Abstract
Electronic health record (EHR) data show promise for deriving new ways of modeling human disease states. Although EHR researchers often use numerical values of laboratory tests as features in disease models, a great deal of information is contained in the context within which a laboratory test is taken. For example, the same numerical value of a creatinine test has different interpretation for a chronic kidney disease patient and a patient with acute kidney injury. We study whether EHR research studies are subject to biased results and interpretations if laboratory measurements taken in different contexts are not explicitly separated. We show that the context of a laboratory test measurement can often be captured by the way the test is measured through time.
We perform three tasks to study the properties of these temporal measurement patterns. In the first task, we confirm that laboratory test measurement patterns provide additional information to the stand-alone numerical value. The second task identifies three measurement pattern motifs across a set of 70 laboratory tests performed for over 14,000 patients. Of these, one motif exhibits properties that can lead to biased research results. In the third task, we demonstrate the potential for biased results on a specific example. We conduct an association study of lipase test values to acute pancreatitis. We observe a diluted signal when using only a lipase value threshold, whereas the full association is recovered when properly accounting for lipase measurements in different contexts (leveraging the lipase measurement patterns to separate the contexts).
Aggregating EHR data without separating distinct laboratory test measurement patterns can intermix patients with different diseases, leading to the confounding of signals in large-scale EHR analyses. This paper presents a methodology for leveraging measurement frequency to identify and reduce laboratory test biases.
Keywords: electronic health record, laboratory testing, bias, confounding, missing data, information theory
1. Introduction
Millions of patients across the United States have extensive medical histories stored in electronic form. This immense amount of electronic health record (EHR) data provides a unique platform to perform large-scale research studies of human health. Through careful analysis of the variables in this vast dataset, researchers can conduct a variety of multifaceted studies such as prediction of future patient health state, evaluation of intervention effectiveness, computational disease modeling, and identification of dangerous drug-drug interactions [1, 2, 3]. Automating feature selection from EHR variables is a difficult task, as the EHR is an inherently biased data source: EHR data are collected with the primary goal of delivering and documenting patient care, not with the primary goal of creating a curated research dataset [4, 5]. Identifying and then mitigating such biases will result in not only the development of more accurate methods for deriving computational models of disease but also in learning better prediction models from EHR data. Currently, laboratory tests are one of the most widely-used features in EHR disease-modeling research and are therefore the focus of this paper.
In this work, we hypothesize that (i) the specific context of a laboratory test order can be derived from EHR-observed measurement patterns, and (ii) that this context can be leveraged for better disease modeling. While a laboratory test’s numerical values can help distinguish healthy from sick patients, test values themselves cannot separate sick patients by their ailment when the test is associated with multiple diseases. For instance, while the numerical results may be comparable, the rate of measurement for a gestational diabetes screening glucose test and a chronic diabetes monitoring glucose test will differ greatly. We predict that how often a laboratory test is ordered within a particular time window can help correctly separate one disease state from another. We further hypothesize that laboratory test measurement patterns provide complementary and independent information from the numerical values indicated by the laboratory tests. We formally explore the relationship between both laboratory test measurement gaps and laboratory test values to determine whether the context in which a laboratory test is ordered alters the way its value should be interpreted, and is therefore a critical feature for disease modeling. While analyses of laboratory measurement patterns have been conducted [6, 7, 8, 9], the analyses and interpretations have focused on resource overutilization and informing clinical practice rather than on EHR-driven research.
Before describing our methods and findings, we provide background on laboratory testing from an informatics standpoint and report on previous work in the emerging research area of EHR bias identification and mitigation.
1.1. Capturing The Context of Laboratory Testing: Reasons for Ordering and Relationship to Numerical Values
At the point of patient care, different laboratory tests are ordered at different rates, often dictated by what physiologic process the test is measuring, and very often there exist multiple reasons for ordering a particular laboratory test. The three most common reasons for ordering a test are (i) diagnosing a condition, (ii) screening for a condition, or (iii) monitoring a pre-existing condition. Some laboratory tests are ordered for one specific, clinical reason: for instance, the prostate-specific antigen (PSA) test is ordered exclusively to screen patients for prostate cancer. Others serve multiple clinical purposes: TSH, for instance, is used both to diagnose and monitor patients with disorders associated with the thyroid hormone. Finally, some tests, such as creatinine, are ordered both for clinical purposes like monitoring chronic disease progression and diagnosing acute conditions, and for healthcare process purposes like following guidelines as part of a routine panel for preventive testing [10]. When hospital protocol dictates measurement times, as is the case with routine preventative panels, creatinine’s measurement patterns arguably reflect healthcare processes more than they reflect the health status of a patient. Thus, the context in which a laboratory test is ordered depends both on its clinical purpose and the surrounding healthcare processes.
Deriving the context of a laboratory measurement is a challenge, however. EHR data lack an explicit indication for why each laboratory test was ordered, and using other dimensions of EHR data for derivation of such information (such as ICD-9 codes and clinical notes) is equally problematic. ICD-9 codes are notoriously non-specific to patient disease state and are often not recorded for all patient ailments[11, 12]. Clinical notes rarely explicitly state the exact reason a test has been ordered.
The specific description of the context in which a laboratory test is measured, therefore, is not included in most computational models of disease. In fact, most often models include only a laboratory test’s numerical value, a range of values[13, 14], or the presence or absence of a laboratory test[15] as features, but no contextual information about the situation surrounding the order. In this paper, we investigate whether aggregating numerical values of laboratory tests taken in multiple separate contexts without explicitly separating the contexts can lead to the confounding of resarch conclusions.
In research with clinical data, there is an implicit assumption that a laboratory test’s numerical value and the rate at which the test is ordered are highly correlated features. This assumption about value and measurement rate correlation likely stems from the existence of value-based guidelines and the widespread expectation that laboratory test values which fall outside of normal ranges prompt intervention and retesting. Value-based guidelines for laboratory test ordering dictate measurement frequency based on a test’s numerical value. For instance, the guideline for performing a diagnostic PSA test states that if a patient’s PSA is slightly over 4.0 ng/mL in the initial measurement, the PSA test should be remeasured within 48 hours to confirm the need for a biopsy. Our work formally investigates the linear and nonlinear relationship between numerical value and measurement patterns in EHR-recorded data.
1.2. EHR biases
EHR data are biased because they are gathered in an uncontrolled environment and are not carefully curated for research purposes. EHR data are noisy, sometimes erroneous, and often sparse[14]. At the same time EHR data contain sometimes conflicting (e.g., notes and coded data provide differing medication lists) and redundant information (e.g., clinicians often copy-and-paste from previous notes). From a temporal standpoint, the EHR contains data about elements that evolve at different time scales and often evolve over time, as treatment affects patient state. Because of these complexities, assessing the impact of EHR biases and correcting for their impact on data-driven methods is an emerging research topic.
Recent research has shown that naïve EHR statistical analyses can lead to the reversals of cause and effect [16], induction of spurious signals [17], large errors when predicting optimal drug dosage [18], cancellation of temporal signals when aggregating different cohorts[19, 20, 21], and model distortion when not accounting for redundancy in the narrative part of the EHR [22].
