Abstract
Background
Design patterns, in the context of software development and ontologies, provide generalized approaches and guidance to solving commonly occurring problems, or addressing common situations typically informed by intuition, heuristics and experience. While the biomedical literature contains broad coverage of specific phenotype algorithm implementations, no work to date has attempted to generalize common approaches into design patterns, which may then be distributed to the informatics community to efficiently develop more accurate phenotype
Methods
Using phenotyping algorithms stored in the Phenotype KnowledgeBase (PheKB), we conducted an independent iterative review to identify recurrent elements within the algorithm definitions. We extracted and generalized recurrent elements in these algorithms into candidate patterns. The authors then assessed the candidate patterns for validity by group consensus, and annotated them with attributes.
Results
A total of 24 electronic Medical Records and Genomics (eMERGE) phenotypes available in PheKB as of 1/25/2013 were downloaded and reviewed. From these, a total of 21 phenotyping patterns were identified, which are available as an online data supplement.
Conclusions
Repeatable patterns within phenotyping algorithms exist, and when codified and cataloged may help to educate both experienced and novice algorithm developers. The dissemination and application of these patterns has the potential to decrease the time to develop algorithms, while improving portability and accuracy.
Keywords: Electronic health record, Phenotype, Algorithms, Software design, Design patterns
Introduction
Electronic health records (EHRs) have been shown to be a valuable source of information for biomedical research, including the definition and identification of clinical phenotypes.[1-5] The increasing use of EHRs[6, 7] has resulted in large quantities of data available for secondary purposes such as research. In order to better handle this growing source of data, we need to improve methods and approaches to phenotype more efficiently.
The electronic Medical Records and Genomics (eMERGE) network has been a leader in the development of phenotype algorithms based on EHR data. In addition to the work done through eMERGE for genome-wide association studies (GWAS),[8-15] there are additional examples of electronic algorithms to mine EHRs for identifying diseases for biomedical research and clinical care[16-20] disease surveillance,[21] pharmacovigilance,[22] as well as for decision support.[23] These studies have provided some guidance on dealing with the challenges of using EHR and claims data.[2, 20, 24-26] This guidance has often been in the context of a single algorithm, although more recent work has begun to address the broader challenges of using EHR data for phenotyping.[1, 27] Additionally, research is being conducted to identify how electronic phenotype algorithms may be represented and made more portable across disparate EHRs,[28, 29] which has the potential to automate approaches to handle the complexities and nuances of EHR data.
A major goal of the current phase of eMERGE is to improve the ease and speed of developing new phenotype definitions. No known work to date, however, has attempted to broadly classify challenges and solutions to using EHR data for the development of electronic phenotype algorithms, or demonstrated an approach to widely disseminate the findings. This knowledge could potentially reduce the time to develop phenotype algorithms, improve portability to other sites and even accuracy by describing experiences developing other algorithms. The primary goal of this paper is to apply lessons from prior work in software design patterns to the problem of defining and disseminating EHR-based phenotype algorithms.
In software engineering, the use of design patterns are frequently used to generate solutions to common problems or scenarios.[30] These patterns are free from any technical implementation details, such as programming language or database platform. Design patterns are not applicable only in the domain of software development. They have roots in architecture,[31] and have recently been applied to the development of ontologies.[32, 33] and health information technology (HIT) solutions.[34, 35] Even though design patterns are used in multiple domains, they share similar constructs that form a basis of overall pattern languages.[36] Generally, design patterns provide: (1) a description of a scenario or problem that exists and that the pattern may address; (2) a template for a solution; and, (3) considerations for when to apply the pattern, or what its implications may be.[30, 31, 36] Design patterns are not intended to capture every possible pattern that may occur in the target domain; rather, they represent best practices and common approaches to solving a problem. In practice, they may be derived from intuition, heuristics and experience.
In order to more widely disseminate solutions to common problems and scenarios found in the development of electronic phenotype algorithms, we propose the creation of “EHR-driven phenotype extraction design patterns”—logical patterns recurring frequently in phenotyping algorithms that are EHR and technology agnostic. This paper presents an initial catalog of such patterns from experiences within the eMERGE network.
Methods
The steps used to define, develop and review phenotype design patterns are shown in Figure 1, and are explained in more detail below.
Figure 1.
