Abstract
Purpose
Currently approved DNA hypomethylating nucleosides elicit their effects in part by depleting DNA methyltransferase I (DNMT1). However, their low response rates and adverse effects continue to drive the discovery of newer DNMT1 depleting agents. Herein, we identified two novel 2′-deoxycytidine (dCyd) analogs, 4′-thio-2′-deoxycytidine (T-dCyd) and 5-aza-4′-thio-2′-deoxycytidine (aza-T-dCyd) that potently deplete DNMT1 in both in vitro and in vivo models of cancer and concomitantly inhibit tumor growth.
Methods
DNMT1 protein levels in in vitro and in vivo cancer models were determined by Western blotting and antitumor efficacy was evaluated using xenografts. Effects on CpG methylation were evaluated using methylation-specific PCR. T-dCyd metabolism was evaluated using radiolabeled substrate.
Results
T-dCyd markedly depleted DNMT1 in CCRF-CEM and Kg1a leukemia and nCI-H23 lung carcinoma cell lines, while it was ineffective in the HCt-116 colon or IgrOV-1 ovarian tumor lines. On the other hand, aza-T-dCyd potently depleted DNMT1 in all of these lines indicating that dCyd analogs with minor structural dissimilarities induce different DNMT1 turnover mechanisms. Although T-dCyd was deaminated to 4′-thio-2′-deoxyuridine, very little was converted to 4′-thio-thymidine nucleotides, suggesting that inhibition of thymidylate synthase would be minimal with 4′-thio dCyd analogs. Both T-dCyd and aza-T-dCyd also depleted DNMT1 in human tumor xenografts and markedly reduced in vivo tumor growth. Interestingly, the selectivity index of aza-T-dCyd was at least tenfold greater than that of decitabine.
Conclusions
Collectively, these data show that 4′-thio modified dCyd analogs, such as T-dCyd or aza-T-dCyd, could be a new source of clinically effective DNMT1 depleting anticancer compounds with less toxicity.
Keywords: DNMT1, DNA methyltransferase, Decitabine, azacytidine, Zebularine, Deoxycytidine
Introduction
DNA methyltransferase I (DNMT1), an enzyme that functions as a maintenance methyltransferase during replication, is essential for cell survival and has also been implicated in tumorigenesis [1]. DNMT1 is viewed as the methyltransferase that contributes to the hypermethylation of cytosines in CpG sites in tumor suppressor genes leading to their silencing in a manner akin to inactivating mutations [2–5]. A number of reports have implicated DNMT1 in cellular transformation and the tumorigenic process [6–8]. Enforcing expression of DNMT1 increases proliferation and induces transformation of NIH 3T3 fibroblasts [6], whereas antagonizing DNMT1 expression by genetic or pharmacological means decreases CpG methylation and increases expression of tumor suppressor genes, which reduces proliferation and cellular transformation [9–11]. Depression of DNMT1 levels has also been shown to suppress tumorigenesis in a manner independent of its DNA methyltransferase activity [12, 13]. Knockout studies reveal that depletion of DNMT1 can cause decreases in cell viability which are preceded by molecular events that are consistent with the activation of DNA damage response [14]. Regardless of the mechanisms by which DNMT1 causes tumorigenesis, it is now recognized that DNMT1 protein stability is increased in cancer cells [15].
Decitabine (aza-dCyd) and azacitidine (aza-Cyd) are two DNA hypomethylating nucleosides that deplete DNMT1 that are currently being used in myelodysplastic syndromes and other hematologic malignancies [15–18]. Two mechanisms are considered to contribute jointly to the hypomethylation induced by these agents. First, chromatin bound DNMT1 enzyme attempts to methylate any aza-dCyd that is incorporated diagonally opposite 5-methylcytosine bases in the parent strand. However, because of the lack of a proton at the N5 position of the aza-substituted pyrimidine ring, the enzyme is not released by β-elimination [19]. Second, the trapping of the enzyme and the covalent complexes that are formed activate the proteasomal degradation of the free enzyme [17, 18]. It has been suggested that it is trapping of chromatin bound DNMT1 molecules that is the primary cause of hypomethylation. However, degradation of free DNMT1 exacerbates the effect by preventing the full methylation of hemi-methylated CpG sites [18].
two other nucleoside analogs, 5-F-2′-deoxycytidine (F-dCyd) and zebularine, have also received considerable preclinical experimental attention, because of their ability to cause depletion of DNMT1 in cancer cells [20, 21]. DNMT1 active site occupancy by the DNA-incorporated analog is essential to cause DNMT1 trapping and the ensuing degradation of the enzyme. Many features of the current agents (such as relatively inefficient incorporation into DNA, DNA synthesis inhibition at high doses, chemical instability of the triazine ring, metabolic instability due to deamination and inhibition of enzymes not related to DNA methylation) result in decreased hypomethylation efficiency and preclude administration for extended schedules at high doses. Thus, there is a need to identify DNMT1 depleting agents that would be more efficacious with less toxicity. The design of new analogs for this purpose must recognize that sequential intracellular steps that lead to DNA incorporation and strand elongation need to be efficient: i.e., modifications in the nucleoside should not: (a) prevent efficient activation to the triphosphate and incorporation into DNA, (b) inhibit extension of the DNA chain after incorporation or (c) prevent extrusion of the base from the double helix or its binding to the active site of DNMT1. Sustained new DNA synthesis of demethylated daughter strands in replicated cells also necessitates that the modified nucleosides or their metabolites should not, in general, abrogate DNA replication by inhibiting other enzymes that are involved in DNA synthesis, such as thymidylate synthase (TS).
As part of the anticancer drug discovery program at Southern Research Institute, we evaluated a dCyd analog, 4′-thio-2′-deoxycytidine (T-dCyd), and found that it was a substrate for dCyd kinase and was efficiently incorporated into DNA without inhibition of DNA synthesis [22]. Results from these studies collectively suggested that the 4′-thio modification to dCyd analogues could affect metabolism in a positive manner resulting in relatively little production of 2′-deoxyuridine or thymidine nucleotides in cells [22]. Furthermore, we recently discovered that treatment with T-dCyd caused DNMT1 depletion in cancer cells, suggesting that it could be a useful epigenetic agent [23]. In this work, we have synthesized T-dCyd and its 5-aza analog, and evaluated their ability to deplete DNMT1 both in vitro and in vivo and investigated their effects on tumor growth in mice.
Materials and methods
Synthesis of T-dCyd and aza-T-dCyd
T-dCyd and aza-T-dCyd were synthesized as described before, and their structures were verified using CHN elemental analysis and NMR spectroscopy [24, 25].
Western blotting assay for DNMT1 depletion
CCRF-CEM, NCI-H23, HCT-116 and IGROV-1 cell lines were obtained from ATCC (Manassas, VA). KG1a cells were obtained from DSMZ GmbH (Braunschweig, Germany). Exponentially growing cells were treated with 4′-thionucleosides or DMSO (vehicle) at indicated concentrations for different periods. Cell lysates were prepared, and typically 25–50 µg of cell lysate protein was run on 7.5 % SDS-PAGE gels and analyzed by Western blotting. DNMT1 antibodies (ab19905 and ab16632) and GAPDH antibody (rabbit mAb 14C10) were from Abcam (Cambridge, MA) and Cell Signaling (Danvers, MA), respectively.
Metabolism of T-dCyd and incorporation into DNA
To study the metabolism of T-dCyd in the HCT-116 colon carcinoma line, cells were incubated with 1 µM [5-3H] T-dCyd (Moravek Biochemicals, Brea, CA) for 1 h. The cell culture medium was removed, cells were detached from the flask by trypsinization, and cell pellets were extracted with acid as described previously [26]. The radioactivity in the acid-insoluble fraction was determined as a measure of the amount of T-dCyd that was incorporated into DNA. The acid-soluble extract was subjected to strong anion exchange (SAX) HPLC to identify intracellular metabolites, and an acid-soluble extract of the cell culture medium was subjected to reverse phase HPLC to identify metabolites in the medium.