One particularly problematic bias inherent to the EHR is the prevalence of data points that are missing not at random[23] (e.g., patients are seen and measured more often when they are sick, and measured less often when they are healthy). Inferring missing information, such as values when the patient is not seen, is a challenging research area. While there have been different approaches to mitigating this type of missingness[24, 14], mostly researchers ignore missing values or interpolate them[25, 26, 27], with some recent work on classifying which variables should be interpolated and which should be ignored[28]. Lin and Haug demonstrated that some missing values are themselves informative by creating Bayesian networks that explicitly model the absence of clinical variables; these models were able to predict medical problems better than those that ignored or interpolated missing values[15]. Our work builds upon Lin and Haug’s findings and focuses on leveraging the temporal missingness within laboratory measurement data. We see the patterns of laboratory test measurement as patterns of missing data. As a way to mitigate the EHR biases, we explore the use of different missing-not-at-random patterns to classify different patient health states and stratify heterogeneous populations into homogenous patient groups.
2. Material and Methods
Our study is carried out in three consecutive tasks, as described below:
Task 1 We explored the correlation of laboratory values to the laboratory test’s time to repeat, examining whether the value and time between consecutive measurements (measurement gap) encode separate information or overlap in information content.
Task 2 To understand the overall dynamics of laboratory tests recorded in the EHR, we categorized types of laboratory measurement patterns, identifying those more likely to cause biases in EHR-based research.
Task 3 We used lipase as a case study for how rates of measurement can be used to account for biases in laboratory test measurement data.
2.1. Ethics statement
This work was approved by the Columbia University Institutional Review Board (IRB #AAAK7201). As it is impractical to collect consent for such a large-scale study and the data is already available through the in-house data warehouse, informed consent was waived by the Institutional Review Board for this retrospective research.
2.2. Clinical Data
We extracted patient records from the NewYork-Presbyterian Hospital (NYPH) clinical data warehouse. We narrowed our population to patients that have visited the NYPH Ambulatory Internal Medicine clinic at least 3 times. The full longitudinal records (i.e., all inpatient and outpatient data points) for these patients were gathered.
Three physicians reviewed and edited a list of frequently measured laboratory tests. They constructed a set of 70 laboratory tests of interest to primary care and internal medicine. We extracted the time series for these tests between September 1990 and September 2010 for all of the patients in the population.
2.3. Task 1: Correlation between Measurement Gap and Numerical Value
We quantified the relationship between value and measurement gap, asking: in the patient population, is there added information in looking at how a patient was measured, not only at the measurement value? Given a particular laboratory test and all patients’ time series for that test, we constructed a joint probability density function (PDF) using a kernel density estimate in Matlab. The PDF consisted of laboratory values and time between consecutive lab measurements (or gaps between measurements) in days.
To assess the degree of correlation between a laboratory test’s numerical values and its measurement gaps, we experimented with (i) linear correlation (estimated at the 95% confidence interval) and an associated p-value and (ii) a non-linear measure of correlation, mutual information (MI) between laboratory test values and gaps between measurements. Mutual information attains a value of zero when the random variables underlying the distributions (values and measurement patterns) are completely independent. Mutual information attains a maximum when the two distributions are deterministic functions of each other. In the latter case, the mutual information is equal to the entropy of the single equivalent distribution. Using the PDF, both the linear correlation and mutual information were calculated for the entire dataset (the full time scale) and separately for measurements with long and short measurement gaps to capture distinct temporal dynamics at different time scales. The confidence interval estimates on the MI were made using a previously described method[29]. By examining many measurement gap histograms, we chose a heuristic cutoff of 3 days for what constitutes a long gap and what constitutes a short gap: the short time scale calculations only look at gaps and values where consecutive measurements are taken no more than 3 days apart and long time scale calculations look at gaps and values for measurement gaps that are longer than 3 days.
2.4. Task 2: Finding Laboratory Test Measurement Motifs
We explored the different types of laboratory test measurement dynamics that exist in the EHR data by creating measurement gap histograms for 70 different laboratory tests. We computed the following quantity: for each laboratory test taken on each patient, we calculated the days between two consecutive measurements of that laboratory test on that patient. A histogram was created for each test, mapping the day gap between consecutive measurements and number of such gaps, when aggregated across the entire patient dataset. For example, if a patient had a creatine test taken on February 3rd and another creatinine test taken on February 5th, the count of creatinine tests with a measurement gap of 2 days would be incremented by one.
We visualized the measurement gap histograms in different coordinate systems to explore the measurement dynamics across laboratory tests. We uncovered differences when examining the histograms in the logarithmic coordinate system. Using log-log coordinates, we visually looked for modes present in the histograms. If there is linearity in a measurement gap histogram when presented in log-log coordinates (i.e., a power-law) that implies scale-free measurement dynamics and that all time scales represent a single context or reason for ordering the laboratory test. If no approximately linear relationship between the frequency of measurement gaps exists, we visually looked for changes (e.g., peaks) that separate the different dynamics patterns; these different patterns may qualitatively imply different contexts of measurement based on either a change in health state or based on the healthcare documentation process. We catalogued the measurement gap histograms based on observed approximate linearity and the presence of peaks in the histograms, as determined by a manual review of the curves.
2.5. Task 3: Studying the Potential Effect of Measurement Motifs on Research
In this task, we focus on a specific laboratory test as a use case for studying the effect of measurement motifs on EHR-driven research.
For the use case, we chose to study lipase and acute pancreatitis. Acute pancreatitis is a well-understood condition and because its diagnosis is largely laboratory-based it is a good test case to validate our hypotheses. Both amylase and lipase tests have been used for acute pancreatitis diagnosis but they are not specific for this condition: both tests are also used for monitoring of chronic pancreatitis and diagnosing pancreatic cancer. We conducted all experiments on both laboratory tests; in this paper, we focus on lipase as recent literature has shown it to have higher diagnostic sensitivity and specificity.[30]. Our results for amylase were similar to those for lipase.
We asked the question: can the known association between an abnormal lipase value and acute pancreatitis be recovered from EHR data? To verify our hypothesis that laboratory measurement dynamics can impact the accuracy of identifying patients with acute pancreatitis, we considered three views of the data, based on the dynamics of lipase measurements within each patient’s record: (i) only visits with short lipase measurement gaps, (ii) only visits with long lipase measurement gaps, and (iii) all visits independent of the length between lipase measurements. In each of these settings, we assessed the association between acute pancreatitis and lipase and studied the properties of visits that belong in the setting using ICD-9 codes and clinical notes. We hypothesize that as acute pancreatitis is an acute disease, visits with short lipase measurement gaps will be more highly associated and relevant to acute pancreatitis.