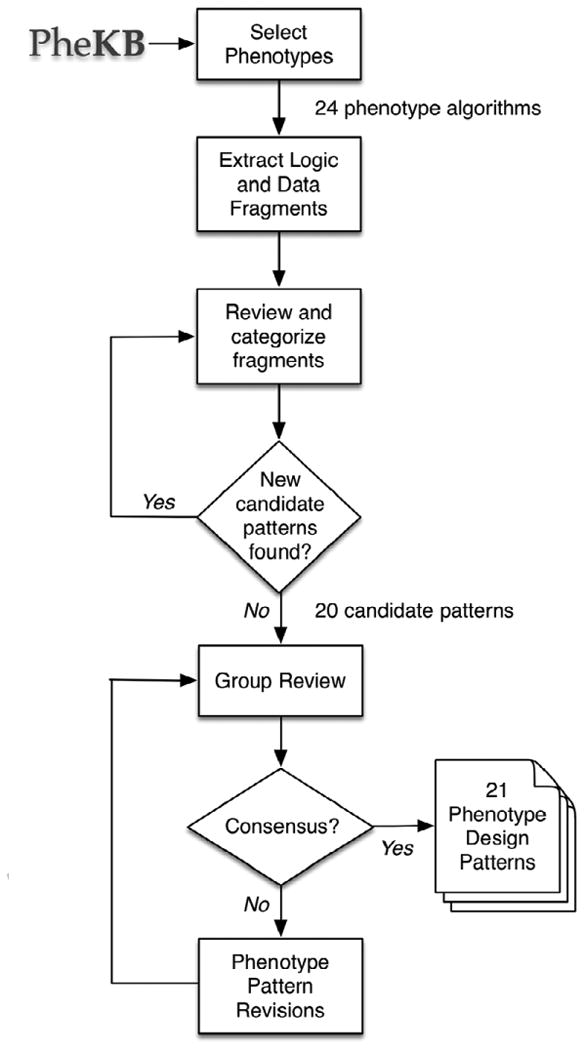
Methods for developing and reviewing phenotype design patterns.
Setting
The eMERGE network[37] is a National Human Genome Research Institute (NHGRI)-sponsored initiative that has demonstrated the feasibility of EHR-derived phenotypes in order to conduct genome-wide association studies (GWAS). Within the network, sites develop and locally validate an EHR-based phenotype algorithm, which are then implemented and validated at one or more additional network sites. While phenotype algorithms themselves are largely recorded as text documents[38] and have to be re-implemented in a format that can be executed at each site, the transfer of phenotypes from one site to other sites with different EHR systems demonstrates the broader application of EHR-derived phenotyping.
Phenotype Selection
Phenotypes created by the eMERGE network are publicly available on the Phenotype KnowledgeBase website (PheKB, http://www.phekb.org), and are classified by the group or consortium under which the algorithm was created, as well as a status to indicate how mature the algorithm is in its development process. For this study, all phenotype algorithms associated with the eMERGE network that were marked with a “Final” or “Validated” status were downloaded on January 25, 2013. The algorithm set consists of both case/control studies (i.e. Cataracts, Resistant Hypertension) as well as quantitative measures (i.e. Red Blood Cell Indices, White Blood Cell Indices). The algorithms were developed, implemented, and validated by chart review by at least one other eMERGE site. Phenotypes that were available in PheKB but had not been validated were considered too preliminary for study, and were excluded. In addition, the selected algorithms within the eMERGE network were not developed independently (sites collaborated on and built new algorithms after having reviewed others), which allowed evaluation of shared experiences as algorithms were developed over time.
Phenotype Algorithm Review
One of the authors (LVR) reviewed each of the phenotype algorithms, and identified unique, discrete fragments in the text definitions that represented the inputs, logic, and constraints within the algorithms. As multiple artifacts can exist for each phenotype algorithm (i.e. chart abstraction forms for validation, data dictionary definitions), only documentation containing a textual description of the algorithm was reviewed.
Coding the algorithms followed a form of discourse analysis, where we identified short text fragments that represent distinct constructs of the algorithm definition. There are various forms of these constructs, such as the sources of data, temporal criteria and Boolean combinations. We extracted these fragments and annotated them with a set of tags to denote the context and use of that fragment in the algorithm. The list of tags was dynamically created as new examples were encountered. In addition, given the repeated nature of types of fragments within an algorithm, only distinct fragments were extracted, and subsequent repeats or similar fragments were ignored. The intent was to study distinct examples per algorithm, and the selection and use of fragments and their tags were used by reviewers independently to aid in identifying candidate patterns. Figure 2 shows as an example the fragments identified in a single phenotype algorithm and the contextual tags applied.
