Abstract
Leucine-rich repeat kinase 2 (LRRK2) is a large, ubiquitous protein of unknown function. Mutations in the gene encoding LRRK2 have been linked to familial and sporadic Parkinson disease (PD) cases. The LRRK2 protein is a single polypeptide that displays GTPase and kinase activity. Kinase and GTPase domains are involved in different cellular signalling pathways. Despite several experimental studies associating LRRK2 protein with various intracellular membranes and vesicular structures such as endosomal/lysosomal compartments, the mitochondrial outer membrane, lipid rafts, microtubule-associated vesicles, the golgi complex, and the endoplasmic reticulum its broader physiologic function(s) remain unidentified. Additionally, the cellular distribution of LRRK2 may indicate its role in several different pathways, such as the ubiquitin-proteasome system, the autophagic-lysosomal pathway, intracellular trafficking, and mitochondrial dysfunction. This review discusses potential mechanisms through which LRRK2 may mediate neurodegeneration and cause PD.
Keywords: LRRK2, Parkinson’s disease, intracellular traffic, quality control mechanisms, mitochondria
1. Gene and protein organization
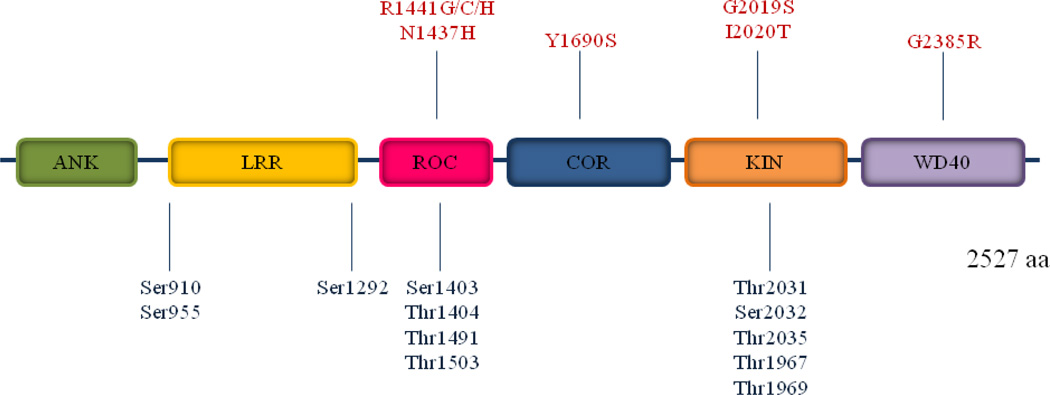
The leucine-rich repeat kinase 2 (LRRK2) gene on chromosome 12 spans a genomic distance of 144 kb and contains 51 exons. LRRK2 is a large cytosolic multidomain protein consisting of 2527 amino acids and a molecular weight of approximately 285 kDa. LRRK2 has multiple functional domains. There is a Ras-of-Complex (ROC) GTPase domain adjacent to a C-terminal-of-ROC (COR) linker region. A serine/threonine protein kinase domain is also present. There are putative protein-protein interaction domains that flank the central ROC-COR-kinase catalytic region like an ankyrin domain. The N-terminal region contains leucine-rich repeat (LRR) motifs, and WD40 repeats near the C-terminus of the protein probably form a beta-propeller structure (Figure 1) (Greggio and Cookson, 2009). The overall structure suggests LRRK2 may act as a scaffold for other proteins, some of which could themselves regulate LRRK2 or else undergo LRRK2-mediated modifications. LRRK2, therefore, may have the ability to integrate and modify multiple signalling pathways.
Figure 1.
Schematic representation of LRRK2. The central region of LRRK2 contains a GTPase domain also called ROC, a COR domain of unknown function and a kinase domain, flanked on either side by multiple protein–protein interaction domains: WD40 domain; ANK, ankyrin repeat domain and LRR, leucine-rich repeats. PD pathogenic mutations are depicted in red on top and phosphorylation sites are depicted in blue below. The scale of each domain or protein is not proportional to its actual size.