2.5.1. Settings
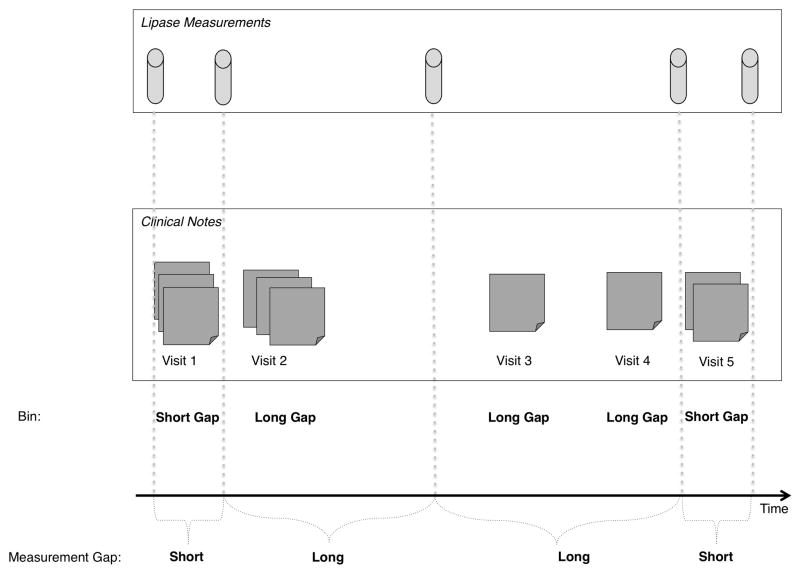
We divided each patient record into individual visits (defining a full in-patient admission as one visit). Each record (represented as a set of visits) was divided into bins of visits with short lipase measurement gaps and visits with long lipase measurement gaps. Figure 1 shows a schematic diagram of an individual’s longitudinal record: visits 1 and 5 belong to the short-gap bin because the lipase measurements were taken in rapid succession, the other visits belong in the long-gap bin because they show a long time between consecutive lipase measurements.
Figure 1. A schematic of a longitudinal record.
A single patient’s longitudinal record is divided up into visits (represented here as a set of notes written during the visit) and visits are binned into short or long gaps with respect to lipase measurements. For instance, the first and fifth visit are binned as short-gap visits, because they contain at least two consecutive laboratory test measurements that occur within a short time period. Visits 2, 3, and 4 are long-gap visits because they occur between two consecutive lipase measurements that are taken over a longer period of time.
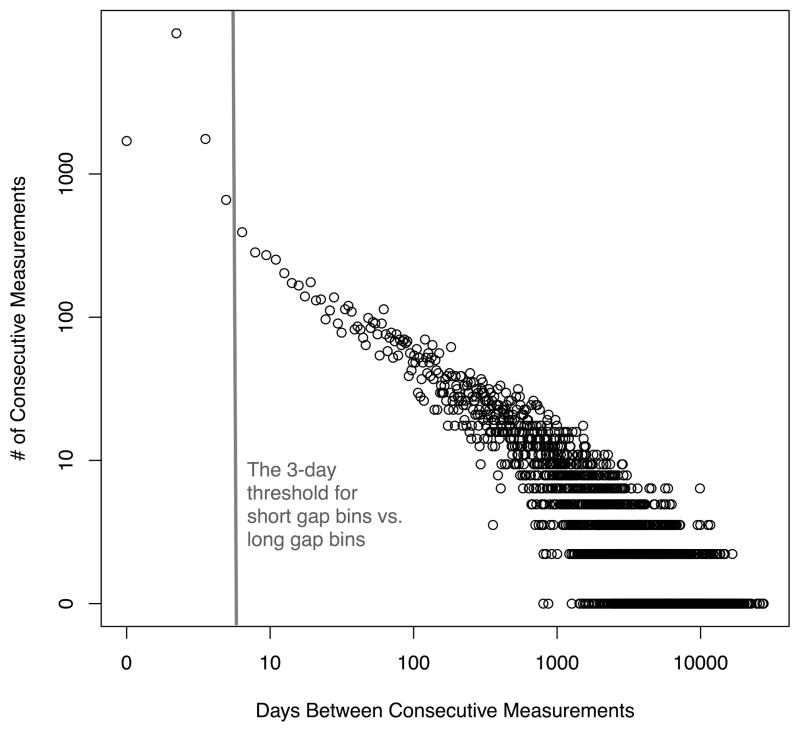
To determine the threshold for how many days define a short-gap and long-gap, we used the laboratory test’s measurement pattern histogram. Under the hypothesis that peaks in the histogram are indicative of the context within which a laboratory test is ordered we use the location of peaks and trends around them as thresholds for separating visits into short- and long-gap bins. This visit-separation method is generally insensitive to the exact threshold and is heuristically defined for each laboratory test as a function of the location and number of peaks in the measurement histogram. The measurement histogram of lipase (Figure 2) had one peak at one day and showed a change in measurement pattern at 3 days (the lipase histogram is only nearly linear after the 3 day gap).
Figure 2. The measurement gap histogram curve for the lipase laboratory test.
The measurement curve is presented on log-log scale as a histogram of the days between consecutive lipase test measurements for each patient, aggregated across the full population. We examined the figure visually and found that the pattern in measurement gap frequency changes at approximately 3 days; after 3 days the histogram curve is nearly linear. We used 3 days as a threshold for separating measurements on a short time scale (0–3 days) from measurements on a long time scale (over 3 days).
As there is only a measurement gap when a patient has two or more tests, only patients with at least two lipase measurements were included in the analysis. Similarly, no visits after the last recorded lipase test were added to either bin. Our visit-binning method is not limited to two (short- and long-gap) bins and there can be more granular bins such as bins of regular weekly visits. In the case of lipase, the measurement pattern histogram illustrated two distinct measurement patterns.
To analyze the differences between bins of short-gap measurements and long-gap measurements, we separated all ICD-9 codes and clinical notes created during short-gap lipase measurements from those collected during long-gap bins. We conducted association studies for lipase and acute pancreatitis in all three settings: (i) only short-gap visits, (ii) only long-gap visits, (iii) all visits.
2.5.2. Analyses
To assess in what ways the visits with short lipase measurement gaps differ from visits with long lipase measurement gaps, we ran three analyses using ICD-9 codes and clinical notes. For all of the following analyses, we used patients who exist in both the short-gap and the long-gap bins. This filtration reduced the confounders and ensured that differences we uncovered were from genuine separate health states. All p-values were Bonferroni-corrected.
Note Types
We looked at note types across the short and long measurement gap bins. Note types can inform the status of the patient. For example, a high frequency of admission and discharge notes indicate many inpatient visits, while primary provider notes are indicative of outpatient doctor visits. We performed a chi-squared test to assess the strength of association between the frequency of each note type and the gap bin; this test was chosen because we are comparing counts across different bins.
Note Content
We analyzed the frequency and coverage of all words across the notes in each bin. Differences in note content indicate differences in topics and hint at different contexts of measurement across gap bins. To correct for the redundancy within notes, we calculated word coverage. Redundancy across notes within a patient was implicitly handled, as individual patient records were divided into both bins. We look at the note content, both frequency and coverage, to check the separation created by long and short lipase gaps. The presence of certain words that relate to specific health states can hint at the level of separation across the gap bins and provide clues as to whether relevant parts of the patient’s record are indeed being separated out.
For each word we calculated:
We performed a chi-squared test to assess the strength of association between the coverage of each word and the gap bin.