Methylation-specific PCR (MSP) analysis of p15 CpG methylation and RT-PCR assay for expression of p15
MSP analysis of CpG methylation in the p15 tumor suppressor gene was accomplished as described previously [27]. Briefly, genomic DNA from KG1a cells treated with various drugs was modified with bisulfite using the EZ DNA Methylation kit from Zymo Research (Irvine, CA). The DNA was then used in PCRs with primer pairs that were specific for either unmethylated or methylated p15 DNA as described [27]. PCRs were loaded onto agarose gels and visualized under UV illumination. For RT-PCR analysis of p15 message, total cellular RNA was made from drug-treated KG1a cells treated using TRI reagent. RT-PCR was accomplished as described previously [28].
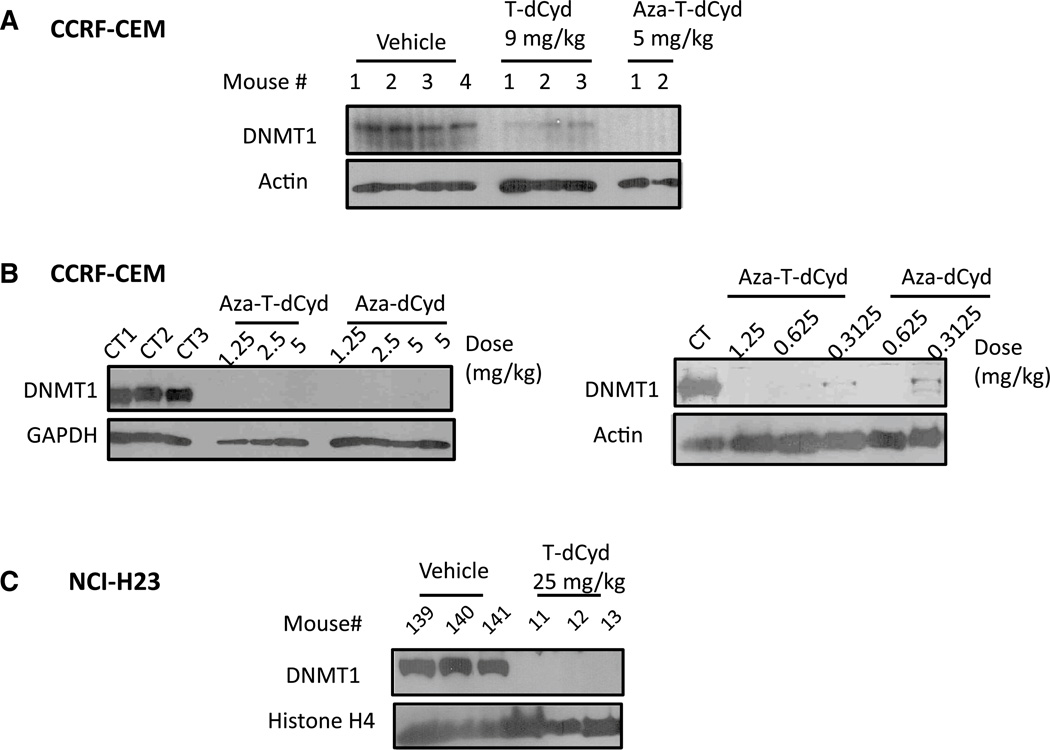
In vivo analysis of DNMT1 depletion in CCRF-CEM and NCI-H23 tumors following administration of 4′-thio nucleosides
Young female athymic nu/nu mice (Charles River Laboratories, Wilmington, MA) were implanted subcutaneously (s.c.) with a 30–40 mg CCRF-CEM tumor fragment on Day 0. T-dCyd was injected intraperitoneally (i.p.) daily for 9 days (q1d × 9) at a dose of 9 mg/kg/dose (Fig. 4a) or at doses of 9, 4.5 or 2.25 mg/kg/dose (data not shown). Aza-T-dCyd was injected i.p. at a dose of 5 mg/kg/dose (q1d × 9) (Fig. 4a) or at doses of 5, 2.5, 1.25, 0.625, 0.3125 mg/kg/dose (Fig. 5b). Aza-dCyd was injected i.p. at doses of 5, 2.5, 1.250, 0.625 or 0.3125 mg/kg/dose (q1d × 9) (Fig. 4b). All treatments began when individual tumor volumes reached 144–270 mm3 (Day 19, Fig. 4a) and 100–144 mm3 (Day 14, Fig. 4b). Mice were observed daily, and their s.c. tumors were collected 24 h after the last treatment and analyzed for DNMT1 as described above. For DNMT1 depletion studies in the NCI-H23 model, mice were injected i.p. daily with T-dCyd for 3 days (q1d × 3) at a dose of 25 mg/kg/dose and tumors were collected 30 min after the last treatment and analyzed for DNMT1 as described above (Fig. 4c).
Fig. 4.
T-dCyd and aza-T-dCyd deplete DNMT1 protein levels in tumors implanted in nude mice, a Mice-bearing subcutaneous CCRF-CEM tumors were injected i.p with T-dCyd for 9 days (qld × 9) at a dose of 9 mg/kg/dose or with aza-T-dCyd at 5 mg/kg/dose. b Aza-T-dCyd or aza-dCyd was injected i.p. at doses of 5, 2.5 or 1.25 mg/ kg/dose (qld × 9) (left panel). In the right panel aza-T-dCyd was injected i.p. at doses of 1.25, 0.625 or 0.3125 mg/kg/dose (qld × 9) and aza-dCyd was injected at doses of 0.625 or 0.3125 mg/kg/dose (qld × 9). Tumors were harvested 24 h after last treatment, and lysates were fractionated on SDS-PAGE gels and were analyzed by Western blotting using the indicated antibodies, c T-dCyd depletes DNMT1 protein levels in NCI-H23 lung tumors implanted in nude mice. Mice implanted with subcutaneous NCI-H23 tumors were injected i.p with T-dCyd for 3 days (qld × 3) at a dose of 25 mg/kg/ dose. Tumors were harvested 3 h after last treatment, and lysates were fractionated on SDS-PAGE gels and were analyzed by Western blotting using the indicated antibodies
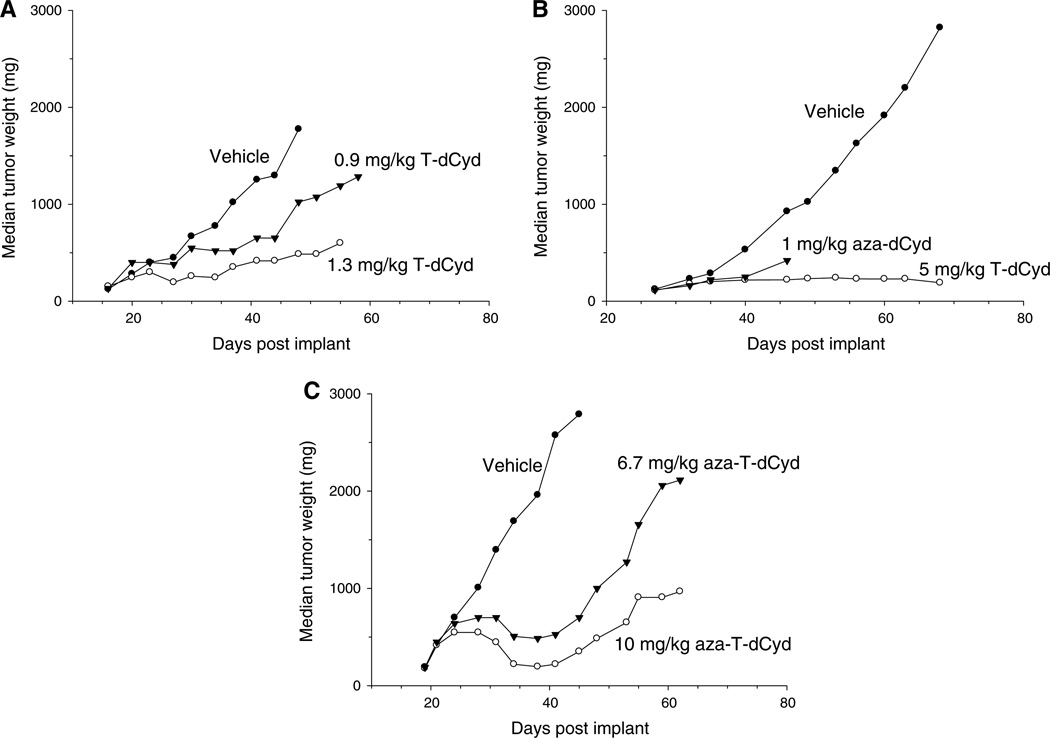
Fig. 5.