Figure 2.
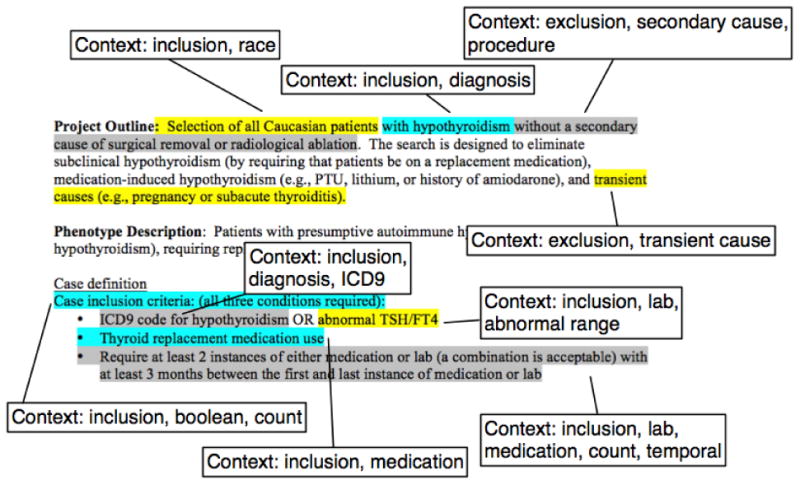
Example annotation and tagging of fragments within the eMERGE Hypothyroidism phenotype algorithm (available from http://www.phekb.org).
Identifying Phenotyping Patterns
Once we coded all of the algorithms, one of the authors (LVR) iteratively reviewed the list of fragments for repeating categories, which represent candidate patterns. If a potential candidate pattern was identified, we sought additional supporting examples in the list of fragments. We accepted a candidate pattern only if it was present in two or more algorithms, to better represent that the approach was repeated and generalizable. The fragment review process was iterative, using the initial contextual tags as high-level categories. Sub-categories were created ad-hoc by subjective assessment of the reviewer, given specific details of the fragments, and new sub-category tags added where identified, with sub-categories representing candidate patterns. This process was performed on the full list of fragments until no new candidate patterns were identified. We then created a template for phenotyping patterns, based on typical representations for software and other design patterns. Table 1 shows the fields in this template.
Table 1. Fields used to define phenotyping patterns.
| Field | Description |
|---|---|
| Name | An abbreviated name to identify the pattern |
| Description | A description of the approach to take to address a scenario encountered in electronic phenotyping. |
| Reasoning | An explanation of why the pattern's approach is used. This helps to provide additional context on the complexities of EHR data. |
| Examples | Example text fragments that represent solutions to address the problem, or particular scenario. |
| Considerations when to use | Specific scenario(s) under which the pattern should be considered for use. |
| Considerations when not to use | Specific scenario(s) under which the pattern may not be needed, or may not be appropriate to use. |
| Related to | Other patterns that this pattern is related to, as well as a brief description of the relationship. |
The candidate patterns were subsequently reviewed by WKT, JAP, ANK, DSC, JP, PLP and GT. Reviewers independently voted to approve, tentatively accept, or reject each candidate pattern, provide justification for tentative acceptance or rejection, and to also provide recommendations to improve or clarify the patterns. Level of agreement between the seven reviewers on the initial vote was measured by Gwet's AC1[39]. Reviewers, given their collective experience with developing phenotype algorithms, were also asked to recommend any candidate patterns (following the template fields shown in Table 1) that they felt should exist and were not included. This expert-based heuristic assessment was used to complement the single-reviewer, example-based approach used to create the initial candidate pattern list. Recommended patterns from the reviewers were assessed against the list of fragments to see if there were supporting examples. Comments and votes from the reviewers were consolidated, and majority-rejected patterns removed. The remaining patterns were considered the list of approved patterns, and their content iteratively refined by all authors.
To further correct for potential bias of a single reviewer, the methods were repeated by another author (WKT). Given the reviewer's involvement in the group review process, the intent of this supplemental review was primarily to account for missed patterns and further refine patterns given new evidence. The second reviewer iteration also started with the identification of fragments from the phenotype algorithm documents, followed by an iterative review to create candidate patterns, and included a joint reconciliation process with the first reviewer to determine if candidate patterns were represented in the first set, or constituted a new pattern.
Following this supplemental review process, all authors performed a final review and refinement step to arrive at the final list of approved patterns.