Association Study
To test whether laboratory measurement dynamics affect a typical EHR association study[31], we conducted a phenome-wide search using the binomial test to find ICD-9 codes associated with an elevated lipase. For every ICD-9 code, we compared its frequency of occurrence with high lipase in all three settings. The binomial test was used to assess the statistical significance of deviations due to high lipase from the expected distribution of ICD-9 codes. The variables were defined as follows, per ICD-9:
3. Results
Our final dataset consisted of 14,141 patients, their notes, ICD-9 codes, and laboratory test times and values for 70 tests, spanning 20 years. On average each patient had 150.4 ICD9 codes [95% CI: 147.7–153.1], 825.8 laboratory tests [95% CI:807.7–847.0], and 133.5 clinical notes [95% CI: 130.8–136.3] over the entire study period.
3.1. Task 1: Correlation between Measurement Gap and Numerical Value
Both correlation metrics, linear and non-linear (through mutual information), between measurement patterns and test values were carried out for the 70 laboratory tests (see Appendix Tables 1 and 2 for full results). When we did not separate by time scale, there was little correlation: with the linear measure, all laboratory tests had a correlation very close to zero. With the mutual information measure, although also very low, a few laboratory tests demonstrated some level of correlation. The highest mutual information was .15, detected for the albumin laboratory test and only nine other tests had a mutual information higher than 0.1 (see Appendix Tables 1 and 2). Overall, these very low correlations indicate that there is separate information encoded in the laboratory test measurement pattern and the laboratory test’s numerical value.
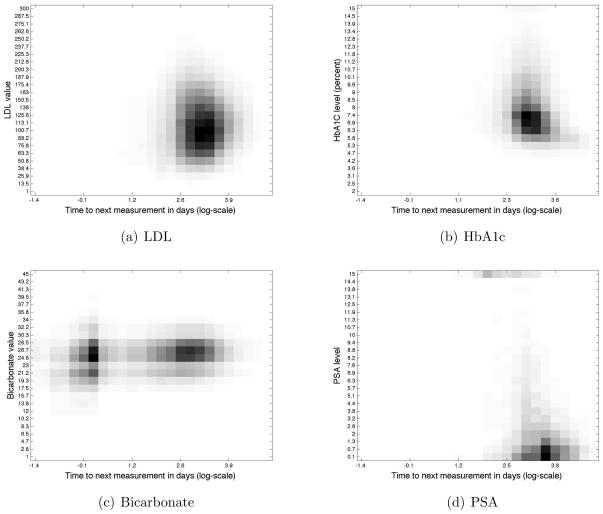
Exploring the correlation statistics separately for different time scales (short and long gaps), some laboratory tests such as LDL displayed no correlation(Figure 3(a)), using either metric, on any time scale, while other laboratory tests such as HbA1c and creatinine, showed some degree of correlation. For LDL, the numerical value does not affect its testing rate. We interpret the absence of correlation in measurement patterns and value as a result of healthcare process, such as adherence to guidelines for testing[32]. HbA1c displays a clear negative linear correlation of −0.193 only on the slow time scale, a higher HbA1c value is correlated to a shorter time until next measurement (Figure 3(b)). Creatinine also displays a clear negative linear correlation of −0.208, but on the short time scale.
Figure 3. Density plots of the PDFs (consisting of laboratory values and time between consecutive measurements) for four laboratory tests shown on the full time scale.
The x axis represents the log time to next measurement in days: log(1 hr)=−1.38, log(1 day)=0, log(1 week)=.85, log(1 month)=1.47, log(1 year)=2.56. Each graph shows different levels of correlation: LDL has no correlation on any time scale as shown by the mostly round ball, HbA1c has a negative correlation as shown by the L-shape of the curve, Bicarbonate separates along two time scales while PSA is almost exclusively measured on the long time scale of over a year between consecutive measurements.
The results from the linear correlation calculations were consistent with earlier work on the relationship between laboratory value and measurement frequency[8]. Weber and Kohane assigned categories to laboratory tests based on how numerical values were perceived (e.g.: “Bad-Good” represented a laboratory test where a low value was bad, and a high value was good). In our work, a positive linear correlation indicates a “Bad-Good” test where a low value prompts rapid retesting and a high value has a longer measurement gap, similarly a negative linear correlation represents a “Good-Bad” test.
The interplay between correlations on the full time scale and separately on the short and long time scales revealed interesting findings about measurement dynamics. For example, bicarbonate showed a positive linear correlation on both long and short time scales (the higher the value, the longer the gap between measurements), but when aggregating the time scales together and computing the total linear correlation, the correlation disappeared (Figure 3(c)). By contrast, the PSA screening test had very similar mutual informations on the full and long time scales. The differences between tests such as bicarbonate and the PSA screening test hint at differences in the contexts in which laboratory tests are ordered (Figure 3(d)). As PSA is measured in a single context, it is not subject to signal dilution due to timescale aggregation. Alternatively, the correlation results for laboratory tests such as bicarbonate, which are measured for multiple reasons and in various contexts, indicate that it is sometimes necessary to separate laboratory measurements by underlying context of measurement.
3.2. Task 2: Laboratory Test Measurement Motifs
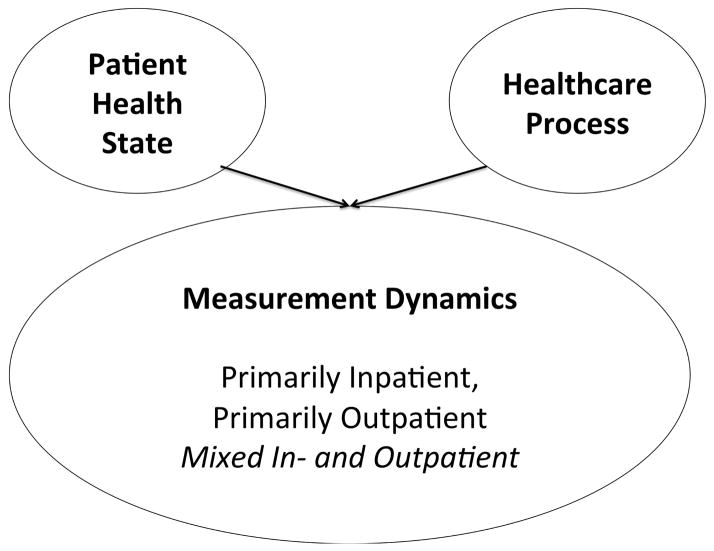
Manual cataloguing of the measurement gap histograms for 70 laboratory tests uncovered a set of three motifs that were most common. The three motifs of laboratory test ordering are influenced by two factors: patient health state and the healthcare process (Figure 4). These two factors contribute to create the shape of the laboratory test’s histogram. Certain histogram motifs highlight the presence of multiple contexts in which the test is being ordered. The histogram shape can determine whether further population stratification is necessary for conducting analyses or whether the laboratory measurements already represent a mostly homogenous patient set. The contributions of the patient health state and the healthcare process is dependent on the laboratory test itself and define the three motifs of test ordering: (i) primarily inpatient, (ii) primarily outpatient, (iii) a mixture of in- and outpatient. Figure 5 shows typical graphs from each of these three categories.
Figure 4. A Bayesian network describing the two factors that influence a laboratory tests measurement pattern.