T-dCyd and aza-T-dCyd are effective at slowing growth of NCI-H23 lung tumors implanted in nude mice, a Mice were treated i.p. with vehicle (saline) or T-dCyd at 1.3 and 0.9 mg/kg/dose three times a day, 4 h apart, daily for 9 days, b Mice were treated i.p. with vehicle (saline) (qld × 5, qlw × 5), 1 mg/kg/dose aza-dCyd (qld × 5, qlw × 3) or 5 mg/kg/dose T-dCyd (qld × 5, qlw × 5). c Aza-T-dCyd is effective at slowing growth of NCI-H23 lung tumors implanted in nude mice. Mice were treated i.p. with vehicle (saline) or aza-T-dCyd at 10 and 6.7 mg/kg/dose daily for 9 days (qld × 9)
Efficacy studies of T-dCyd, aza-T-dCyd and aza-dCyd in human NCI-H23 lung tumor xenografts
Studies in nontumored mice were conducted prior to the efficacy studies to determine the maximally tolerated dose of each compound on each schedule. Although different doses and schedules were evaluated in these studies, all of the experiments were conducted with doses that were at the maximally tolerated dose on the particular schedule that was used.
For the experiments shown in Fig. 5a–c, mice were implanted s.c. with a 30–40 mg human NCI-H23 lung tumor fragment. Tumor implantation day was designated Day 0. In Fig. 5a, young male athymic nu/nu mice were injected i.p. with T-dCyd (0.9 or 1.3 mg/kg/dose dissolved in saline) three times a day, 4 h apart (q4h × 3), for nine consecutive days (q1d × 9). The vehicle-treated control group had 12 mice, and two treatment groups had 6 mice each. Treatments began when individual tumor volumes were approximately 140 mm3 (Day 16). There were no deaths in this experiment, and there were two tumor-free survivors in both T-dCyd treated groups, at the end of the experiment (Day 58).
In Fig. 5b, young female athymic nu/nu mice were injected i.p. with 5 mg/kg/dose of T-dCyd (dissolved in 0.9 % saline) daily for five consecutive days (q1d × 5) for five cycles or 1 mg/kg/dose of aza-dCyd (dissolved in 0.9 % saline) daily for five consecutive days for three cycles. Mice did not receive drug 2 days between each round of therapy. The vehicle-treated control group had 20 mice, and the two treatment groups had 14 mice each. Treatments began when individual tumor volumes were approximately 120 mm3 (Day 27).
In Fig. 5c, young female athymic nu/nu mice were injected i.p. with aza-T-dCyd (6.7 or 10 mg/kg/dose dissolved in saline) (q1d × 9). The vehicle-treated control group had 20 mice, and two treatment groups had 6 mice each. Treatments began when individual tumor volumes were approximately 150 mm3 (Day 19). There was one death in each treatment group (Day 31 in the 10 mg/kg group and Day 36, in the 6.7 mg/kg group).
Results
Comparison of the effects of dCyd analogs on viability in cancer cell lines
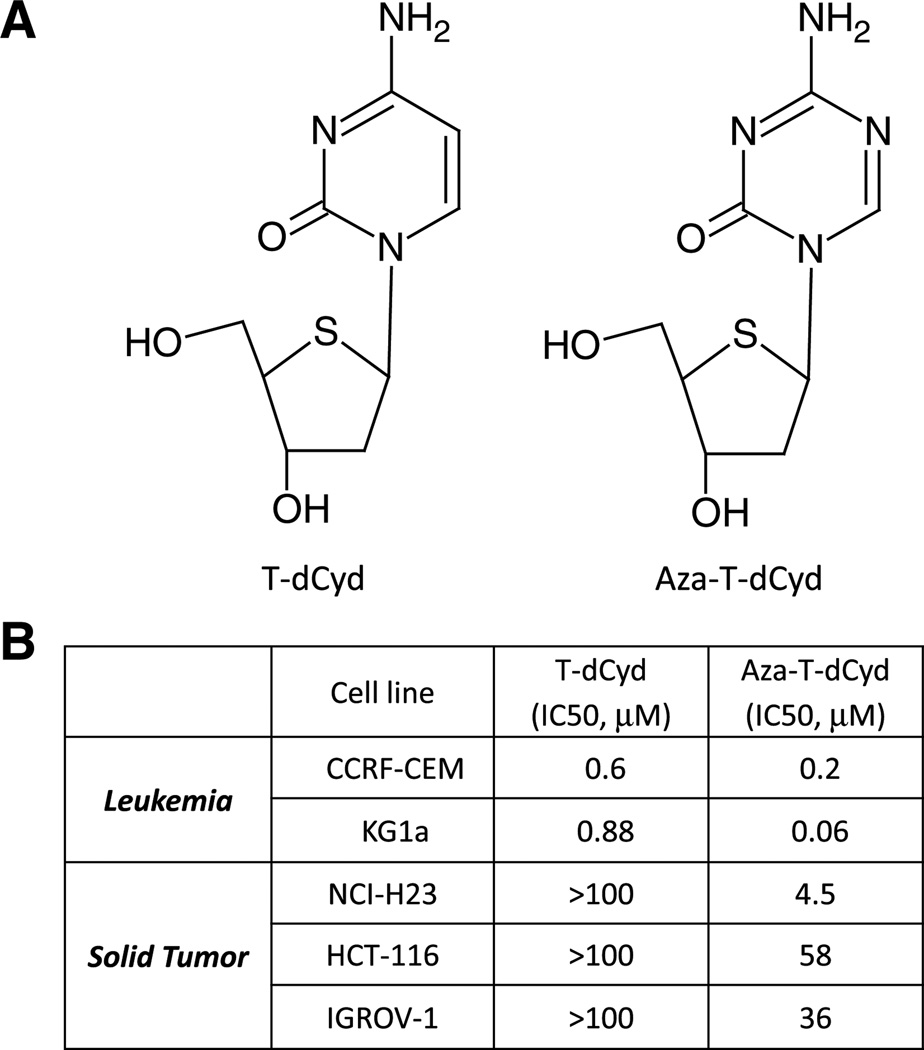
4′-Thio analogs differ from natural nucleosides in that the 4′ oxygen atom in the deoxyribose ring is replaced by a sulfur atom. We first examined two leukemia lines for their sensitivities to two 4′-thio analogs that differ only at the five position of the cytosine ring (Fig. 1a). T-dCyd contains a natural cytosine nucleobase, whereas in aza-T-dCyd, a nitrogen atom replaces the carbon atom at this position. Cells were incubated with these agents for 72 h, and cytotoxicity was determined using the Cell Titer Glo assay. Dose–response curves were established, and IC50s were determined. Both leukemia lines (CCRF-CEM and KG1a) were sensitive to T-dCyd and aza-T-dCyd with IC50s below 1 µM (Fig. 1b). Aza-T-dCyd was 3–15-fold more potent than T-dCyd. Aza-T-dCyd also decreased viability in the NCI-H23 lung carcinoma, HCT-116 colon carcinoma and IGROV-1 ovarian carcinoma with IC50s of 4.5, 58 and 36 µM, respectively, whereas T-dCyd had less than 50 % effect on viability in these cell lines at 100 µM (Fig. 1b).
Fig. 1.