Result
In total, 24 eMERGE algorithms from PheKB marked as “Final” or “Validated” were reviewed. The list of algorithms reviewed is shown in Table A1 of the online data supplement. For these algorithms, the initial reviewer identified 340 fragments. Of the 340 fragments, 35.6% (n=121) of the fragments were selected, having met the selection criteria. These selected fragments were used as evidence to construct an initial set of 19 candidate patterns. The initial level of agreement between the 7 reviewers on acceptance of the candidate patterns was 69.7%. No candidate patterns were rejected at the end of the review process. In the course of the group review, one new pattern was recommended by reviewers, with supporting examples found amongst the 24 phenotype algorithms. The secondary reviewer produced 402 fragments, from which 14 candidate patterns were identified. From this set, 13 were deemed supportive of the initial reviewer's 19 candidate patterns, and included a new pattern not previously identified. The second reviewer did not explicitly identify all used fragments, and so no usage statistics from the second review are reported.
In total, 21 phenotype patterns were identified, and were placed into one of five categories. The list of pattern names, with abbreviated descriptions and associated benefits, is shown in Table 2. The full listing of patterns is available as an online data supplement.
Table 2. List of phenotyping patterns derived from a review of 24 eMERGE phenotype algorithms.
| Pattern Name | Description | Benefits |
|---|---|---|
| Anchor Date | Define a static date around which all queries and validations are anchored. | Makes results reproducible, such that ongoing changes in a person's disease state do not invalidate existing validations. |
| Composition of Algorithms | Using an algorithm that was created and validated as a component of another algorithm. | Promotes creation and reuse of validated phenotypes to be used in other phenotypes, without re-creating, or using sub-optimal, definitions. |
| Consolidate Multiple Values | For quantities represented by repeated or multiple measures, provide a single computed value to represent the multiple values. | Simplifies analysis and the amount of data that needs to be managed. |
| Account for Data Outliers | Filter out noisy or incorrect values to help ensure calculations (including average) are not skewed. | Improves accuracy of the phenotype definition. |
| Ad Hoc Categories | Group codes, medications, etc. into ad hoc categories that are not part of a standard terminology or protocol. | Simplifies the definition of a phenotype algorithm. |
| Multi-Mode Sources | Account for information collected in multiple formats or sources across an EHR over time. | Can improve accuracy of the phenotype definition by including all sources of data where information may solely be recorded. |
| Established Patient | Make sure the patient is seen within the healthcare system at a regular enough basis so that the information pertinent to the algorithm would be on record. | Improves accuracy by making sure enough data is present to make an accurate determination about disease state. |
| Confirm Variable Was Checked | Make sure the patient has been seen by a healthcare professional, and that the encounter would be sufficient enough to measure the absence or presence of a disease or other observation. | As patients who are not checked for a disease may in fact have that disease, which could confound analyses, ensures that patients have been checked. |
| Qualifiers for Evidence | Require additional qualifiers, such as severity, to exist before accepting a clinical observation. | May improve accuracy, and also allows for stratification of disease based on its progression, state, and/or severity. |
| Rule of N | Require at least N independent pieces of evidence substantiating a condition or event to reduce the chance that extraneous or incidental data is over interpreted as indicating the condition or event is present. | Improves accuracy by correcting for data that could have been recorded without sufficient context. |
| Use Distinct Time Intervals | When requiring a count of items, make sure they happen on different dates and/or times, optionally with some time interval between them. | Allows for a more specific phenotype definition by setting a time window in which a disease should have progressed, or ensures that observations are spaced apart to indicate an ongoing condition. |
| Credentials of the Actor | Require that a person with appropriate authority (e.g., a physician with specific credentials or practicing in a particular specialty department) recorded the clinical data. | Increases confidence or precision in a diagnosis if a specialist has recorded it. |
| Establish Assertion Status | Determine if assertion qualifiers affect the meaning of medical observations (e.g., the meaning of assertions about “cough” vary depending on whether they are qualified by negation, uncertainty, hypothetical, historical references). | Improves accuracy by establishing context around observations. |
| Medications Likely Taken | Require more assurance that a patient was actually taking the medication, such as through claims data or having multiple prescriptions over time. | Improves accuracy of phenotypes that rely on medication usage to confirm absence and/or presence of the phenotype. |
| Medication Details | When checking for medications, it may be necessary to look at dose, frequency and/or route. | Allows more precise definition of phenotype where medication attributes are important. |
| Evolving Reference Standards | Use ranges of dates in which vocabulary codes or lab ranges are valid, if the underlying standards are known to have changed. | Increases accuracy of the phenotype by ensuring the right code is used. |