1.1 GTPase and kinase functions
LRRK2 belongs to the ROCO family. Roco family members contain both small GTPase and kinase domains, as well as a COR region. Most ROCO proteins also have either an LRR domain, a WD40 domain, or both (Bosgraaf and Van Haastert, 2003). Its structure is shared by one mammalian homolog, LRRK1, which also has a kinase and GTPase domain (Greggio and Singleton, 2007).
The kinase domain of LRRK2 apparently shares homology with mixed-lineage kinase (MLKs) and receptor interacting protein kinase (Gandhi et al., 2009; West et al., 2005). MLKs are part of the mitogen activated protein kinase (MAPK) family, and act as MAPK kinase kinases (MAPKKKs) to initiate and transduce a wide range of cell death-relevant responses (Gallo and Johnson, 2002). How and whether LRRK2 acts as a MAPKKK is unclear because the mechanisms through which it is activated, as well as its downstream kinase effects, are not well-characterized (Biskup and West, 2009; Melrose, 2008).
Dimerization is a common phenomenon among protein kinases. Dimerization can help mediate auto-regulation and modulate downstream signalling molecules. LRRK2 appears to exist as a homodimeric protein. This was first suggested by co-immunoprecipitation experiments using tagged full-length protein or LRRK2 fragments (Gloeckner et al., 2009). Studies using LRRK2 fragments or using the LRRK2 prokaryotic homolog indicate that dimerization occurs in the ROC-COR region. The crystal structure of the ROC domain also shows LRRK2 exists as a dimer, and that the COR domain that follows the ROC domain may function as a hinge between the ROC and kinase domains (Deng et al., 2008; Greggio et al., 2008; Jorgensen et al., 2009). However, neither ROC deletion by itself nor N-terminal deletion (including the ANK and LRR domains) prevents dimerization. On the other hand, a WD40 truncated-LRRK2 could not be dimerized. Together, these studies suggest that while the ROC and WD40 domains mediate LRRK2 dimerization, the WD40 domain is more crucial (Lee et al., 2012).
While specific mutations can destabilize LRRK2 fragment dimers (Deng et al., 2008; Gotthardt et al., 2008), the extent to which full length LRRK2 dimerization is affected in PD is unclear. How perturbed dimerization may affect LRRK2 function is similarly uncertain, but hypotheses exist. One hypothesis is that LRRK2 kinase activity is dimerization-dependent (Greggio et al., 2006; Sen et al., 2009; Smith et al., 2006). In support of this, Berger and colleagues found that LRRK2 dimers, which are enriched in cell membranes, have more kinase activity than monomeric LRRK2 (Berger et al., 2010; Greggio and Cookson, 2009).
Less is known about LRRK2’s GTPase capacity. LRRK2 was first identified as an authentic GTPase in 2007. GTPase activity appears to be mediated by the ROC domain, which binds and hydrolyses GTP in a manner similar to that of the Ras-related small GTPase, Rac1. The ROC domain, therefore, may function in a manner typical of Ras-related small GTPases (Guo et al., 2007).
It was initially suspected that the LRRK2 GTPase domain might regulate LRRK2 kinase activity (Bosgraaf and Van Haastert, 2003). In support of this, ROC domain mutations that prevent GTP binding also inactivate kinase function (Cookson, 2010). With GTP bound, the switch region on the outside of the domain assumes an active state, which increases kinase activity. With GDP bound, the tertiary structure of the switch domain assumes an inactive state, which decreases kinase activity. Biosa and co-workers further demonstrated that normal kinase activity requires guanine nucleotide binding and, to a lesser extent, GTP hydrolysis. Interestingly, guanine nucleotide binding but not GTP hydrolysis appears to regulate LRRK2 dimerization, structure, and stability (Biosa et al., 2013). Two independent groups, however, reported neither GTP nor GDP binding affect recombinant LRRK2 kinase activity (Liu et al., 2010; Taymans et al., 2011).