Characterization of T-dCyd and aza-T-dCyd. a Structures of T-dCyd and aza-T-dCyd, b cells were treated with T-dCyd or aza-T-dCyd for 72 h and cell viability was measured using the Cell Titer Glo assay. Mean IC50 values of three experiments in the indicated cancer cell lines are shown (SD < 10 %)
T-dCyd and aza-T-dCyd deplete DNMT1 protein in cancer cells
A previous report had demonstrated that an oligonucleotide containing T-dCyd was successful in inhibiting methyltransferase activity of M. Hha I DNA methyltransferase in vitro, despite containing an unmodified cytosine, which flips out of the double helix and engages the active site of the enzyme in a manner similar to that seen with the natural nucleoside, dCyd [29]. We surmised therefore that since our previous studies had demonstrated that T-dCyd was phosphorylated and incorporated into DNA in human cells [22], it would likely occupy the active site of mammalian DNMT1 as well. Since DNA demethylators destabilize and deplete the DNMT1 protein [17, 18], we assessed whether treatment of human cancer cells with T-dCyd analogs would demonstrate a similar effect.
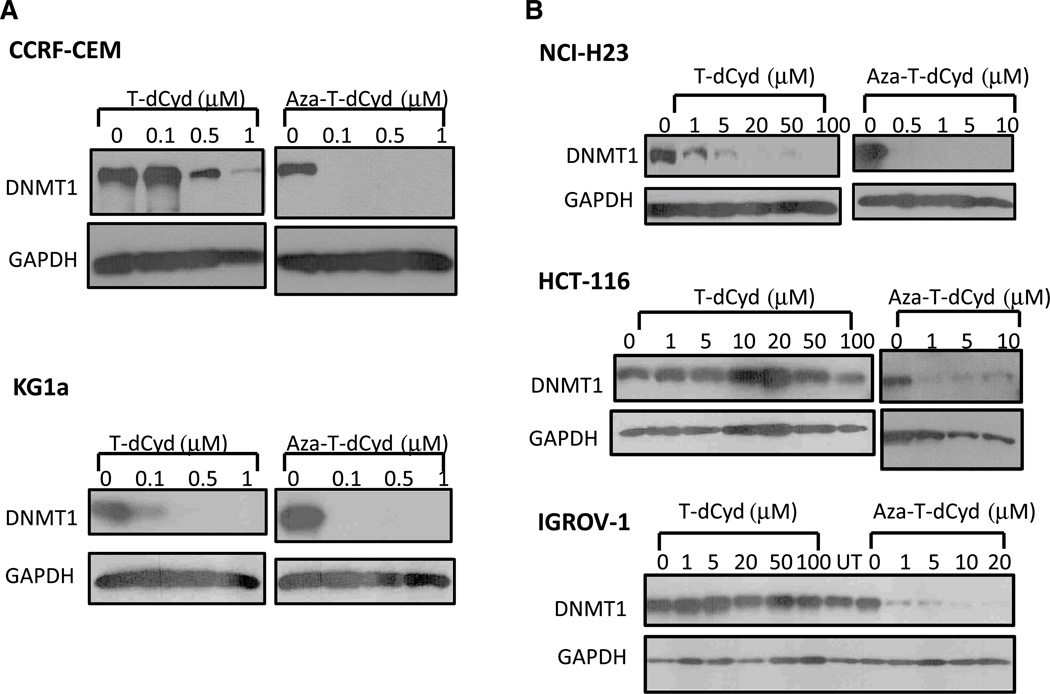
We first determined the protein levels of DNMT1 in CCRF-CEM leukemia cells treated for 96 h with T-dCyd or aza-T-dCyd at the indicated concentrations (Fig. 2a). We initially chose leukemia cells since clinically used drugs acting via this mechanism have been approved for myelodysplastic syndromes. As shown in Fig. 2, treatment with T-dCyd and aza-T-dCyd resulted in marked depletion of DNMT1 in CCRF-CEM cells at sub-micromolar concentrations. Both agents also markedly depleted DNMT1 in KG1a myeloid leukemia cells (Fig. 2a). We also determined the protein levels of DNMT1 in three solid tumor cell lines (NCI-H23 lung, HCT-116 colon and IGROV-1 ovarian), treated with T-dCyd or aza-T-dCyd at the indicated doses (Fig. 2b). Both agents also markedly depleted DNMT1 in NCI-H23 cells (Fig. 2b). DNMT1 levels were also depleted in HCT-116 and IGROV-1 when exposed to aza-T-dCyd at concentrations of 1–10 µM, whereas T-dCyd was not efficacious up to 100 µM (Fig. 2b). The effect of aza-T-dCyd on DNMT1 levels in these solid tumor cell lines was observed at doses that were far below the IC50 for cytotoxicity.
Fig. 2.
T-dCyd and aza-T-dCyd induce DNMT1 depletion in leukemia cells and in solid tumor cells. Exponentially growing leukemia cells (a CCRF-CEM, upper panel and KG1a, lower panel) or solid tumor cells (b NCI-H23 lung carcinoma, HCT-116 colon carcinoma or IGROV-1 ovarian carcinoma cells) were exposed to indicated concentrations of either T-dCyd or aza-T-dCyd for 96 h. Cell lysates were fractionated on SDS-PAGE gels and were analyzed by Western blotting using antibodies against DNMT1 or GAPDH
T-dCyd and aza-T-dCyd induce CpG demethylation and re-expression of the p15 tumor suppressor gene
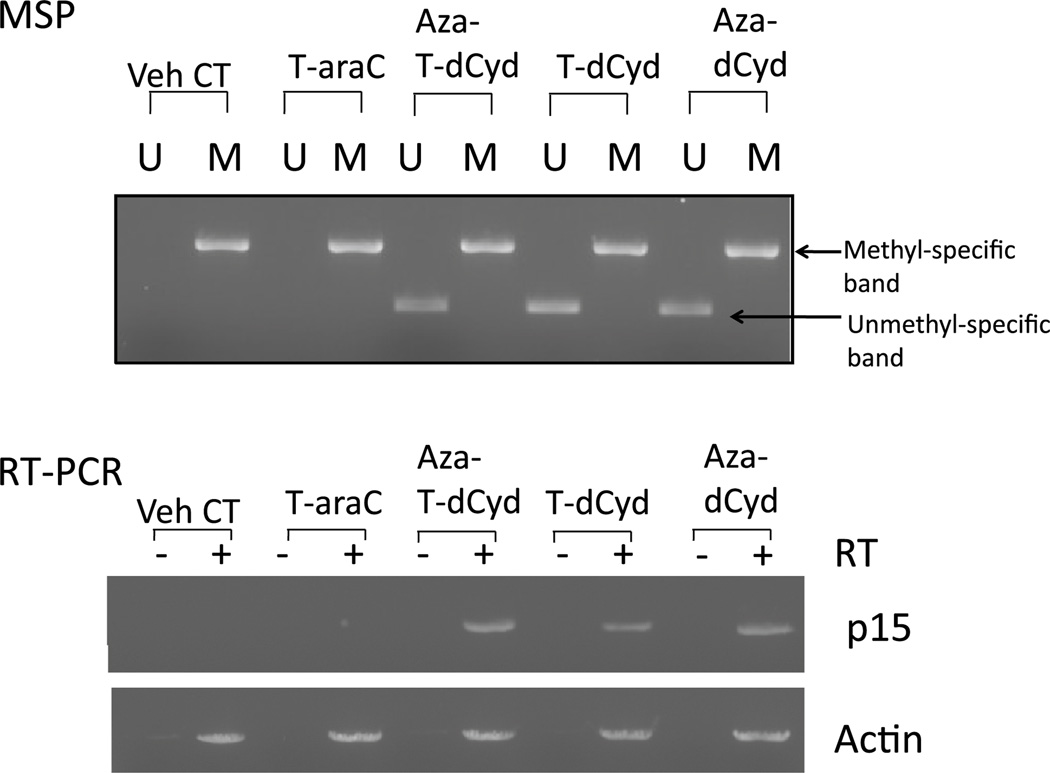
Previous studies have demonstrated that aberrant methylation patterns of the p15 tumor suppressor CpG island occur in acute myeloid leukemia and that this aberrant pattern was recapitulated in KG1a myeloid leukemia cells [28, 30, 31]. We first determined the methyl-CpG status of the hypermethylated CpG island in the p15 tumor suppressor gene of these cells. KG1a cells were exposed to either vehicle, T-dCyd (3 µM), aza-T-dCyd (1 µM), aza-dCyd (1 µM) or 4′-thioarabinofuranosyl cytosine (T-araC; 0.3 µM) for 72 h. Methylation-specific PCR (MSP) using bisulfite-modified DNA with primers specific for unmethylated DNA and methylated DNA demonstrated that T-dCyd and aza-T-dCyd were capable of demethylating the p15 CpG similar to the clinically used aza-dCyd (Fig. 3). However, treatment with T-araC (a cytotoxic 4′-thionucleoside analog with a natural cytosine base but containing a 4-thioarab-inose sugar [22]), did not demethylate the p15 CpG indicating that the 4-thio-2-deoxyribose was essential for this effect. Incorporation of T-araC into DNA is known to result in profound inhibition of DNA synthesis [36], due to its ability to cause DNA chain termination [37].