| Transient Condition Caveats | For patients having transient conditions (e.g., pregnancy), take into account how those transient conditions may alter the interpretation of proximal clinical observations. | Improves accuracy of analysis by removing variables that may be confounded by some condition. |
| Medical Setting of Action | Explicitly require that data be collected (or not be collected) in a particular setting of interest (i.e. inpatient, outpatient). | Improves accuracy of the phenotype by using the encounter setting to add context to the interpretation of an observation. |
| Context of Evidence | Consider the context or setting in which a clinical observation is made. For example, when interpreting clinical text mentions of particular conditions, take into account how its interpretation may vary depending on the section of a report in which it appears (e.g., Past Medical History, Problem List, or Family History). | Improve accuracy by looking at the context of how something is recorded. |
| Temporal Dependencies | Consider the relationship over time between different events and/or ages at which events occurred. | Provides more complete phenotype definitions where progression and temporal dependencies are important. |
| Inception of Condition | Explicitly define the date to use when determining the onset of a condition using multiple sources of information (medications, labs, diagnosis codes). | Provides consistency in the results of a phenotype algorithm. |
All of the reviewed phenotype algorithms implemented at least one pattern, with the High-Density Lipoproteins (HDL) algorithm having the most patterns (n=10). Across all of the 24 reviewed algorithms, there was an average of 5 patterns per algorithm (standard deviation 2.7). For the 21 identified phenotype design patterns, there was an average of 5.7 algorithms that implemented each pattern (standard deviation 2.8), with the “Rule of N” pattern identified in 13 of the 24 algorithms, and the “Composition of Algorithms” and “Ad Hoc Categories” patterns being identified in the minimum two algorithms. A listing of all phenotype design patterns and the phenotype algorithms they are found in is available in Table A2 of the online data supplement.
Discussion
Our major finding is that identifiable design patterns occur frequently in phenotype algorithms. We have identified and validated 21 unique phenotype design patterns, which we have defined to be: a) present in two or more separate phenotype algorithms (recurring), b) free of any EHR, institution or technology specific details (generalizable), and c) applicable in certain situations, with guidance on when to use (contextual). This affirms experimentally what has been believed anecdotally in the phenotyping community—that, while each phenotype may be unique, effective phenotype algorithms frequently contain similar patterns of clinical variables. Furthermore, this is consistent with the concept of design patterns in general—identifying solutions where a feeling of “deja-vu” exists around a problem.[30]
The need for phenotype design patterns stems from the challenges of using EHR data. The EHR is not always an explicit representation of the patient's health at a point in time, due in part to limited data collected or lack of context surrounding its interpretation.[27, 40] One lesson from eMERGE is that the process of developing a phenotype algorithm is best done as an iterative process, involving a diverse team that understands how the information is captured clinically, how it is represented by the EHR, and how that data should be extracted and interpreted. In the course of developing these phenotype algorithms, eMERGE sites typically describe the pros and cons of different approaches they took in order to prevent other sites from making similar mistakes, or to save them time in arriving at an optimal approach.
As an example, the Height phenotype algorithm was noted as implementing the Multi-Mode Sources, Account for Data Outliers and Temporal Dependencies patterns. Briefly, the Height algorithm is looking for adult height measurements for patients that would be unaffected by factors such as disease or medication. The Multi-Mode Sources pattern was employed to use both structured data and NLP. The use of NLP was recommended to more accurately identify vertebral compression fractures, for which ICD-9 codes did not adequately cover. The Account for Data Outliers pattern was applied to correct for data errors in the EHR, such as inches being recorded in a field designated for centimeters (50 inches recorded as 50 cm). Finally, the Temporal Dependencies pattern was applied to exclude height measurements that followed an event that would affect height (i.e. lower limb amputations) and confound analysis if included.
As with design patterns in other domains, the phenotype design patterns presented here represent a level of subjective assessment, although rooted in the review of actual phenotype algorithms. Invariably, there will be some disagreement on what constitutes a phenotype pattern, and what constitutes “common sense” or a basic approach to phenotype algorithm development (similar disagreements exist in the software development community[41]). This was demonstrated by the lower rate of agreement for acceptance of certain patterns amongst the reviewers, with several comments raised if a pattern was too simple. For example, at face value “Composition of Algorithms” may appear to recommend the creation of reusable functions— novice common practice in software development. However, upon closer inspection, the intent of that pattern prescribes consideration for reuse in the development process (much less commonly done), and closer inspection to identify previously hidden “sub-algorithms” that may be extracted, refined and curated for reuse. The descriptions associated with the proposed phenotype design patterns attempt to justify why the pattern exists, but may require refinement or expansion over time.