It is possible that GTPase activating proteins (GAPs) and GTP exchange factors may help regulate LRRK2 function. ArfGAP1 has been shown to enhance the LRRK2 GTPase in Drosophila melanogaster (Xiong et al., 2012). Interestingly, the same authors had previously suggested that LRRK2 toxicity in yeast can be modulated by altering GTPase activity (Xiong et al., 2010). It is also possible that the kinase domain of LRRK2 instead regulates the ROC domain, and that the LRRK2 kinase activity alters LRRK2 GTPase activity (Cookson, 2010; Greggio and Cookson, 2009; Webber et al., 2011). Relevant to this possibility are data that show the LRRK2 kinase domain phosphorylates its ROC domain at several sites (Greggio et al., 2009; Kamikawaji et al., 2009).
While the LRRK2 GTPase can function independently, kinase activity seems to require a functional GTPase domain. Moreover, it is very likely that LRRK2 GTPase has additional independent targets (Ray and Liu, 2012). GTPase activity may also be regulated by LRRK2 dimerization or through recruitment of other cellular proteins (Gotthardt et al., 2008). Overall, data suggest that the LRRK2’s ROC–COR-kinase portion probably constitutes its key regulatory region.
1.2 Phosphorylation and autophosphorylation
LRRK2 kinase activity was first measured by quantitating LRRK2-mediated transphosphorylation of myelin basic protein (MBP). However, this assay was not optimal given the LRRK2’s low catalytic activity, and also because MBP is a substrate for other serine/threonine kinases (Zhao et al., 2012). More optimal substrates include moesin (also called LRRKtide), which LRRK2 phosphorylates at its Thr558 site (Jaleel et al., 2007). Moesin belongs to a group of proteins collectively called the ERM (ezrin/radixin/moesin) proteins. These proteins influence cytoskeletal dynamics by anchoring the cytoskeleton to the plasma membrane (Mangeat et al., 1999). A protein that binds eukaryotic initiation factor 4E (eIF4E), eIF4E binding protein (4E-BP) is also phosphorylated by LRRK2 (Imai et al., 2008). The 4E-BP-eIIF4E complex promotes translation through its binding to capped mRNA species.
The search for LRRK2 kinase substrates facilitated identification of LRRK2-IN-1, the first selective LRRK2 inhibitor (Deng et al., 2011). LRRK2-IN-1, interestingly, eliminates LRRK2 phosphorylation at two sites, Ser910 and Ser935, which do not appear to arise as a consequence of autophosphorylation but appear instead to be phosphorylated by PKA suggesting that PKA is a potential upstream regulatory kinase (Li et al., 2011). Ser910/Ser935 phosphorylation mediates LRRK2’s interaction with another protein, 14-3-3, so the loss of this phosphorylation disrupts 14-3-3 and LRRK2 binding. This, in turn, causes LRRK2 to accumulate within cytoplasmic pools, as opposed to adopting a more even localization (Nichols et al., 2010). Therefore, LRRK2 Ser910/Ser935 phosphorylation, LRRK2 binding to 14-3-3, and the LRRK2 cytoplasmic distribution pattern can be used to monitor LRRK2 activity (Figure 1) (Dzamko et al., 2010; Doggett et al., 2012). Other LRRK2 inhibitors, such as the small molecule inhibitor, GNE-7915, have been identified. GNE-7915 can cross the blood brain barrier, which makes it advantageous for animal-based studies (Estrada et al., 2012).These molecules that inhibit LRRK2 kinase activity were shown to arrest neurodegeneration using different LRRK2 PD models which makes them valuable to clarify the pathways in which LRRK2 play a role. Indeed in various neuronal cell culture systems kinase activity inhibition through site-directed mutagenesis or pharmacologically was shown to attenuate neurotoxicity (Liu et al., 2011a; Luerman et al., 2013; Yao et al., 2013).
While Ser910/Ser935 is not a consequence of autophosphorylation, LRRK2 autophosphorylation sites do exist (Gloeckner et al., 2010). In the presence of ATP and Mg2+, efficient autophosphorylation occurs (Greggio, 2012). Autophosphorylation, therefore, can be used as a surrogate measure of kinase activity (Smith et al., 1993). Autophosphorylation sites localize primarily to the GTPase domain, which suggests some degree of kinase-GTPase inter-regulation occurs (Greggio et al., 2009).