Fig. 3.
Effect of T-dCyd and aza-T-dCyd on p15 CpG methylation and its expression, a MSP analysis: Exponentially growing KG1a were exposed to 3 µM T-dCyd, 1 µM aza-T-dCyd, 1 µM aza-dCyd or 0.3 µM T-araC for 96 h. Genomic DNA was isolated and amplification of bisulfite-treated genomic DNA was accomplished as described in the text. Primer sets used for amplification of DNA are designated as unmethylated (U) and methylated (M). Amplified DNA was run in ethidium bromide stained gels, b RT-PCR analysis of pl 5 expression. Exponentially growing KG1a cells were exposed to 3 µM T-dCyd, 1 µM aza-T-dCyd, 1 µM aza-dCyd or 0.3 µM T-araC for 96 h and total RNA was prepared and reverse transcribed. RT-PCRs for p15 detection was accomplished with first-strand cDNA samples (or negative controls that lacked RT, reverse transcriptase) in ethidium bromide stained agarose gels
Next, we examined the effects of T-dCyd and aza-T-dCyd on the expression of the p15 gene in KG1a cells using a semi-quantitative RT-PCR analysis (Fig. 3) to assess whether treatment with T-dCyd and aza-T-dCyd could cause re-expression of the p15 gene as a result of the depletion of DNMT1. These data demonstrate that both T-dCyd and aza-T-dCyd were as effective as aza-dCyd in inducing re-expression of p15. T-araC treatment did not induce p15 re-expression, indicating that a 4′-thio in the context of arabinose was not sufficient for this effect (Fig. 3).
T-dCyd is activated and incorporated into HCT-116 DNA despite being deaminated
Previously we have shown in CCRF-CEM cells that T-dCyd is readily activated to T-dCTP [22]. However, since T-dCyd is a good substrate for cytidine deaminase and CCRF-CEM cells do not express this enzyme, we evaluated the metabolism of T-dCyd in HCT-116 cells (a cell line that expresses high levels of cytidine deaminase) to understand how expression of cytidine deaminase affects T-dCyd metabolism. All metabolic products were determined in HCT-116 cell cultures after a 1-h incubation with 1 µM [5-3H]dCyd, 1 µM [5-3H]T-dCyd or 1 µM [5-3H]T-dCyd plus 100 µM tetrahydrouridine (THU, a potent inhibitor of cytidine deaminase) (Table 1). The radioactivity in the medium was analyzed by reverse phase HPLC to measure the amount of parent compound, its deaminated product and tritiated H2O. Because the [3H] label is at the five position in the cytosine ring, it is released as [3H]H2O, if the 5-position of T-dUMP or dUMP is methylated by TS, and no radioactive thymidine nucleotides will be produced in cells treated with 5-3H labeled dCyd or its analogues. The acid-soluble extract of the HCT-116 cells was analyzed by SAX HPLC to measure the amount of tritiated intracellular metabolites, and the incorporation of radioactivity into acid-insoluble material was determined as a measure of its incorporation into DNA. In this experiment, we recovered more than 97 % of the original radioactivity in the various metabolic products. Data shown in Table 1 demonstrate that after incubation for 1 h only 64 % of the T-dCyd remained in the culture medium: 34 % of the radioactivity was 4′-thio-2′-deoxyuridine (T-dUrd) in the medium, and 1.5 % of the T-dCyd had been activated to T-dCTP (0.2 %) and incorporated into the DNA (1.3 %) (Table 1). A small peak of radioactivity (0.02 % of the total radioactivity) was detected in the monophosphate region of the SAX HPLC column that was identified as T-dUMP, because its formation was inhibited by the addition of THU, a potent inhibitor of cytidine deaminase. Also very little, if any, [3H] labeled H2O (0.21 %) was detected in this experiment. The addition of THU to cells treated with T-dCyd totally inhibited the appearance of T-dUrd in the medium, but had very little effect on the incorporation of T-dCyd into DNA or the level of the acid-soluble metabolites of T-dCyd.
Table 1.
Metabolism of dCyd or T-dCyd in HCT-116 cells
| Parent | Percent of total radioactivity recovered |
DNA | Total | ||||
|---|---|---|---|---|---|---|---|
| dUrd or T-dUrd | H2O | dUMP or T-dUMP | TP | ||||
| dCyd | 56 | 28 | 12 | 0.01 | 0.05 | 4 | 100 |
| T-dCyd | 64 | 34 | 0.2 | 0.02 | 0.2 | 1.3 | 100 |
| T-dCyd + THU | 97 | 0.8 | 0.1 | ND | 0.2 | 1.0 | 100 |
HCT-116 cells were incubated with 1 µM [5-3 H]dCyd, 1 µM [5-3H]T-dCyd or 1 µM [5-3H]T-dCyd plus 100 µM THU for 1 h at 37 °C, and the complete metabolism of each compound was determined. The culture medium was analyzed for the original compound, its deaminated product, and H2O using reverse phase HPLC. An acid-soluble extract of the cell pellet was analyzed by SAX HPLC for nucleotides, and the radioactivity incorporated into acid-insoluble material was determined as a measure of incorporation of compound into DNA. More than 97 % of the radioactivity added to the culture was recovered at the 1-h time point
ND not detected
With respect to dCyd metabolism, 40 % of the dCyd was deaminated and only 4 % was activated by dCyd kinase to dCTP and incorporated into DNA. Twelve percent of the radioactive dCyd metabolites were detected in the cell culture medium as [3H]H2O and 28 % was [3H]dUrd. The small amount of radioactivity associated with the [3H] T-dUMP peak and [3H]H2O in cells treated with T-dCyd (0.23 %) versus that seen in cells treated with [3H]dCyd (12 %) is in agreement with previous studies that indicate T-dUrd is a very poor substrate for thymidine kinase [24, 32].
Because T-dCyd even at high concentrations did not deplete DNMT1 levels in HCT-116 cells and T-dCyd is rapidly deaminated by HCT-116 cells, we evaluated the rate of disappearance of T-dCyd from the medium of HCT-116 cells treated with 10 or 100 µM T-dCyd. The half-life of T-dCyd was 1.2 h in cell cultures incubated with 10 µM T-dCyd, and it was 5 h in cell cultures incubated with 100 µM T-dCyd. At both concentrations, there was no T-dCyd detected in the culture medium 24 h after the addition of T-dCyd. These results initially suggested that the lack of DNMT1 depletion activity of T-dCyd in HCT-116 cells could be due to its rapid deamination. However, we found that treatment with T-dCyd in the presence of 100 µM THU also did not affect DNMT1 levels (not shown). This fact, coupled with our metabolic studies that show that T-dCyd is activated and incorporated into the DNA in HCT-116 cells (Table 1), suggest that the lack of DNMT1 depletion activity of T-dCyd is not due to its deamination.