Many of the proposed design patterns represent different levels of abstraction with how they may be applied. For example, the “Composition of Algorithms” pattern describes strategies for how an algorithm would be structured. The “Rule of N” pattern is applied directly within the algorithm logic. While no formal classification scheme is proposed to address this within our study, general categories (which are used in the online supplement) can provide a logical grouping.
In addition, as with other design patterns, some of the patterns are closely related, possibly representing a specific instantiation of a pattern for a type of clinical information, or multiple patterns often being used in conjunction. For example, during the review process, the “Confirm Variable Was Checked” pattern—which establishes reasonable evidence that a clinical measure would be recorded—was initially seen as simply checking if something exists (a very basic operation). The justification given for this pattern is that while the output is a Boolean indicator, it provides specific guidance on how to assess multiple sources of information (encounters, appointments, physician details, departments, etc.) to arrive at that decision.
The phenotype algorithms selected for review were specific to the eMERGE network, and were intended to identify phenotypes for GWAS. Algorithms developed for different use cases might have a slightly different focus, and hence different patterns of varying complexity. However, these represent phenotypes spanning a variety of disease types and implemented across multiple institutions against a variety of vendor and home-grown EHRs. While it is possible that phenotype algorithms developed by other institutions, or algorithms developed for other purposes may differ, the algorithms represented a reasonable sample of data types extracted from diverse EHRs. Also, the assignment of design patterns always involves an inherent level of subjectivity. The requirement that examples (represented by the document fragments) exist in at least two algorithms for candidate patterns was one approach to account for this, and the resulting patterns were reviewed by a panel of domain experts who were intimately familiar with the phenotype algorithms, having developed and/or implemented these algorithms at their respective sites. As a result, every algorithm and pattern mapping was vetted by two or more experts familiar with the algorithm. However, the initial use of a single reviewer is a limitation. Although another reviewer did replicate the methods and identified a new phenotype pattern, it was done after the group review process, which would bias the overall collection of patterns found by the second reviewer. This represents one approach to identifying phenotype algorithm patterns and other techniques warrant exploration, such as the Delphi method to avoid potential bias.
The optimal use of design patterns in phenotyping deserves some discussion. While we observed variability in the number of design patterns used in each phenotype algorithm reviewed, and that each algorithm in our sample contained at least one pattern, there is no prescriptive number of phenotype design patterns that should be applied when developing an algorithm. In any domain, patterns may be used at inappropriate times due to inexperience or by cognitive biases.[42, 43] It is important to understand the context in which a pattern should be applied, and be able to justify why a pattern was applied. It is recommended that the pattern definition be read in its entirety, and consideration given if the scenarios described by the pattern seem applicable to the situation at hand. Also, the creation of phenotype algorithms is an iterative process, and in some cases patterns should only be applied after initial versions of the algorithm deem it necessary (e.g., adopt a Rule of N only if single observations introduce excessive false positive error). Finally, conducting validations during the development of phenotype algorithms is strongly encouraged as a best practice,[44] and will help to further validate how a phenotype design pattern may improve results, quantify the amount of improvement it offers, or reveal when marshaling a particular pattern may be appropriate.
Conclusion
Existing EHR-based phenotype algorithms reflect a wealth of experience and knowledge about the secondary use of EHR data. The development of a list of phenotype design patterns based on existing phenotype algorithm definitions from the eMERGE network should help both novice and experienced data analysts navigate the nuances and complexities of working with EHR data for algorithm development. Their use also has the potential to conserve algorithm development time while improving accuracy and portability. The set of patterns presented here is intended as a starting point for articulating and documenting generalizable patterns useful in phenotype development, and we expect members of the broader biomedical informatics community to augment and refine it.
Supplementary Material
Highlights.