Kamikawaji and co-workers identified Ser1403, Thr1404, Thr1410, and Thr1491 in the ROC domain, as well as Thr1967 and Thr1969 in the kinase domain, are functionally relevant autophosphorylation sites (Figure 1). Thr1410 autophoshorylation increases kinase activity, while Thr1491 autophosphorylation reduces kinase activity (Table I). Alanine substitution at Thr1967 decreases kinase activity, whereas alanine substitution at Thr1491 increases kinase activity (Kamikawaji et al., 2009). Three other putative autophosphorylation sites (Thr2031, Ser2032, and Thr2035) have also been identified within the activation segment of the LRRK2 kinase domain (Figure 1) (Li et al., 2010a).
Table I.
Overview of LRRK2 cellular phosphorylation and autophosphorylation with corresponding kinase and GTPase activity.
| LRRK2 residues | Kinase Activity | References | |
|---|---|---|---|
| Phosphorylation | Ser935 | ↑ | Dzamko et al., 2010 |
| Ser910 | ↑ | Dzamko et al., 2010 | |
| autophosphorylation | Ser1403 (ROC domain) | ? | Kamikawaji et al., 2009 |
| Thr1404 (ROC domain) | ? | Kamikawaji et al., 2009 | |
| Thr1410 (ROC domain) | ↑ | Kamikawaji et al., 2009 | |
| Thr1491 (ROC domain) | ↓ | Kamikawaji et al., 2009 | |
| Thr1967 (kinase domain) | ↓ | Kamikawaji et al., 2009 | |
| Thr1969 (kinase domain) | ↑ | Kamikawaji et al., 2009 | |
| Thr2031 (kinase domain) | ? | Li et al., 2010a | |
| Ser2032 (kinase domain) | ? | Li et al., 2010a | |
| Thr2035 (kinase domain) | ? | Li et al., 2010a | |
| Thr1503 (ROC domain) | ↑ | Webber et al., 2011 | |
| Ser1292 (junction of the LRR and the ROC) | ↑ | Sheng et al., 2012 | |
Removal of a conserved autophosphorylation site at Thr1503 decreases both GTP-binding and kinase activity, which suggests autophosphorylation at this amino acid potentiates LRRK2 kinase activity (Table I and Figure 1). However, Thr1503 phosphorylation was not observed in transgenic mouse and cell lines with increased kinase function (Webber et al., 2011). More recently it was demonstrated that LRRK2 autophosphorylation at Ser1292 occurs in vivo and that this is enhanced by several familial PD mutations including R1441G, R1441C, G2019S, and I2020T (Figure 1). Converting Ser1292 to alanine (phospho-deficient mutant) mitigates the effects of LRRK2 PD mutations on neurite outgrowth in cultured rat embryonic primary neurons. This suggests that Ser1292 autophosphorylation may reflect LRRK2 kinase activity in vivo, and that Ser1292 phosphorylation may mediate the effects of some PD mutations (Table I) (Sheng et al., 2012). Finally, the LRRK2 G2019S mutation associates with both increased autophosphorylation and substrate phosphorylation, which may at least partly explain this mutation’s pathogenecity (Luzon-Toro et al., 2007).
Many questions relevant to LRRK2 remain. For example, to date no LRRK2 phosphatases have been identified. Further insight into LRRK2 phosphorylation and autophosphorylation will no doubt help us better understand LRRK2 function and dysfunction, and help development more accurate and sensitive LRRK2 kinase assays.
1.3 Mutations
The LRRK2 coding sequence includes 7500 nucleotides. There are a number of known variants, many of which are not linked currently to any disease and are probably phenotypically inconsequential (Paisan-Ruiz et al., 2008). Other polymorphisms, though, may act as PD risk factors. Indeed, two genome-wide association studies reported certain LRRK2 polymorphisms associate with an increased risk of sporadic PD (Satake et al., 2009; Simon-Sanchez et al., 2009). LRRK2, therefore, seems to act as a “deterministic” gene that causes autosomal dominant PD in the presence of certain mutations, and as a susceptibility gene for sporadic PD (Mata et al., 2012). Even with deterministic mutations, though, penetrance is still incomplete (Hulihan et al., 2008; Latourelle et al., 2008), individuals have survived into their 80s without developing clinical parkinsonism (Kay et al., 2005).