T-dCyd and aza-T-dCyd deplete DNMT1 in vivo in tumors implanted in immunodeficient mice
Mice-bearing CCRF-CEM tumors (approximately 200 mg) were treated with either 9 mg/kg of T-dCyd (i.p. qld × 9) or 5 mg/kg aza-T-dCyd (i.p. qld × 9). One day after the end of therapy, the tumors were removed and the level of DNMT1 protein was determined. As seen in Fig. 4a, DNMT1 levels were decreased in tumors of mice that were treated with both T-dCyd and aza-T-dCyd, although aza-T-dCyd was more effective than T-dCyd. In this experiment, treatment with either T-dCyd or aza-T-dCyd was well tolerated. The mice in the T-dCyd treatment group had a maximum loss in mean body weight of 13 %, whereas there was no loss in mean body weight in mice treated with aza-T-dCyd. Previous studies had determined that these doses were the maximum tolerated doses (MTDs) on the nine-day treatment schedule. In a repeat experiment, mice were treated with 2.25, 4.5 or 9 mg/kg T-dCyd (i.p. qld × 9), and depletion of DNMT1 levels was again observed at the 9 mg/kg dose but not at the two lower doses (not shown), which indicated that treatment with T-dCyd resulted in the depletion of DNMT1 near its MTD. However, with aza-T-dCyd DNMT1, protein levels were depleted at all doses (1.25, 2.5 or 5 mg/kg aza-T-dCyd given i.p. qld × 9) (Fig. 4b, left panel), which indicated that there was excellent selectivity with aza-T-dCyd (at least tenfold). We also evaluated the effect of T-dCyd treatment (25 mg/kg, qd × 3) on DNMT1 levels in NCI-H23 lung carcinoma xenografts and found that DNMT1 levels were completely depleted in these tumors as well (Fig. 4c).
The effect of aza-dCyd (1.25, 2.5 or 5 mg/kg aza-dCyd given i.p., qld × 9) was also determined (Fig. 4b, left panel). Treatment with aza-dCyd also resulted in depletion of DNMT1 in the tumors, although we found that it was much more toxic to the mice than aza-T-dCyd. Treatment of mice with 2.5 and 5 mg/kg aza-dCyd killed both mice in each treatment group, and treatment with 1.25 mg/kg aza-dCyd killed one of two mice in this treatment group. We then further reduced the doses administered to subsequent mice (Fig. 4b, right panel). Results indicated that tumor DNMT1 remained completely depleted in mice treated with either aza-T-dCyd or aza-dCyd even at a dose of 0.625 mg/ kg, although this dose killed one of two mice in the aza-dCyd group. At a dose of 0.3125 mg/kg, there was no acute toxicity with either agent; however, tumor DNMT1 depletion was not complete. These results indicated that aza-T-dCyd depleted DNMT1 as well as aza-dCyd but it was much better tolerated than aza-dCyd, which suggests that it may be a better clinical agent than aza-dCyd due to a wider therapeutic index.
Antitumor efficacy of T-dCyd, aza-T-dCyd and aza-dCyd in NCI-H23 lung tumors implanted in immunodeficient mice
The efficacy of T-dCyd in NCI-H23 xenografts using two different treatment schedules was evaluated in these studies. In Fig. 5a, T-dCyd was injected three times daily for nine consecutive days. Both, the 1.3 and 0.9 mg/kg doses (total daily dose of 3.9 and 2.7 mg/kg/day, respectively) inhibited NCI-H23 tumor growth in a dose-dependent manner. In addition, two of the six mice in each of the T-dCyd treatment groups were tumor-free on Day 58 (day of study termination). Both doses were tolerated without deaths. Animals experienced a maximum mean body weight loss of 17 and 8 %, respectively (data not shown).
In Fig. 5b, we compared the antitumor efficacy of T-dCyd with that of aza-dCyd using the NCI-H23 tumor xenograft model. The compounds were administered once daily for five consecutive days followed by 2 days of rest, which was repeated two times (qld × 5, qlw × 3) for aza-dCyd and four times for T-dCyd (qld × 5, qlw × 5). Although neither aza-dCyd nor T-dCyd caused tumor regressions, they both produced full tumor growth stasis. However, aza-dCyd was very toxic at this dose and schedule with all but one mouse succumbing to toxicity before the 3 weeks of therapy could be finished. Most of the aza-dCyd-treated mice died from drug toxicity (days of death: Day 35; ten mice on Day 40; with three surviving mice killed on Day 46 for sample collections). T-dCyd was much better tolerated, but two of the ten mice were lost to toxicity near or shortly after the end of the fifth treatment cycle (treatment ended Day 57; days of death 56, 60), and three mice were harvested for sample collections on Day 61 to assess pharmacodynamic marker status. The remaining five mice survived to the end of the experiment (Day 82), with small tumors that remained in stasis.
Aza-T-dCyd at a dose of either 6.7 or 10 mg/kg/dose, given qld × 9, was also effective against NCI-H23 tumor xenografts (Fig. 5c). There was only one death in each aza-T-dCyd treatment group and treatment at the lower dose resulted in one complete tumor regression out of six animals. Surviving animals experienced a maximum mean body weight loss of 11 and 14 %, respectively (data not shown).
Aza-T-dCyd is more stable than aza-dCyd
Because aza-dCyd is known to be unstable in aqueous solutions due to hydrolytic cleavage of the triazine ring [33], we evaluated the chemical stability of both aza-dCyd and aza-T-dCyd. The compounds were dissolved in phosphate buffered saline (pH 6.7) and incubated at 37 °C. Samples were removed at various times after the addition of compound, and the amount of parent compound was immediately determined by HPLC as described [22]. Under these conditions, the half-life of aza-dCyd was 13 ± 1 h, whereas with aza-T-dCyd, it was 45 ± 6 h (mean and standard deviation from three separate experiments), which indicated that the half-life of aza-T-dCyd in solution was approximately three times longer than that of aza-dCyd. Similar results were obtained at pH 6.2 and 7.2, but at pH 7.7, the rate of disappearance of aza-T-dCyd (half-life of 12 h) was similar to that of aza-dCyd (half-life of 8 h). T-dCyd is stable in aqueous solutions indefinitely.
Plasma half-life of T-dCyd
The pharmacokinetic parameters of T-dCyd and dCyd were determined in nontumored male athymic nude mice. Either 100 mg/kg [3H]T-dCyd or [3H]dCyd was injected i.p. into 30 mice (10 µCi per injection). Three mice from each treatment group were euthanized at 5, 15, 30, 60 and 120 min after the injection of compound, and plasma was obtained and analyzed by reverse phase HPLC for parent compound and deaminated product as described [22]. The peak plasma concentration of T-dCyd was approximately 100 µM, which occurred between 5 and 10 min after injection of T-dCyd, and its apparent half-life was 10–15 min. T-dUrd was the primary plasma metabolite which reached plasma concentrations as high as 50 µM 15 min after injection of T-dCyd and was still at 10 µM 2 h after injection of T-dCyd. This result indicated that the plasma half-life of T-dUrd (approximately 45 min) was much longer than that of T-dCyd. The rate of disappearance of T-dCyd was similar to that of dCyd (half-life of 16 min). We detected dUrd in the plasma of mice treated with dCyd, but it rapidly disappeared at a rate similar to that of dCyd. A considerable amount of radioactivity was detected in the void volume of the HPLC column in the plasma of mice treated with [3H]dCyd, which was not observed in mice treated with [3H]T-dCyd. This metabolite(s) has not been identified, but because dUrd is a substrate for thymidine phosphorylase, it is likely that this radioactivity is uracil or its catabolic products. The fact that this metabolite(s) was not detected in mice treated with T-dCyd and the long plasma half-life of T-dUrd indicates that T-dUrd is at best a poor substrate for thymidine phosphorylase. This experiment was repeated one time with similar results.