Defined the concept of a “phenotype design pattern” to aid in developing phenotype algorithms
Evaluated 24 phenotype algorithms created by the electronic Medical Record and Genomics (eMERGE) network
Identified 21 phenotype design patterns from the corpus of eMERGE algorithms
Acknowledgments
Funding: The eMERGE Network was initiated and funded by National Human Genome Research Institute through the following grants: U01HG006828 (Cincinnati Children's Hospital Medical Center/Harvard); U01HG006830 (Children's Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health); U01HG006382 (Geisinger Health System); U01HG006375 (Group Health Cooperative); U01HG006379 (Mayo Clinic); U01HG006380 (Mount Sinai School of Medicine); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University); and U01HG006385 (Vanderbilt University serving as the Coordinating Center).
JAP, JCD, JP, LVR and WKT received additional support from NIGMS grant R01GM105688-01. LVR and JBS received additional support from NCATS grant 8UL1TR000150-05. JCD received additional support from NLM grant R01LM010685.
Footnotes
Competing Interests: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS computational biology. 2012 Dec;8(12):e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden DM, Xu H, Denny JC, Wilke RA. Electronic medical records as a tool in clinical pharmacology: opportunities and challenges. Clinical pharmacology and therapeutics. 2012 Jun;91(6):1083–86. doi: 10.1038/clpt.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome research. 2009 Sep;19(9):1675–81. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nature reviews Genetics. 2012;13(6):395–405. doi: 10.1038/nrg3208. 06//print. [DOI] [PubMed] [Google Scholar]
- 5.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nature reviews Genetics. 2011 Jun;12(6):417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 6.Charles D, King J, Patel V, Furukawa M. ONC Data Brief, no 9. Washington, DC: Office of the National Coordinator for Health Information Technology; Mar, 2013. Adoption of Electronic Health Record Systems among U.S. Non-federal Acute Care Hospitals: 2008-2012. [Google Scholar]
- 7.Jamoom E, Beatty P, Bercovitz A, Woodwell D, Palso K, Rechtsteiner E. NCHS Data Brief, no 98. Hyattsville, MD: National Center for Health Statistics; Jul, 2012. Physician Adoption of Electronic Health Record Systems: United States, 2011. [PubMed] [Google Scholar]
- 8.Crosslin DR, McDavid A, Weston N, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Human genetics. 2012 Apr;131(4):639–52. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010 Nov 16;122(20):2016–21. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. American journal of human genetics. 2011 Oct 7;89(4):529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association : JAMIA. 2012 Mar-Apr;19(2):212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PloS one. 2010;5(9) doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2012 Mar-Apr;19(2):225–34. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner SD, Berg RL, Linneman JG, et al. Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PloS one. 2011;6(5):e19586. doi: 10.1371/journal.pone.0019586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waudby CJ, Berg RL, Linneman JG, et al. Cataract research using electronic health records. BMC ophthalmology. 2011;11:32. doi: 10.1186/1471-2415-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll RJ, Eyler AE, Denny JC. Naive Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2011;2011:189–96. [PMC free article] [PubMed] [Google Scholar]
- 17.Giampietro PF, McCarty C, Mukesh B, et al. The role of cigarette smoking and statins in the development of postmenopausal osteoporosis: a pilot study utilizing the Marshfield Clinic Personalized Medicine Cohort. Osteoporosis international : a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA. 2010 Mar;21(3):467–77. doi: 10.1007/s00198-009-0981-3. [DOI] [PubMed] [Google Scholar]
- 18.Ho ML, Lawrence N, van Walraven C, et al. The accuracy of using integrated electronic health care data to identify patients with undiagnosed diabetes mellitus. Journal of evaluation in clinical practice. 2012 Jun;18(3):606–11. doi: 10.1111/j.1365-2753.2011.01633.x. [DOI] [PubMed] [Google Scholar]
- 19.Kudyakov R, Bowen J, Ewen E, et al. Electronic health record use to classify patients with newly diagnosed versus preexisting type 2 diabetes: infrastructure for comparative effectiveness research and population health management. Population health management. 2012 Feb;15(1):3–11. doi: 10.1089/pop.2010.0084. [DOI] [PubMed] [Google Scholar]