LRRK2 mutations typically cause a PD-typical phenotype that is characterized by tremor, rigidity, bradykinesia, and postural instability (Haugarvoll and Wszolek, 2009). Autopsies of such patients show prominent loss of melanised dopamine neurons in the substantia nigra pars compacta. The age onset is variable, but symptoms generally develop in the sixth or seventh decade.
LRRK2 mutations, and especially the G2019S mutation, are observed in both autosomal dominant and apparent sporadic PD patients. Associated histologic changes can include either Lewy Bodies (LBs) and tau inclusions. G2019S mutation carriers typically show LB pathology, whereas R1441C, Y1699C, and I2020T mutation carriers do not (Poulopoulos et al., 2012).
Six missense mutations in different domains clearly segregate with disease (Table II and Figure 1). A group of LRRK2 mutations are found within the ROC-COR domains. The R1441 position in the ROC/GTPase, when mutated to cysteine, glycine or histidine (R1441C/G/H) causes familial PD, as does the Y1699C mutation in the adjacent COR domain. The most common mutation, G2019S, is found in the kinase domain, and the original Japanese family has an I2020T mutation located in the kinase domain. These LRRK2 PD pathogenic mutations disrupt the modification of LRRK2 cellular phosphorylation sites. Ser910, Ser935, Ser955 and Ser973 are fully phosphorylated in the presence of G2019S, while these sites are hypophosphorylated in the presence of R1441G/C, Y1699C, and I2020T mutations (Zhao et al., 2012).
Table II.
Overview of LRRK2 mutations with corresponding kinase and GTPase activity.
| LRRK2 Mutations |
LRRK2 Domain |
Kinase Activity |
GTPase Activity | References | |
|---|---|---|---|---|---|
| GTP Hydrolysis |
GTP Binding |
||||
| R1441C | GTPase | ? | ↓ | ? | Kumar and Cookson, 2011; West et al., 2007 |
| R1441G | GTPase | ? | ↓ | = | Kumar and Cookson, 2011; West et al., 2007 |
| R1441H | GTPase | ? | ? | ? | Rudenko et al., 2012 |
| Y1699C | COR | = | ↓ | = | Kumar and Cookson, 2011; West et al., 2007 |
| G2019S | Kinase | ↑ | = | = | Greggio and Cookson, 2009 |
| I2020T | kinase | ? | ? | ? | Rudenko et al., 2012 |
| I2012T | Kinase | ? | ? | ? | Rudenko et al., 2012 |
| N1437H | GTPase | ? | ? | ↑ | Rudenko et al., 2012 |
| G2385R | C-terminal WD40 | ↓ | ? | ? | Rudenko et al., 2012 |
The G2019S mutation substitutes a serine for a highly conserved glycine located in subdomain VII of the kinase domain. This mutation increases kinase activity by increasing the catalytic rate of the enzyme; it does not enhance substrate affinity (West et al., 2005). The G2019S mutation facilitates substrate access, thereby leading to a toxic increase in kinase activity. G2019S is the only mutation that consistently shows increased kinase activity (Greggio and Cookson, 2009). Other mutations, including the R1441C mutation in the ROC domain as well as the Y1699C and I2020T mutations, have only variably been found to increase kinase activity. In these studies, discrepancies may have arisen due to the use of different substrates in the kinase assays, or because of differences in the way the enzyme was collected (Jaleel et al., 2007; Seol, 2010; West et al., 2007).Overall, though, in experimental models, changes in LRRK2 kinase activity appear to be toxic, and seem to induce degeneration of dopamine neurons (Greggio et al., 2006; Iaccarino et al., 2007; Jorgensen et al., 2009; Smith et al., 2006; West et al., 2007).
Mutations located in or next to the GTPase domain, including R1441C, R1441G, and Y1699C increase the steady-state levels of GTP-bound LRRK2. This probably results as a consequence of reduced GTP hydrolysis (Guo et al., 2007; Kumar and Cookson, 2011; Li et al., 2007; West et al., 2007).
A C-terminal WD40 domain variation that substitutes arginine for glycine (G2385R) increases PD risk and reduces kinase activity. Another mutation, the N1437H mutation that causes PD in a large Norwegian kindred, seems to increase GTP binding, which could suggest an overall reduction in GTPase activity (Table II and Figure 1) (Rudenko et al., 2012).
