS isolation of thymic stromal subsets and transcriptional profiling
Thymi from 1, 3, and 6-month-old male C57Bl/6J mice were enzymatically digested and FACS purified by double sorting to >95% purity on a FACSAria (BD Biosciences). Stromal subsets were collected directly in TRIzol (Life Technologies), and RNA was purified, amplified, and hybridized on Mouse Genome 430 2.0 arrays (Affymetrix). Data were uploaded to GExC (Seita et al., 2012) for RMA normalization (McCall et al., 2010). Gene expression data from thymic stromal subsets and previous thymocyte subsets (GSE34723) are available in GExC (https://gexc.stanford.edu/model/475) and GEO (GSE56928). See Supplemental Experimental Procedures.
Bioinformatics analyses
Identification of DEGs was performed in R using "limma" and "affy" packages, and the Molecular Signature Database (MSigDB) was queried for GSEA. See Supplemental Experimental Procedures.
RT-PCR on the Fluidigm platform
FACS purified stromal subsets were subject to high-throughput qPCR on the Fluidigm platform, using manufacturer’s reagents and procedures (Fluidigm). See Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We thank Ellen Richie and Nancy Manley for helpful discussions, and Elizabeth Zuo for microarray hybridization. This work was supported by the Cancer Prevention and Research Institute of Texas (R1003; L.I.R.E) and the NIH/NIAID (R01AI104870–01A1; L.I.R.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
SK and DP performed bioinformatics analyses, with advice from VI and LE, and wrote the manuscript with LE. SK and HC generated fluidigm data with guidance from JK. HS and LE generated primary data. JS provided advice on GExC.
References
- Anderson G, Takahama Y. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 2012;33:256–263. doi: 10.1016/j.it.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, Boehmer von H, Bronson R, Dierich A, Benoist C, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Balciunaite G, Keller MP, Balciunaite E, Piali L, Zuklys S, Mathieu YD, Gill J, Boyd R, Sussman DJ, Holländer GA. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol. 2002;3:1102–1108. doi: 10.1038/ni850. [DOI] [PubMed] [Google Scholar]
- Berent-Maoz B, Montecino-Rodriguez E, Dorshkind K. Genetic regulation of thymocyte progenitor aging. Seminars in Immunology. 2012;24:303–308. doi: 10.1016/j.smim.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn CC, Augustine CL, Li R, Harvey RP, Malin MA, Boyd RL, Miller JF, Morahan G. The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc Natl Acad Sci USA. 1996;93:5742–5746. doi: 10.1073/pnas.93.12.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Scimone ML, Schaerli P, Grabie N, Lichtman AH, Andrian von UH. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol. 2006;7:1092–1100. doi: 10.1038/ni1385. [DOI] [PubMed] [Google Scholar]
- Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the aged thymus by a single transcription factor. Development. 2014;141:1627–1637. doi: 10.1242/dev.103614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Cui X, Sempowski GD, Chao NJ. Growth hormone accelerates immune recovery following allogeneic T-cell-depleted bone marrow transplantation in mice. Exp. Hematol. 2003;31:953–958. doi: 10.1016/s0301-472x(03)00196-6. [DOI] [PubMed] [Google Scholar]
- Chen L, Xiao S, Manley NR. Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood. 2009;113:567–574. doi: 10.1182/blood-2008-05-156265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinn IK, Blackburn CC, Manley NR, Sempowski GD. Changes in primary lymphoid organs with aging. Seminars in Immunology. 2012;24:309–320. doi: 10.1016/j.smim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY, Carter C, Yu BP, Leeuwenburgh C. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res. Rev. 2009;8:18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. Journal of Experimental Medicine. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- Derbinski J, Pinto S, Rösch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K, Ellis DK, Saraf K, Jelinsky SA, Lin L-L. Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative crosstalk with bacterial lipopolysaccharide. J Immunol. 2008;180:3520–3534. doi: 10.4049/jimmunol.180.5.3520. [DOI] [PubMed] [Google Scholar]