The extent to which each factor contributes, changes which motif the test belongs to, thereby changing how it can be used in research settings. The “mixed in- and outpatient” motif represents laboratory tests taken across patients with multiple health states. Laboratory tests with mixed motifs may contribute to biased results when computing over the multiple health states as one population of patients.
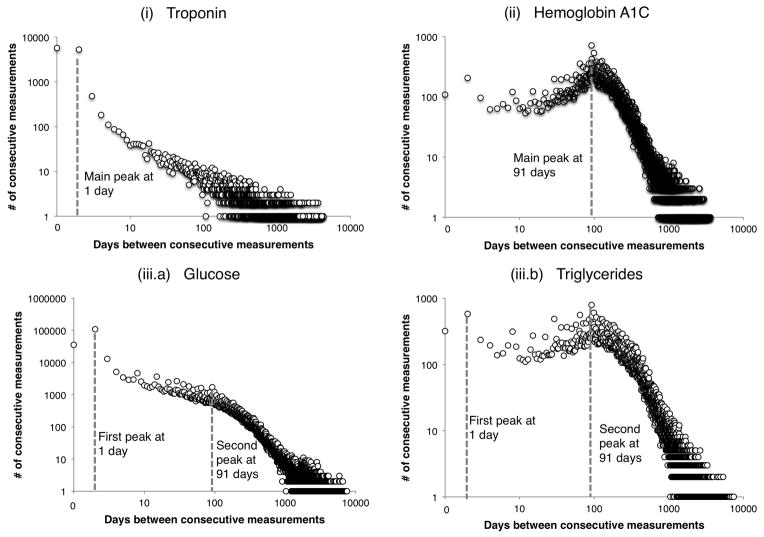
Figure 5. Representative examples of the three measurement gap motifs identified.
Each laboratory test motif is presented on a log-log scale as a histogram of the days between consecutive test measurements for each patient, aggregated across the full population. (i) Troponin represents a primarily inpatient laboratory test, with a peak at 0 days and displays an approximately linear relationship in the coordinate system; (ii) HbA1c is an example of a primarily outpatient laboratory test, showing a highly peaked distribution around 91 days; (iii.a) Glucose represents a mixture of in- and outpatient measurements, evidenced by the complex histogram: a high peak at on a short time scale (less than 10 days) and another peak at long time scales (multiple months); (iii.b) Triglycerides is another example of mixed laboratory test dynamics but with a slightly different mixture type: triglycerides has a high outpatient component and shows two different time scale peaks with a large quantity of measurements on the long time scale.
In general, laboratory tests that show peaks at very short time gaps in their measurement histograms are representative of tests taken during inpatient stays; laboratory tests with measurement graphs that peak at longer gaps of a few months are representative of measurements obtained during an outpatient visit. For some tests, the documentation (outpatient vs. inpatient) reason is aligned with the clinical reason; related to the fact that the numerical value obtained from a particular test is valid for a specific time period only. For example, troponin levels are representative of a patient state at the hour level, and thus their measurements are on the timescale of days. Because troponin is measured for patients suspected of suffering a myocardial infarction, and such a diagnosis has a high rate of inpatient admission, the troponin measurement dynamics are representative of an inpatient stay. Troponin’s measurement dynamics represent a primarily inpatient laboratory test, motif (i).
Other laboratory tests such as microalbumin and HbA1c change at a slower time scale and are ordered primarily in outpatient settings; as the values change slowly, there is no need to repeat their measurement during a short-term hospital admission. Therefore, HbA1c and microalbumin measurement dynamics represent primarily outpatient visits, motif (ii).
Laboratory tests that follow motif (iii) represent a set of tests whose measurement dynamics result from a mixture of both clinical and documentation reasons. For instance, glucose changes rapidly and is widely used in inpatient settings to monitor short-time scale changes but is also a regular test performed during outpatient visits to monitor chronic diabetics. The glucose dynamics are evident by the histogram diagram in Figure 5 where there is a fast time scale peak at 1 day, and a smaller slow time scale peak at 91 days. The peak at 91 days shows quarterly patient monitoring. Many other laboratory tests have motif (iii) measurement dynamics: for example, creatinine displays an almost identical histogram as glucose because they belong on the same basic metabolic panel and lipase along with amylase also have a mixture motif because of their use in both inpatient settings for acute events and outpatient settings for long-term monitoring.
Triglycerides is also a laboratory test with a mixture motif but with very different mixture weight (iii.b). This type of mixture laboratory test represents a dynamic that is also a result of mixed documentation and clinical reasons but with much heavier weight on the outpatient component mixing. Most of the population receives triglycerides at 3-month time scales to assess heart health but a small subset of patients have their triglycerides monitored on a much shorter time scale, these are ICU patients with feeding tubes. The large portion of outpatient testing is seen in the triglycerides measurement gap histogram because the peak at 91 days is at a similar height as the peak at 1 day.
These three laboratory measurement motifs determine how to use different laboratory tests in EHR research. The tests for which clinical and documentation reasons align (laboratory tests used almost exclusively for in-patients or outpatients) represent a homogenous set of contexts, or patient states. Thus, with laboratory tests in motif (i) or (ii), aggregating across patient values is a safe approach. In contrast, the laboratory tests with the mixed measurement motifs, might represent several separate patient states (such as patients receiving triglyceride measurements as outpatient patients and patients receiving triglyceride measurements as ICU patients). Aggregating patient’s values without separating the different patient state contexts in a large-scale study may introduce biases. The next section shows results of selecting patient cohorts by relying on a laboratory test with mixed dynamics (lipase) and how it impacts disease modeling (acute pancreatitis).
3.3. Task 3: Measurement Patterns Highlight Clinical State
We considered the histograms of measurement dynamics as plots of missing measurements. We presumed that for laboratory tests in the mixed motif category (iii), the data are missing not at random and may be informative of the patient’s health state. Following intuition from Little’s pattern mixture models[33] we hypothesized that different missing data patterns (or varying gaps between measurements) define different health states.
We explored this idea using lipase and hypothesized that (i) lipase measurement dynamics indicate two distinct missing values patterns, each representative of a clinical condition, rather than a documentation state; and (ii) separating the dataset by missingness patterns (visits with short gaps between measurements vs. visits with long gaps between measurements) helps recover the association between elevated lipase and the health state of acute pancreatitis.
3.3.1. Note Types
The note-type analysis indicated a significant difference between the common note types used in the short-gap and long-gap bins. The note types in Table 1 showed that lipase measurement dynamics can highlight true clinical differences, rather than documentation differences between inpatient and outpatient visits.
Table 1.