It is still unclear whether a common pathogenic mechanism for all PD-inducing mutations or PD-associated polymorphisms exists (Tsika and Moore, 2012) and if so what that mechanism might be. Many investigators have thus far focused on kinase function, although changes in GTP binding, constitutive phosphorylation, impaired dimerization, or excess protein degradation may all play a role (Lee et al., 2012; Rudenko et al., 2012). Interestingly, a recent study shed light to a new hypothesis for LRRK2 toxicity. In this study the authors observed that blocking LRRK2 kinase activity reduced mutant LRRK2 toxicity and this was correlated with a decrease in LRRK2 levels indicating that LRRK2 levels are more important than kinase activity per se in predicting toxicity (Skibinski et al., 2014). Given the complexity of the LRRK2 protein however, several aspects of LRRK2 function might be targeted, such as, inhibition of kinase activity and GTP binding, preservation of constitutive phosphorylation of LRRK2, disruption of LRRK2 dimerization or LRRK2 degradation (Lee et al., 2012; Rudenko et al., 2012).
2. Role in cell function
LRRK2 is expressed in most organs including brain, heart and liver. Particularly high levels are observed in kidney and lung (Paisan-Ruiz et al., 2004; Zimprich et al., 2004). It is also expressed in some immune cells (Kubo et al., 2010; Maekawa et al., 2010). In the brain LRRK2 is observed primarily in neurons of the olfactory bulb, striatum, cortex, hippocampus, midbrain, brainstem, and cerebellum (Biskup et al., 2006; Higashi et al., 2007a). Levels are relatively low in substantia nigra and ventral tegmental area dopaminergic neurons, as compared to the higher levels that are seen in the striatum, cerebral cortex, cerebellum, and hippocampus (Han et al., 2008; Higashi et al., 2007b). Additionally, expression can be experimentally altered; MPTP, a mitochondrial toxin that destroys dopamine neurons, induces an acute increase in LRRK mRNA (Hurley et al., 2007).
Within cells, LRRK2 associates with various intracellular membranes and vesicular structures including endosomes, lysosomes, multivesicular bodies, the mitochondrial outer membrane, lipid rafts, microtubule-associated vesicles, the golgi complex, and the endoplasmic reticulum (Cookson, 2010; Tan and Jankovic, 2006). This distribution could reflect a functional role in multiple pathways, including regulation of autophagy, microtubule dynamics, and mitochondrial function. To date, investigators have largely attempted to define LRRK2-related physiology by studying the consequences of increased and decreased LRRK2 expression.
2.1 Dopamine homeostasis and vesicle trafficking
The different LRRK2 transgenic mouse models generated thus far do not faithfully recapitulate a PD phenotype. While some transgenic models exhibit DA neuron death, neurodegeneration is limited and not obviously progressive. Transgenic mice develop only mild motor deficits (Rudenko and Cookson, 2010). Moreover, LRRK2 knockout mice have an intact dopaminergic system (Andres-Mateos et al., 2009; Hinkle et al., 2012).
LRRK2 localizes to vesicles, where it interacts with vesicular proteins (Biskup et al., 2006). In presynaptic vesicles, LRRK2 silencing leads to a redistribution of vesicles, alters recycling dynamics, and increases vesicle kinetics. This suggests LRRK2 may play a role in synaptic vesicle trafficking (Piccoli et al., 2011). Shin and colleagues further report that LRRK2, in conjunction with Rab5b, contributes to synaptic function by modulating synaptic vesicle endocytosis (Shin et al., 2008). Moreover, overexpression or knockdown of endogenous LRRK2 in primary neuronal cells significantly impairs synaptic vesicle endocytosis, a phenomenon that can be reversed with co-expression of Rab5b (Heo et al., 2010a).
LRRK2 phosphorylates EndoA, an evolutionary conserved protein that is critically involved in synaptic vesicle endocytosis. LRRK2-mediated EndoA phosphorylation profoundly affects membrane tubulation and membrane association, which suggests that in synapses LRRK2, through its role as a kinase, facilitates efficient vesicle formation (Matta et al., 2012).