- Eaton SM, Maue AC, Swain SL, Haynes L. Bone marrow precursor cells from aged mice generate CD4 T cells that function well in primary and memory responses. The Journal of Immunology. 2008;181:4825–4831. doi: 10.4049/jimmunol.181.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancke B, Suter M, Hochrein H, O'Keeffe M. M-CSF: a novel plasmacytoid and conventional dendritic cell poietin. Blood. 2008;111:150–159. doi: 10.1182/blood-2007-05-089292. [DOI] [PubMed] [Google Scholar]
- Foster K, Sheridan J, Veiga-Fernandes H, Roderick K, Pachnis V, Adams R, Blackburn C, Kioussis D, Coles M. Contribution of neural crest-derived cells in the embryonic and adult thymus. J Immunol. 2008;180:3183–3189. doi: 10.4049/jimmunol.180.5.3183. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Garfin PM, Min D, Bryson JL, Serwold T, Edris B, Blackburn CC, Richie ER, Weinberg KI, Manley NR, Sage J, et al. Inactivation of the RB family prevents thymus involution and promotes thymic function by direct control of Foxn1 expression. J Exp Med. 2013;210:1087–1097. doi: 10.1084/jem.20121716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DHD, Seach N, Ueno T, Milton MK, Liston A, Lew AM, Goodnow CC, Boyd RL. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 2006;108:3777–3785. doi: 10.1182/blood-2006-02-004531. [DOI] [PubMed] [Google Scholar]
- Griffith AV, Fallahi M, Nakase H, Gosink M, Young B, Petrie HT. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity. 2009;31:999–1009. doi: 10.1016/j.immuni.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith AV, Fallahi M, Venables T, Petrie HT. Persistent degenerative changes in thymic organ function revealed by an inducible model of organ regrowth. Aging Cell. 2012;11:169–177. doi: 10.1111/j.1474-9726.2011.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, Minato N. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol. 2007;8:304–311. doi: 10.1038/ni1438. [DOI] [PubMed] [Google Scholar]
- Haynes L, Maue AC. Effects of aging on T cell function. Curr Opin Immunol. 2009;21:414–417. doi: 10.1016/j.coi.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Klein L, Hinterberger M, Rohrscheidt von J, Aichinger M. Autonomous versus dendritic cell-dependent contributions of medullary thymic epithelial cells to central tolerance. Trends Immunol. 2011;32:188–193. doi: 10.1016/j.it.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Klug DB, Carter C, Crouch E, Roop D, Conti CJ, Richie ER. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc Natl Acad Sci USA. 1998;95:11822–11827. doi: 10.1073/pnas.95.20.11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L-J, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, Bösl MR, Holländer GA, Hayashi Y, Malefyt R, de W, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med. 2011;208:383–394. doi: 10.1084/jem.20102327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Park J, Foss D, Goldschneider I. Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. Journal of Experimental Medicine. 2009;206:607–622. doi: 10.1084/jem.20082232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Luo M, Xie W, Wells JM, Goodheart MJ, Engelhardt JF. Sox17 modulates Wnt3A/ -catenin-mediated transcriptional activation of the Lef-1 promoter. AJP: Lung Cellular and Molecular Physiology. 2010;299:L694–L710. doi: 10.1152/ajplung.00140.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HE, Goldberg GL, Chidgey A, Van den Brink MRM, Boyd R, Sempowski GD. Thymic involution and immune reconstitution. Trends Immunol. 2009;30:366–373. doi: 10.1016/j.it.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–2584. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min D, Panoskaltsis-Mortari A, Kuro-O M, Holländer GA, Blazar BR, Weinberg KI. Sustained thymopoiesis and improvement in functional immunity induced by exogenous KGF administration in murine models of aging. Blood. 2007;109:2529–2537. doi: 10.1182/blood-2006-08-043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey PJ, Charrier K, Alpert A, Bressler L. In vivo administration of IL-1 induces thymic hypoplasia and increased levels of serum corticosterone. J Immunol. 1988;141:1456–1463. [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S-I, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Nikolich-Žugich J, Li G, Uhrlaub JL, Renkema KR, Smithey MJ. Age-related changes in CD8 T cell homeostasis and immunity to infection. Seminars in Immunology. 