Note types indicative of healthcare setting (in vs. outpatient) are spread across short and long gap bins, hinting that the measurement gap-based separation is not representative of healthcare, but rather of health states. This table shows the top 10 normalized differences in % of total note types in each bin, each has a highly statistically significant difference in % total note types.
| Frequency of Note Types in Each Bin | |||||
|---|---|---|---|---|---|
| 0–3 day measurement gap | 3+ day measurement gap | ||||
|
| |||||
| Note Type | Raw Frequency | % Total Note Types | Raw Frequency | % Total Note Types | Difference in % |
|
| |||||
| Signout | 6269 | 0.144 | 21345 | 0.039 | 0.105 |
|
| |||||
| Miscellaneous Nursing Note | 4443 | 0.102 | 14944 | 0.027 | 0.075 |
|
| |||||
| 12-Lead Electro-cardiogram | 2126 | 0.049 | 9217 | 0.017 | 0.032 |
|
| |||||
| X-Ray of Chest, Portable | 1460 | 0.034 | 3901 | 0.007 | 0.027 |
|
| |||||
| Discharge Summary | 783 | 0.018 | 3030 | 0.006 | 0.012 |
|
| |||||
| Progress Note | 837 | 0.019 | 3937 | 0.007 | 0.012 |
|
| |||||
| Adult Social Work Progress Note | 662 | 0.015 | 2383 | 0.004 | 0.011 |
|
| |||||
| Physical Therapy | 605 | 0.014 | 2325 | 0.004 | 0.010 |
|
| |||||
| Admission Note | 579 | 0.013 | 2154 | 0.004 | 0.009 |
|
| |||||
| Respiratory Care Patient Assessment | 512 | 0.012 | 1434 | 0.003 | 0.009 |
There are note types that are only written during inpatient stays: an admission note, a signout note during a hospital shift change, and a discharge note. If separating visits based on lipase measurement dynamics was separating on a purely documentation basis with inpatient visits in the short-gap bin and outpatients in the long-gap bin, we would expect these inpatient-specific notes to be exclusively present in the short-gap bin. Instead, there are more inpatient notes in the longer gap bin but the coverage of inpatient notes is larger in the 0–3 day bin; the signout, admission, discharge account for 14%, 2%, and 1% of the total note-types, respectively. The long-gap bin contains a large amount of inpatient data demonstrating that the laboratory test measurement dynamics are able to to isolate visits based on health not hospital status in the short-gap bin. The note types present in the short-gap bin are relevant for diseases associated with lipase measurement as well. Elevated lipase often leads to testing for inflamed pancreas by ordering Ultrasound scans, CT scans, ERCP or Chest X-Rays. Common note-types for all 4 procedures were significantly more frequent in the 0–3 gap bin and ranked in the top 10% of Table 1. The results from the note type analysis suggest that lipase measurement dynamics are able to separate by patient health status and find specific visits that more likely pertained to acute pancreatitis events.
3.3.2. Note Content
The words with the largest difference in coverage between the short-gap and long-gap bins are very relevant to pancreatitis (Table 2): both lipase and amylase can be used to diagnose pancreatitis, Librium is an anti-anxiety drug often given to alcoholic patients with withdrawal symptoms, and alcoholic patients often have pancreatitis. There are more references to “pancreatitis” in the long-gap bin but the normalized frequency and coverage of the word is much higher in the short-gap bin. This indicates that the notes written during shorter gaps in measurement are more focused on the pancreatitis diagnosis.
Table 2.
Words associated with a pancreatitis health state are more frequently found in the short gap bins, suggesting that the separation is grouping notes written during visits that are more concentrated on the pancreatitis diagnosis into the 0–3 day measurement gap. The raw, normalized, and coverage frequencies of words in each gap sorted by the difference in coverage between the two bins is shown in this table. The words with a larger than 1% difference are shown, each of the words shown is highly associated with the 0–3 gap.
| Frequency of Words in Each Bin | |||||||
|---|---|---|---|---|---|---|---|
| 0–3 day measurement gap | 3+ day measurement gap | ||||||
|
| |||||||
| Words | Raw Frequency | % Total Words | % Notes containing that Word | Raw Frequency | % Total Words | % Notes containing that Word | Difference in Coverage |
|
| |||||||
| pancreatitis | 10732 | 0.14 | 6.94 | 13303 | 0.01 | 1.41 | 5.52 |
|
| |||||||
| lipase | 4908 | .06 | 4.28 | 9728 | 0.01 | 1.50 | 2.79 |
|
| |||||||
| amylase | 3855 | .05 | 3.48 | 98246 | 0.01 | 1.30 | 2.19 |
|
| |||||||
| withdrawal | 4139 | .05 | 2.80 | 12562 | 0.01 | 1.28 | 1.52 |
|
| |||||||
| librium | 3303 | .04 | 2.00 | 6064 | 0.00 | 0.59 | 1.42 |
|
| |||||||
| pancreatic | 4393 | .06 | 2.76 | 15992 | 0.01 | 1.38 | 1.38 |
|
| |||||||
| epigastric | 3668 | .05 | 2.92 | 15767 | 0.01 | 1.89 | 1.04 |
The word “pancreatic” modified “cancer” with a high prevalence in both gaps. This results from many patients with long-term pancreatic disorders experiencing acute episodes during their illness. We found that the word “cancer” has a coverage and frequency about 3 times higher in the long-gap bin than in the short-gap bin.
3.4. Recommendation for EHR Research with Laboratory Measurements
Knowing that acute pancreatitis is associated with high lipase levels, we used the binomial test to investigate whether separating visits by lipase measurement gaps highlight this association more prominently. In each setting, we performed a phenome-wide analysis to see the association between high lipase and ICD-9 577.0 (Acute Pancreatitis).
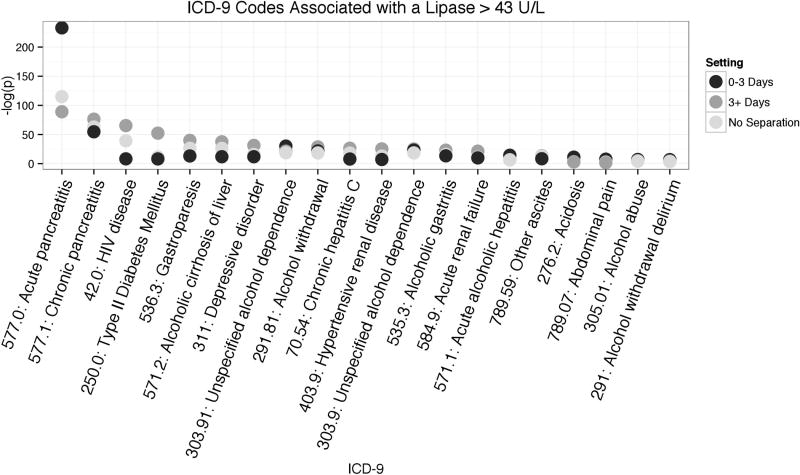
In the 0–3 day gap setting, the binomial test found the top association to be acute pancreatitis with an extremely significant Bonferroni corrected p-value of < 1 × 10−234. In the other two settings (greater than 3 day gap, and no separation by gaps), acute pancreatitis was also found to be the top association but with a much smaller p-value (Figure 6). The long-gap setting had the smallest association and therefore the no separation setting also showed a much lower p-value. These p-value differences demonstrate that signal dilution is a consequence of ignoring measurement frequency bias during phenome-wide analyses. The process of binning laboratory values by their gaps between measurements (Table 3) can reduce confounding by not mixing different patient health states.
Figure 6. The results of a binomial association test between high lipase and ICD-9 codes.
The binomial test was performed in all three settings (short gaps between measurements of 0–3 days, long gaps of more than 3 days, and all visits regardless of gaps between lipase measurements). The top 20 most significant associations are shown. For illustration purposes, the ICD-9 codes are sorted by association to high lipase in the 3+ days gap.