In regard to dopamine signaling, LRRK2 mutations have been shown to affect activity-dependent DA neurotransmission and catecholamine release (Tong et al., 2009). This manifests as reduced levels of extracellular striatal dopamine and reduced levels of dopamine metabolites. Motor deficits in these mice are minor, but these deficits nevertheless respond to L-dopa (Chen et al., 2012; Li et al., 2010b; Li et al., 2009; Maekawa et al., 2012; Melrose et al., 2010). Expression of G2019S LRRK2 induces an age-dependent loss of nigrostriatal dopamine neurons. The brains of aged G2019S mice also show perturbed autophagy and abnormal mitochondria. Cultured dopamine neurons from these mice display markedly reduced neurite complexity (Ramonet et al., 2011). Reduced striatal DAT levels in LRRK2 transgenic mice have also been reported, which could reflect dysfunction or degeneration of dopamine neuron terminals (Chen et al., 2012). Impaired dopamine re-uptake can occur (Zhou et al., 2011); this change, along with reduced DAT levels, suggests impaired dopamine re-uptake could contribute to altered dopamine levels - in LRRK2 transgenic mice.
2.2 Relationship to α-synuclein
Potential interactions between α-synuclein (SNCA) and LRRK2 have been considered and their functions may have some degree of functional overlap. For example, they both play a role in microtubule assembly and microtubule dynamics (Parisiadou and Cai, 2010).
LRRK2 localizes to a subset of brainstem-type LBs, but not to cortical-type LBs, NFTs, other tau inclusions, or TDP-43-positive inclusions. Approximately 20–100% (mean=60%) of SNCA-positive LBs contain LRRK2 (Perry et al., 2008). It is reported that 10–80% of classic LBs are rimmed by LRRK2 (Alegre-Abarrategui et al., 2008; Vitte et al., 2010). In post-mortem brains with LB pathology, LRRK2 co-immunoprecipitates with SNCA (Yacoubian et al., 2010). In cultured cells transfected to overexpress both LRRK2 and SNCA, under conditions that promote oxidative stress LRRK2 and SNCA physically associate with each other (Qing et al., 2009b). Increasing striatal SNCA expression also concomitantly raises LRRK2 mRNA, which suggests LRRK2 and SNCA levels may be co-regulated (Westerlund et al., 2008).
The question of whether LRRK2 might phosphorylate SNCA has been considered. Experimental data does suggest LRRK2 and its putative kinase domain-containing fragments can phosphorylate recombinant SNCA at S129. This modification is believed to promote the formation of SNCA filaments and oligomers, and is enriched in LBs. Accordingly, G2019S LRRK2 phosphorylates SNCA at S129 more avidly than WT LRRK2 (Qing et al., 2009a).
It was demonstrated that in PD affected brain regions such as the amygdala and anterior cingulate cortex, but not in the unaffected visual cortex, LRRK2 levels positively correlate with SNCA phosphorylation and aggregation (Guerreiro et al., 2012). In cell culture models of SNCA over-expression, concomitant overexpression of LRRK2 facilitates SNCA release and its subsequent uptake by neighboring cells (Kondo et al., 2011). Overexpression of WT, G2019S, and R1441C LRRK2 increases SNCA expression through an extracellular signalregulated kinase pathway (ERK)-mediated pathway (Carballo-Carbajal et al., 2010).
Expressing LRRK2 in the A53T SNCA transgenic mouse PD model accelerates neurodegeneration and enhances SNCA aggregation. This is believed to arise through effects on microtubule dynamics, Golgi bodies, and the ubiquitin-proteasome pathway (UPS) (Lin et al., 2009). Loss of LRRK2, on the other hand, seems to cause both accumulation and aggregation of SNCA, as well as other ubiquitinated proteins, presumably as a consequence of impaired autophagy-lysosome pathway function (Tong et al., 2010).