- 20.Singh B, Singh A, Ahmed A, et al. Derivation and validation of automated electronic search strategies to extract Charlson comorbidities from electronic medical records. Mayo Clinic proceedings Mayo Clinic. 2012 Sep;87(9):817–24. doi: 10.1016/j.mayocp.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubberke ER, Nyazee HA, Yokoe DS, et al. Implementing automated surveillance for tracking Clostridium difficile infection at multiple healthcare facilities. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2012 Mar;33(3):305–8. doi: 10.1086/664052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon D, Park MY, Choi NK, Park BJ, Kim JH, Park RW. Detection of adverse drug reaction signals using an electronic health records database: Comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm. Clinical pharmacology and therapeutics. 2012 Mar;91(3):467–74. doi: 10.1038/clpt.2011.248. [DOI] [PubMed] [Google Scholar]
- 23.Wright A, Pang J, Feblowitz JC, et al. Improving completeness of electronic problem lists through clinical decision support: a randomized, controlled trial. Journal of the American Medical Informatics Association : JAMIA. 2012 Jul-Aug;19(4):555–61. doi: 10.1136/amiajnl-2011-000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen-Dicker J, Klompas M. Comparison of electronic laboratory reports, administrative claims, and electronic health record data for acute viral hepatitis surveillance. Journal of public health management and practice : JPHMP. 2012 May-Jun;18(3):209–14. doi: 10.1097/PHH.0b013e31821f2d73. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman D. Pitfalls of using administrative data for research. Digestive diseases and sciences. 2010 Jun;55(6):1506–8. doi: 10.1007/s10620-010-1246-x. [DOI] [PubMed] [Google Scholar]
- 26.Jensen PN, Johnson K, Floyd J, Heckbert SR, Carnahan R, Dublin S. A systematic review of validated methods for identifying atrial fibrillation using administrative data. Pharmacoepidemiology and drug safety. 2012 Jan;21(Suppl 1):141–7. doi: 10.1002/pds.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2013 Jan 1;20(1):117–21. doi: 10.1136/amiajnl-2012-001145. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. Journal of biomedical informatics. 2012 Aug;45(4):763–71. doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2012;2012:911–20. [PMC free article] [PubMed] [Google Scholar]
- 30.Gamma E, Helm R, Johnson R, Vlissides J. Design patterns: elements of reusable object oriented software. Addison-Wesley Longman Publishing Co., Inc.; 1995. [Google Scholar]
- 31.Alexander C, Ishikawa S, Silverstein M. A Pattern Language: Towns, Buildings, Construction. USA: Oxford University Press; 1977. [Google Scholar]
- 32.Hoehndorf R, Ngonga Ngomo AC, Pyysalo S, Ohta T, Oellrich A, Rebholz-Schuhmann D. Ontology design patterns to disambiguate relations between genes and gene products in GENIA. Journal of biomedical semantics. 2011;2(Suppl 5):S1. doi: 10.1186/2041-1480-2-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortensen JM, Horridge M, Musen MA, Noy NF. Applications of ontology design patterns in biomedical ontologies. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2012;2012:643–52. [PMC free article] [PubMed] [Google Scholar]
- 34.Timpka T, Eriksson H, Ludvigsson J, Ekberg J, Nordfeldt S, Hanberger L. Web 2.0 systems supporting childhood chronic disease management: a pattern language representation of a general architecture. BMC medical informatics and decision making. 2008;8:54. doi: 10.1186/1472-6947-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkinson MD, Vandervalk B, McCarthy L. The Semantic Automated Discovery and Integration (SADI) Web service Design-Pattern, API and Reference Implementation. Journal of biomedical semantics. 2011;2(1):8. doi: 10.1186/2041-1480-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salingaros NA. The structure of pattern languages. arq: Architectural Research Quarterly. 2000;4(02):149–62. [Google Scholar]
- 37.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC medical genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2011;2011:274–83. [PMC free article] [PubMed] [Google Scholar]
- 39.Gwet K. Handbook of Inter-Rater Reliability: How to Estimate the Level of Agreement Between Two or Multiple Raters. Gaithersburg, MD: STATAXIS Publishing Company; 2001. [Google Scholar]
- 40.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. Journal of the American Medical Informatics Association : JAMIA. 2013 Jan 1;20(1):144–51. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Budgen D. A survey of experienced user perceptions about software design patterns. Information and Software Technology. 2013;55(5):822–35. 5// [Google Scholar]
- 42.Laurier W, Poels G. Research Note: Ontology-Based Structuring of Conceptual Data Modeling Patterns. IGI Global. 2012:50–64. [Google Scholar]
- 43.Tversky A, Kahneman D. Judgment under Uncertainty: Heuristics and Biases. Science (New York, NY) 1974 Sep 27;185(4157):1124–31. doi: 10.1126/science.185.4157.1124. [DOI] [PubMed] [Google Scholar]
- 44.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. Journal of the American Medical Informatics Association : JAMIA. 2013 Mar 26; doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