Table 3.
The actionable measurement gap separation method for finding and removing a confounding bias in laboratory test EHR data.
| Measurement Gap Separation Method | ||
|---|---|---|
| Step | Action | Motivation |
|
| ||
| 1 | Plot a histogram of the frequency and measurement gap in log-log coordinates. | The histogram provides a method to visually examine the laboratory tests measurement dynamics. |
|
| ||
| 2 | Examine the modality of the plot; looking for multi-modality. | If the histogram is multi-modal, it may imply a difference in patient health states or a healthcare process bias. |
|
| ||
| 3 | If there are multiple peaks, define a measurement gap threshold to separate the peaks. | This separation defines multiple settings for the EHR experiment, creating sets of homogenous data points with respect to their measurement gaps. |
|
| ||
| 4 | Perform the EHR experiment separately for each setting. | Separately performing experiments for different settings may remove confounding bias. |
Our results also demonstrate the generalizability of this measurement gap separation method. The measurement bins, created based on lipase measurement dynamics, were able to differentiate levels of association in other diseases as well. Type II diabetes (which may reflect clinicians screening type II diabetics with high triglycerides for pancreatitis) and HIV were differently associated with each setting. Without the lipase measurement-based separation of visits these disease associations are confounded by the bias of short-term lipase measurements. Interestingly, the ICD-9 for chronic pancreatitis is similarly associated with all three settings. The consistency of chronic pancreatitis is from patients having acute episodes during their chronic illness, conversely, not all acute pancreatitis patients have chronic pancreatitis. This asymmetry is demonstrated in the association patterns for acute and chronic pancreatitis.
4. Discussion
EHR research studies rely heavily on laboratory tests and their numerical values. We studied how laboratory test’s pattern of measurements may provide additional information to a laboratory test’s values. We discovered there is very limited correlation between how often a test is ordered and the value of the test. This lack of correlation implies that the value and measurement gap are informationally orthogonal to each other and are both important features to include when looking at laboratory tests, and specifically when using laboratory tests as features to represent patient disease state. In addition, there is evidence of different correlation results when examining laboratory test values on different time scales, showing that temporality plays a crucial role in the use of clinical laboratory test data.
We found evidence that measurement patterns of laboratory tests are dictated by clinical and physiological knowledge, but often confounded by the healthcare process, such as hospital documentation practices whether from workflows or guidelines.
One clear artifact of hospital document patterns was revealed by examining the measurement gap histograms of 70 laboratory tests. Many laboratory test histograms have peaks at exactly 91 days, although their histogram height is varied. Our hospital serves a highly captive and sick population of patients in the surrounding neighborhoods. As the population is sick, 3 month checkups are a common practice and as the population lives nearby, the general adherence to a strict 3 month (91 day) schedule is high. It is clear that this 91 day peak is caused by the operations of the hospital and makeup of the population – not the clinical state of each individual patient. Although this particular healthcare process bias is specific to our institution, we postulate similar types of biases exist across the country and should be mitigated before using the EHR-recorded data for research.
Upon cataloguing all 70 laboratory tests we uncovered three types of measurement dynamics motifs, one which represents “mixed” laboratory tests where clinical factors and documentation standards are misaligned.
4.1. Separation of Mixed Motif Laboratory Tests by Measurement Pattern Mitigates EHR Laboratory Bias
Laboratory tests with multiple ordering reasons, such as those used for both diagnosis and monitoring, present challenges to EHR-based research. When using laboratory values without accounting for the laboratory test’s frequency of measurement, confounders can dilute the results of a study. The dynamics of laboratory measurement gaps across a population can reveal different measurement patterns present in the data; examining the measurement patterns of a particular test and then performing patient record decomposition in a strategic manner reduces signal dilution. For example, filtering the full patient cohort by visits within a particular measurement pattern of missingness will provide a more focused dataset of patient states.
We demonstrated our method of visit separation through measurement gap analysis on lipase as a specific use case. Using note types, note content, and ICD-9 codes we showed that our method could group inpatient visits pertinent to a particular clinical condition (acute pancreatitis in our example) away from inpatient visits pertinent to other clinical reasons. We also performed a secondary analysis to test that separating on hospital status does not yield the same results as separating by measurement gap. When looking exclusively at inpatient visits (without accounting for measurement gap), the association between high lipase and acute pancreatitis was only as high as the “3+ Days” association found using the measurement gap separation method (Figure 6). Therefore, we infer that the measurement gap separation method can indeed separate on health status, not simply healthcare process.
The work presented in this paper is highly relevant to researchers working on cohort identification algorithms, especially with the recent push for more automated ways to perform high-throughput phenotyping[34, 35, 36]. We present the stratification of an individual’s medical record by laboratory test measurement frequency as a new conceptual paradigm for studying EHR data with EHR-recorded laboratory tests.
4.2. Limitations
This study is carried out on a single institution. While the population under study was large, future work is to replicate these findings on a population from a different institution. We also acknowledge the limitation of using only one test to demonstrate laboratory test biases, but we present the lipase and acute pancreatitis association study as a single proof-of-concept example to exhibit the potential importance of separating laboratory test values by measurement pattern. Furthermore, the study is limited due to changing hospital documentation and clinical best practices. These changes may dictate the frequency tests should be ordered and their purpose (such as the transition from amylase to lipase as the recommended diagnostic test for acute pancreatitis), leading to variable results across different time periods. We conducted an informal investigation and found few differences in dynamics across time periods, although not enough to affect the overall results of the study.
5. Conclusion
For the re-use of clinical data to facilitate novel data-driven informatics research, understanding salient features and correcting for EHR biases is a necessary step. We show that surprisingly, there is often little shared information between laboratory test values and the laboratory test’s rate of measurement in time. Further, measurement patterns are useful features to use in disease modeling and they can result from a combination of hospital workflow practices and clinical states. When the clinical and documentation biases are not in concert (as is often the case with laboratory tests used for multiple purposes), EHR-driven association studies may produce biased results. We catalogued the measurement dynamics of laboratory tests into three motifs, one of which which has mixed patterns of measurement and is prone to biases. Finally, we demonstrate how to control for the biases by disambiguating patient health states based on laboratory measurement frequency, using the laboratory test lipase and acute pancreatitis as an illustrative example.
Supplementary Material
Acknowledgments
This work is supported by National Science Foundation IGERT #1144854 (RP), National Library of Medicine award R01 LM06910 (DA), National Library of Medicine award R01 LM010027 (NE), and National Science Foundation award #1344668 (NE).
The authors are grateful to Dr. George Hripcsak, Dr. Hojjat Salmasian, and Dr. Adler Perotte for their meaningful discussions and Dr. Janet Kayfetz for her helpful comments, as well as two anonymous reviewers for their valuable comments on earlier versions of this paper. The authors would also like to thank Dr. David Sontag for the inspiring discussions about laboratory measurements in the EHR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rimma Pivovarov, Email: rp2521@columbia.edu.
David J. Albers, Email: dja2119@cumc.columbia.edu.
Jorge L. Sepulveda, Email: jls2282@columbia.edu.