Discussion
In this report, we have demonstrated that both T-dCyd and aza-T-dCyd effectively deplete DNMT1 in cancer cells resulting in the re-expression of the tumor suppressor gene p15. Both compounds depleted DNMT1 in human tumor xenografts as well and were effective in inhibiting tumor growth in these models. It is particularly interesting that treatment with T-dCyd resulted in good depletion of DNMT1, even though it has an unmodified cytosine nucleobase (Fig. 1). Kumar et al. [29] had shown that T-dCyd when used in an oligonucleotide replacing the target dCyd strongly inhibited the methylation reaction by M. HhaI methyltransferase in solution. Since the presence of sulfur at the 4 position of the sugar was not incompatible chemically with the known mechanism for C5-cytosine methylation, and crystal structures of the complex did not reveal substantial deformations, these investigators suggested a greatly slowed reaction [29]. Further studies showed that T-dCyd significantly increases the off-rate of the bound oligonucleotide, suggesting that the substrate-enzyme complex forms with M. HhaI methyltransferase but fails to become fully stabilized. Thus, it is likely that a similar mechanism of action of T-dCyd vis-á-vis the human DNMT1 could result in transient enzyme-substrate complexes that initiate events that destabilize the free DNMT1 protein by targeting it to the proteasome.
In our studies, we found that aza-T-dCyd was a more potent DNMT1 depleting agent than T-dCyd and that it was also efficacious in cell lines where T-dCyd was rather ineffective (Fig. 2), even though T-dCyd was converted to its triphosphate and incorporated into DNA (Table 1). This might be expected since the nitrogen in place of C5 in the cytosine ring in aza-T-dCyd could behave in a similar fashion to that of the clinically approved aza-nucleosides, i.e., it facilitates nucleophilic attack at the C6 of the cytosine, but then renders the enzyme–DNA complex irreversible, resulting in proteasomal degradation of the free DNMT1 protein. In these studies, incubation of HCT 116 cells with T-dCyd (1 µM) resulted in approximately 25 pmol of T-dCyd incorporated into DNA in a 1-h period. Previous studies [22] had indicated that incubation of CEM cells with T-dCyd (0.1 µM) resulted in approximately 5 pmol of T-dCyd incorporated into DNA in a 1-h period. Adjusting for the concentration differences in these experiments indicates that similar amounts of T-dCyd was incorporated into DNA in both sensitive (CEM) and insensitive (HCT 116) cell lines; a conclusion that is supported by studies showing that dCyd kinase activity was similar in the two cell lines (data not shown). Therefore, the amount of T-dCyd incorporated in the DNA cannot explain the different DNMT1 depleting activity observed in these two cell lines. Since T-dCyd has an unmodified cytosine base, it is likely that the mechanism of DNMT1 turnover induced by this compound is distinct and less efficient than the one induced by azacytosine nucleosides. It is possible that mechanistic differences in DNMT1 turnover between different cell lines could determine whether or not treatment with T-dCyd will result in depletion of DNMT1, which could explain some of the variable response that is observed with T-dCyd. It is also likely that the covalent trapping of DNMT1 on DNA containing aza-T-dCyd results in specific events that turn on DNMT1 turnover while also turning on other pathways in order to remove these bulky adducts that ultimately lead to cell death. The pathways turned on by T-dCyd incorporated DNA on the other hand could be different and dependent on other events. For instance, it was recently shown that the lysine methyltransferase SET7 colocalizes with DNMT1 and monomethylates it, which leads it to proteasome-mediated degradation [34]. Therefore, one could speculate that in certain cell types, such as the leukemia lines tested herein, DNMT1 might be more permissive for rapid degradation due to such additional uncharacterized modifications. One could further envisage a situation wherein the presence of lysine methylation or another modification on DNMT1 in a certain cell type could be a major determinant of whether a particular analog such as T-dCyd can initiate its degradation.
The results of our current and prior work [22] indicate that T-dCyd has favorable metabolic features that maximize its inhibition of DNMT1 activity and minimize its conversion to metabolites that result in off target toxicities. T-dCyd is a good substrate for dCyd kinase, and it is incorporated into DNA at a rate that was similar to that of the natural nucleoside, dCyd, and that significant incorporation of T-dCyd into DNA occurred at concentrations that did not result in the inhibition of cell growth [22]. This result suggests that similar to 4′-thio-thymidine triphosphate [35], T-dCTP is a good alternative substrate for DNA polymerases and that its incorporation into DNA does not result in the inhibition of subsequent DNA chain elongation. T-dCMP is also a very poor substrate for dCMP deaminase which indicates that this important metabolic pathway in dCyd metabolism will not siphon off T-dCMP to dUrd and thymidine nucleotides which could potentially inhibit other enzymes (such as TS) causing toxicities unrelated to the inhibition of DNMT1. Although T-dCyd treatment would generate T-dUrd in animals, our studies in HCT-116 cells indicated that T-dUrd is a poor substrate for thymidine kinase and is not very toxic to mammalian cells. These results suggest that the metabolism of other dCyd analogs that incorporate the 4′-thio modification (aza-T-dCyd) would also be limited and would minimize production of nucleotide metabolites that could inhibit other enzymes involved in DNA metabolism (DNA polymerases, ribonucleotide reductase, TS, etc.).
Although the effect of these agents on human tumor xenografts in mice is encouraging, one cannot definitively conclude from the results shown in Figs. 4 and 5 that the effect seen on tumor growth (Fig. 5) is entirely due to depletion of DNMT1. It is possible that the depletion of DNMT1 is only partly responsible for the antitumor activity observed with these compounds, and future studies will be needed to determine whether depletion of DNMT1 is responsible for the effect of tumor growth. Based on the proposed mechanism of action (inhibition of DNA methylation of daughter strands), DNMT1 depleting agents might not be immediately toxic to tumor cells. It should take two or more generations of tumor cell growth for the depletion of DNMT1 to result in the expression of tumor suppressor genes to exert a negative effect on tumor growth. The fact that treatment with aza-T-dCyd resulted in depletion of DNMT1 at concentrations that were far below the IC50 supports this hypothesis. The metabolism and mechanism of action of all of these agents, including aza-dCyd and aza-Cyd, have not been thoroughly evaluated, and therefore, it is possible that their observed antitumor activity in mice is due to other mechanisms as well. Furthermore, human tumor xenografts may not be the best model for use in the evaluation of DNMT1 targeting agents, because it is possible that these tumors have been selected to rapidly proliferate in mice and that the re-expression of tumor suppressor genes may not be sufficient to stop their proliferation in mice, whereas re-expression of tumor suppressor genes in actual patient cancers would be highly effective.
It is important to note that it was not clear, a priori, whether an aza-cytosine nucleoside with a 4′ thio moiety would prove to be an effective compound, even though both aza-dCyd and T-dCyd result in good depletion of DNMT1. It is not possible to predict the biological activity of a compound that combines two or more modifications in other nucleoside analogs, because nucleosides must undergo several anabolic reactions in cells before they can be incorporated into nucleic acids and the combined modifications create a new molecule that interacts with each of these enzymes in an unknown manner. There are many examples where small structural modifications to nucleoside analogs result in unexpected biological activity either good or bad. A guideline in the design of new nucleoside analogs that has been borne out by decades of studies is that they should only include small structural changes, and as few changes as possible in comparison with the natural molecule. However, our studies herein lead us to conclude that despite the 4′ thio moiety aza-T-dCyd is a reasonable substrate for these reactions. In other words, it is sufficiently activated and incorporated into DNA without inhibition of DNA synthesis, and once incorporated, it is recognized by and forms a complex with DNMT1 resulting in a potent DNMT1 depletion response.