2012;24:356–364. doi: 10.1016/j.smim.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Ohigashi I, Nakagawa Y, Takahama Y. Cytokine crosstalk for thymic medulla formation. Curr Opin Immunol. 2011;23:190–197. doi: 10.1016/j.coi.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Ortman CL, Dittmar KA, Witte PL, Le PT. Molecular characterization of the mouse involuted thymus: aberrations in expression of transcription regulators in thymocyte and epithelial compartments. Int. Immunol. 2002;14:813–822. doi: 10.1093/intimm/dxf042. [DOI] [PubMed] [Google Scholar]
- Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- Plotkin J, Prockop SE, Lepique A, Petrie HT. Critical role for CXCR4 signaling in progenitor localization and T cell differentiation in the postnatal thymus. J Immunol. 2003;171:4521–4527. doi: 10.4049/jimmunol.171.9.4521. [DOI] [PubMed] [Google Scholar]
- Proietto AI, van Dommelen S, Zhou P, Rizzitelli A, D'Amico A, Steptoe RJ, Naik SH, Lahoud MH, Liu Y, Zheng P, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci USA. 2008;105:19869–19874. doi: 10.1073/pnas.0810268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repass JF, Laurent MN, Carter C, Reizis B, Bedford MT, Cardenas K, Narang P, Coles M, Richie ER. IL7-hCD25 and IL7-Cre BAC transgenic mouse lines: new tools for analysis of IL-7 expressing cells. Genesis. 2009;47:281–287. doi: 10.1002/dvg.20497. [DOI] [PubMed] [Google Scholar]
- Ribeiro AR, Rodrigues PM, Meireles C, Di Santo JP, Alves NL. Thymocyte selection regulates the homeostasis of IL-7-expressing thymic cortical epithelial cells in vivo. The Journal of Immunology. 2013;191:1200–1209. doi: 10.4049/jimmunol.1203042. [DOI] [PubMed] [Google Scholar]
- Robles AI, Larcher F, Whalin RB, Murillas R, Richie E, Gimenez-Conti IB, Jorcano JL, Conti CJ. Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc Natl Acad Sci USA. 1996;93:7634–7638. doi: 10.1073/pnas.93.15.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, Ehrlich LIR, Fathman JW, Dill DL, Weissman IL. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS ONE. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempowski GD, Hale LP, Sundy JS, Massey JM, Koup RA, Douek DC, Patel DD, Haynes BF. Leukemia Inhibitory Factor, Oncostatin M, IL-6, and Stem Cell Factor mRNA Expression in Human Thymus Increases with Age and Is Associated with Thymic Atrophy. The Journal of Immunology. 2000;164:2180–2187. doi: 10.4049/jimmunol.164.4.2180. [DOI] [PubMed] [Google Scholar]
- Sutherland JS, Goldberg GL, Hammett MV, Uldrich AP, Berzins SP, Heng TS, Blazar BR, Millar JL, Malin MA, Chidgey AP, et al. Activation of Thymic Regeneration in Mice and Humans following Androgen Blockade. The Journal of Immunology. 2005 doi: 10.4049/jimmunol.175.4.2741. [DOI] [PubMed] [Google Scholar]
- van Tienen FHJ, Laeremans H, van der Kallen CJH, Smeets HJM. Wnt5b stimulates adipogenesis by activating PPARgamma, and inhibiting the beta-catenin dependent Wnt signaling pathway together with Wnt5a. Biochem. Biophys. Res. Commun. 2009;387:207–211. doi: 10.1016/j.bbrc.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Wurbel M-A, Philippe J-M, Nguyen C, Victorero G, Freeman T, Wooding P, Miazek A, Mattei M-G, Malissen M, Jordan BR, et al. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol. 2000;30:262–271. doi: 10.1002/1521-4141(200001)30:1<262::AID-IMMU262>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Yano M, Kuroda N, Han H, Meguro-Horike M, Nishikawa Y, Kiyonari H, Maemura K, Yanagawa Y, Obata K, Takahashi S, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J Exp Med. 2008;205:2827–2838. doi: 10.1084/jem.20080046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm Y-H, Kanneganti T-D, Vandanmagsar B, Zhu X, Ravussin A, Adijiang A, Owen JS, Thomas MJ, Francis J, Parks JS, et al. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell Rep. 2012;1:56–68. doi: 10.1016/j.celrep.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook EC, Krishack PA, Zhang S, Zeleznik-Le NJ, Firulli AB, Witte PL, Le PT. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood. 2011;118:5723–5731. doi: 10.1182/blood-2011-03-342097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.