Noémie Elhadad, Email: noemie.elhadad@columbia.edu.
References
- 1.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods of Information in Medicine. 2009;48(1):38–44. [PubMed] [Google Scholar]
- 2.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC medical genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Chused A, Elhadad N, Friedman C, Markatou M. Automated knowledge acquisition from clinical narrative reports. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2008. pp. 783–7. [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2013;20(1):117–21. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersh WR, Weiner MG, Embi PJ, Logan JR. Caveats for the Use of Operational Electronic Health Record Data in Comparative Effectiveness Research. Medical care. 2013 doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyon AW, Higgins T, Wesenberg JC, Tran DV, Cembrowski GS. Variation in the frequency of hemoglobin A1c (HbA1c) testing: population studies used to assess compliance with clinical practice guidelines and use of HbA1c to screen for diabetes. Journal of diabetes science and technology. 2009;3(3):411–7. doi: 10.1177/193229680900300302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena S, Anderson DW, Kaufman RL, Hannah JA, Wong ET. Quality assurance study of cardiac isoenzyme utilization in a large teaching hospital. Archives of pathology & laboratory medicine. 1993;117(2):180–3. [PubMed] [Google Scholar]
- 8.Weber GM, Kohane IS. Extracting Physician Group Intelligence from Electronic Health Records to Support Evidence Based Medicine. PLoS ONE. 2013;8(5):e64933. doi: 10.1371/journal.pone.0064933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Walraven C. Population-based Study of Repeat Laboratory Testing. Clinical Chemistry. 2003;49(12):1997–2005. doi: 10.1373/clinchem.2003.021220. [DOI] [PubMed] [Google Scholar]
- 10.McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods. Saunders; 2011. [Google Scholar]
- 11.Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Medical care. 2005;43(5):480–5. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 12.Farzandipour M, Sheikhtaheri A, Sadoughi F. Effective factors on accuracy of principal diagnosis coding based on International Classification of Diseases, the 10th revision (ICD-10) International Journal of Information Management. 2010 [Google Scholar]
- 13.Chen DP, Dudley JT, Butte AJ. Latent physiological factors of complex human diseases revealed by independent component analysis of clinar-rays. BMC Bioinformatics. 2010;11(Suppl 9):S4. doi: 10.1186/1471-2105-11-S9-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasko TA, Denny JC, Levy MA. Computational Phenotype Discovery Using Unsupervised Feature Learning over Noisy, Sparse, and Irregular Clinical Data. PLoS ONE. 2013;8(6):e66341. doi: 10.1371/journal.pone.0066341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JH, Haug PJ. Exploiting missing clinical data in Bayesian network modeling for predicting medical problems. Journal of Biomedical Informatics. 2008;41(1):1–14. doi: 10.1016/j.jbi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Hripcsak G, Albers DJ, Perotte A. Exploiting time in electronic health record correlations. Journal of the American Medical Informatics Association : JAMIA. 2011;18 (Suppl 1):i109–15. doi: 10.1136/amiajnl-2011-000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albers DJ, Hripcsak G. A statistical dynamics approach to the study of human health data: Resolving population scale diurnal variation in laboratory data. Physics Letters A. 2010;374(9):1159–64. doi: 10.1016/j.physleta.2009.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagreiya H, Altman RB. The utility of general purpose versus specialty clinical databases for research: Warfarin dose estimation from extracted clinical variables. Journal of Biomedical Informatics. 2010;43(5):747–51. doi: 10.1016/j.jbi.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albers DJ, Hripcsak G, Schmidt M. Population Physiology: Leveraging Electronic Health Record Data to Understand Human Endocrine Dynamics. PLoS ONE. 2012;7(12):e48058. doi: 10.1371/journal.pone.0048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albers DJ, Hripcsak G. Using time-delayed mutual information to discover and interpret temporal correlation structure in complex populations. CHAOS. 2012;22(1):013111. doi: 10.1063/1.3675621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albers D, Hripcsak G. Estimation of time-delayed mutual information and bias for irregularly and sparsely sampled time-series. 2011. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen R, Elhadad M, Elhadad N. Redundancy in electronic health record corpora: analysis, impact on text mining performance and mitigation strategies. BMC Bioinformatics. 2013;14(1):10. doi: 10.1186/1471-2105-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–92. [Google Scholar]
- 24.Schafer JL, Graham JW. Missing data: Our view of the state of the art. Psychological Methods. 2002;7(2):147–77. [PubMed] [Google Scholar]
- 25.Farhangfar A, Kurgan LA, Pedrycz W. A Novel Framework for Imputation of Missing Values in Databases. IEEE Transactions on Systems, Man, and Cybernetics - Part A: Systems and Humans. 2007;37(5):692–709. [Google Scholar]
- 26.Abdala OT, Saeed M. Estimation of missing values in clinical laboratory measurements of ICU patients using a weighted K-nearest neighbors algorithm. Computers in Cardiology. 2004:693–6. [Google Scholar]
- 27.Hug CW. PhD thesis. MIT; 2006. Predicting the risk and trajectory of intensive care patients using survival models. [Google Scholar]
- 28.Cismondi F, Fialho AS, Vieira SM, Reti SR, Sousa JMC, Finkelstein SN. Artificial Intelligence in Medicine. Artificial intelligence in medicine. 2013;58(1):63–72. doi: 10.1016/j.artmed.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Albers DJ, Hripcsak G. Estimation of time-delayed mutual information and bias for irregularly and sparsely sampled time-series. Chaos, solitons, and fractals. 2012;45(6):853–60. doi: 10.1016/j.chaos.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banks PA, Freeman ML the Practice Parameters Committee of the American College of Gastroenterology. Practice Guidelines in Acute Pancreatitis. The American Journal of Gastroenterology. 2006;101(10):2379–400. doi: 10.1111/j.1572-0241.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- 31.Warner JL, Alterovitz G. Phenome Based Analysis as a Means for Discovering Context Dependent Clinical Reference Ranges. AMIA Proceedings. 2012;2012:1441. [PMC free article] [PubMed] [Google Scholar]
- 32.Grundy SM, Bilheimer D, Chait A, Clark LT. Report of the National Cholesterol Education Program (NCEP) Expert Panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 1993 [Google Scholar]
- 33.Little RJ. Pattern-mixture models for multivariate incomplete data. Journal of the American Statistical Association. 1993;88(421):125–34. [Google Scholar]
- 34.Chen Y, Carroll RJ, Hinz ERM, Shah A, Eyler AE, Denny JC, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. Journal of the American Medical Informatics Association : JAMIA. 2013 doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei WQ, MM, TaoPhD C, JiangPhD G., MD CGC, DrPH A High Throughput Semantic Concept Frequency Based Approach for Patient Identification: A Case Study Using Type 2 Diabetes Mellitus Clinical Notes. AMIA Proceedings 2010. :1–5. [PMC free article] [PubMed] [Google Scholar]
- 36.Lussier YA, Liu Y. Computational Approaches to Phenotyping: High-Throughput Phenomics. Proceedings of the American Thoracic Society. 2007;4(1):18–25. doi: 10.1513/pats.200607-142JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.