It was of considerable interest that aza-T-dCyd was much better tolerated than aza-dCyd even though it had similar effects on DNMT1 depletion in tumor cells. This result indicated that the selectivity index of aza-T-dCyd (ratio of MTD to that of minimal DNMT1 depleting dose) was at least tenfold greater than that of aza-dCyd. The reason why aza-T-dCyd was less toxic than aza-dCyd is not understood, but suggests that aza-dCyd elicits off target effects that aza-T-dCyd does not. It is possible that, as with T-dCyd, the 4′-thio modification in aza-T-dCyd limits its conversion to dUrd nucleotides via dCMP deaminase. Further studies are necessary to directly compare the metabolism and mechanism of action of these two agents to understand exactly how they differ. Although the DNMT1 depleting agents are only approved for use in the treatment of hematologic diseases, the rationale for re-expression of tumor suppressor genes is also relevant to solid tumors (4). Solid tumors are typically not very sensitive to nucleoside analogs, because many express low levels of dCyd kinase activity. Since our studies indicate that aza-T-dCyd is as good as aza-dCyd in depletion of DNMT1 but is much better tolerated, they suggest that ten times as much aza-T-dCyd will be activated in solid tumor cells increasing the chance that DNMT1 depleting strategy could also be applied to the treatment of solid tumors. Regardless, the greater selectivity of aza-T-dCyd in depletion of DNMT1 with respect to aza-dCyd indicates that this agent should be evaluated in humans as a potential DNMT1 depleting agent.
Acknowledgments
We thank Dr. Mark Suto for careful review of the manuscript. We also thank Dr. Joel Morris for T-dCyd synthesis and Dr. Robert J. Kinders for providing tumor lysates from T-dCyd-treated mice. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. NCI-Frederick and Southern Research Institute are accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the “Guide for Care and Use of Laboratory Animals” (National Research Council; 2010; National Academy Press; Washington, D.C.). Work described in this report was funded by NIH Grant # P01 CA34200, NCI contract N01-CO-12400, the ADDA (Alabama Drug Discovery Alliance) and the UAB Center for Clinical and Translational Science under 5UL1 RR025777.
Footnotes
Conflict of interest None.
Contributor Information
Jaideep V. Thottassery, Email: thottassery@southernresearch.org, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA; University of Alabama at Birmingham, Comprehensive Cancer Center, Birmingham, AL, USA.
Vijaya Sambandam, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA.
Paula W. Allan, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA
Joseph A. Maddry, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA
Yulia Y. Maxuitenko, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA
Kamal Tiwari, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA.
Melinda Hollingshead, Biological testing Branch, NCI at Frederick, Frederick, MD, USA.
William B. Parker, Email: parker@southernresearch.org, Drug Discovery Division, Southern research Institute, 2000 ninth avenue South, Birmingham, AL 35205, USA; University of Alabama at Birmingham, Comprehensive Cancer Center, Birmingham, AL, USA.
References
- 1.Brown KD, Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat Genet. 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer— a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Issa JP, Herman J, Bassett DE, Jr, Nelkin BD, Baylin SB. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vertino PM, Issa JP, Pereira-Smith OM, Baylin SB. Stabilization of DNA methyltransferase levels and CpG island hypermethylation precede SV40-induced immortalization of human fibroblasts. Cell Growth Differ. 1994;5:1395–1402. [PubMed] [Google Scholar]
- 8.Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–390. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 9.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 10.Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 11.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, Baylin SB. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 12.Milutinovic S, Knox JD, Szyf M. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAFl/CIPl) J Biol Chem. 2000;275:6353–6359. doi: 10.1074/jbc.275.9.6353. [DOI] [PubMed] [Google Scholar]
- 13.Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- 14.Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, Egorin MJ. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine. Cancer Chemother Pharmacol. 2008;62:363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
- 21.Marquez VE, Barchi JJ, Jr, Kelley JA, Rao KV, Agbaria R, Ben-Kasus T, Cheng JC, Yoo CB, Jones PA. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. The magic of its chemistry and biology. Nucleosides, Nucleotides Nucleic Acids. 2005;24:305–318. doi: 10.1081/ncn-200059765. [DOI] [PubMed] [Google Scholar]
- 22.Parker WB, Shaddix SC, Rose LM, Waud WR, Shewach DS, Tiwari KN, Secrist JA., III Metabolism of 4′-thio-beta-D-arabinofuranosylcytosine in CEM cells. Biochem Pharmacol. 2000;60:1925–1932. doi: 10.1016/s0006-2952(00)00520-7. [DOI] [PubMed] [Google Scholar]
- 23.Thottassery JV, Tiwari KN, Westbrook L, Secrist JA, III, Parker WB. Novel 2′-deoxycytidine analogs as DNA demethylation agents. Proc Am Assoc Cancer Res. 2011;71:2537. doi:10.1158/1538-7445.AM2011-2537. [Google Scholar]
- 24.Secrist JA, III, Tiwari KN, Riordan JM, Montgomery JA. Synthesis and biological activity of 2′-deoxy-4′-thio pyrimidine nucleosides. J Med Chem. 1991;34:2361–2366. doi: 10.1021/jm00112a007. [DOI] [PubMed] [Google Scholar]
- 25.Tiwari KN, Cappellacci L, Montgomery JA, Secrist JA., III Synthesis and anti-cancer activity of some novel 5-aza-cytosine nucleosides. Nucleosides, Nucleotides Nucleic Acids. 2003;22:2161–2170. doi: 10.1081/ncn-120026872. [DOI] [PubMed] [Google Scholar]
- 26.Someya H, Shaddix SC, Tiwari KN, Secrist JA, III, Parker WB. Phosphorylation of 4′-thio-beta-D-arabinofuranosylcytosine and its analogs by human deoxycytidine kinase. J Pharmacol Exp Ther. 2003;304:1314–1322. doi: 10.1124/jpet.102.045435. [DOI] [PubMed] [Google Scholar]
- 27.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodge JE, Munson C, List AF. KG-1 and KG-1a model the p15 CpG island methylation observed in acute myeloid leukemia patients. Leuk Res. 2001;25:917–925. doi: 10.1016/s0145-2126(01)00053-4. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by Hhal methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, Baylin SB. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 31.Dodge JE, List AF, Futscher BW. Selective variegated methylation of the p15 CpG island in acute myeloid leukemia. Int J Cancer. 1998;78:561–567. doi: 10.1002/(sici)1097-0215(19981123)78:5<561::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 32.Verri A, Focher F, Duncombe RJ, Basnak I, Walker RT, Coe PL, de Clercq E, Andrei G, Snoeck R, Balzarini J, Spadari S. Anti-(herpes simplex virus) activity of 4′-thio-2′-deoxyuridines: a biochemical investigation for viral and cellular target enzymes. Biochem J. 2000;351(Pt 2):319–326. [PMC free article] [PubMed] [Google Scholar]
- 33.Rogstad DK, Herring JL, Theruvathu JA, Burdzy A, Perry CC, Neidigh JW, Sowers LC. Chemical decomposition of 5-aza-2′-deoxycytidine (Decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chem Res Toxicol. 2009;22:1194–1204. doi: 10.1021/tx900131u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker WB, Shaddix SC, Rose LM, Tiwari KN, Montogmery JA, Secrist JA, III, Bennett LL., Jr Metabolism and metabolic actions of 4′-thiothymidine in L1210 cells. Biochem Pharmacol. 1995;50:687–695. doi: 10.1016/0006-2952(95)00178-3. [DOI] [PubMed] [Google Scholar]
- 36.Someya H, Waud WR, Parker WB. Long intracellular retention of 4′-thio-arabinofuranosylcytosine 5′-triphosphate as a critical factor for the anti-solid tumor activity of 4′-thio-arabinofuranosylcytosine. Cancer Chemother Pharmacol. 2006;57:772–780. doi: 10.1007/s00280-005-0126-0. [DOI] [PubMed] [Google Scholar]
- 37.Richardson KA, Vega TP, Richardson FC, Moore CL, Rohloff JC, Tomkinson B, Bendele RA, Kutcha RD. Polymerization of the triphosphates of araC, 2′,2′-difluorodeoxycytidine (dFdC) and OSI-7836 (T-araC)by human DNA polymerase alpha and DNA primase. Biochem Pharmacol. 2004;68:2337–2346. doi: 10.1016/j.bcp.2004.07.042. [DOI] [PubMed] [Google Scholar]