In contrast to these mouse studies, basal ganglia and limbic cortex SNCA aggregation was found to be lower in LRRK2 G2019S carriers than it was in persons with idiopathic PD (Mamais et al., 2013). Furthermore, it was recently shown that by removing endogenous SNCA, LRRK2 dependent neurodegeneration and the levels of diffuse mutant LRRK2 are dramatically reduced (Skibinski et al., 2014). Overall, studies such as these suggest LRRK2 and SNCA can interact, although the true nature of this putative relationship remains unclear.
2.3 Relationship to mitochondria
LRRK2 is present in mitochondria (Biskup et al., 2006). In C. Elegans, over-expressing LRRK2 enhanced survival following exposure to two different mitochondrial toxins, whereas knockdown of lrk-1 (the endogenous C. elegans ortholog of LRRK2) reduced survival (Saha et al., 2009). Similarly, Drosophila that express mutant LRRK2 display increased sensitivity to rotenone-mediated complex I inhibition (Ng et al., 2009). WT LRRK2, but not the mutants forms, attenuate H2O2-induced oxidative stress, thereby suggesting a protective role for LRRK2 (Liou et al., 2008).
No structural mitochondria abnormality is observed in mouse neurons that overexpress either WT or mutant LRRK2 (Lin et al., 2009). In skin biopsies from human LRRK2 G2019S carriers, however, mitochondrial function and morphology are perturbed characterized by reduced mitochondrial membrane potential, reduced intracellular ATP levels, mitochondrial elongation, and increased mitochondrial interconnectivity (Mortiboys et al., 2010). In contrast to this, in cortical neurons LRRK2 G2019S overexpression increases DLP-1 activity and promotes mitochondrial fission (Niu et al., 2012; Wang et al., 2012).
In fibroblasts and neuroblastoma cells, the LRRK2 G2019S mutation associates with a decreased mitochondrial membrane potential and lower cell ATP levels. These changes appear to be kinase-dependent (Papkovskaia et al., 2012). SN4741 dopaminergic cells overexpressing either WT or G2019S LRRK2 are more susceptible to H2O2-induced cell death (Heo et al., 2010b), and WT LRRK2 expression increases cell reactive oxygen species (ROS) levels (Niu et al., 2012). LRRK2 mutations reduce activity of peroxiredoxin 3, an antioxidant enzyme located within mitochondria. This effect appears to be phosphorylation-dependent (Angeles et al., 2011).
Induced pluripotent stem cells (iPSCs) derived from fibroblasts obtained from LRRK2 carriers also produce more ROS than iPSCs derived from control subject fibroblasts. Mitochondria in iPSCs prepared from LRRK2 carriers also show less mitochondrial respiration, an increased mitochondrial proton leak, and reduced mitochondrial movement (Cooper et al., 2012).
Mouse cortical neurons that express LRRK2 G2019S or R1441C mutations show impaired calcium homeostasis and mitochondrial degradation. Calcium chelators and voltage-gated L-type calcium channel inhibitors mitigate mitochondrial degradation (Cherra et al., 2013).
Taking into account that mitochondrial dysfunction is widely recognized as a trigger of parkinsonism it is interesting to note that LRRK2 affects mitochondrial dynamics and morphology, mitochondrial calcium buffering, ROS levels and mitochondrial membrane potential highlighting once again LRRK2 intrinsic role in PD.
2.4 Relationship to the cytoskeleton
Ultrastructural analyses indicate that LRRK2 interacts with microtubules in a well-ordered, periodic fashion. This suggests the presence of LRRK2-binding sites on microtubules or microtubule-bound proteins (Kett et al., 2012). To this end, LRRK2 has been shown to bind α/β-Tubulin heterodimers and interact with microtubules (Gandhi et al., 2008). Gillardon further showed that recombinant human LRRK2 phosphorylates β-tubulin purified from bovine brain, that this phosphorylation is enhanced three-fold in the presence of theG2019S mutation, and that a LRRK2-related increase in β-tubulin phosphorylation increases microtubule polymerization (Gillardon, 2009b). LRRK2 and microtubules also interact in both neuronal and non-neuronal cells (Caesar et al., 2013). The R1441G mutation appears to interfere with this interaction, possibly by affecting tubulin acetylation and microtubule stability (Law et al., 2013).
Mice that overexpress either mutant or WT LRRK2 show elevated brain α/β-tubulin polymerization. This leads to microtubule overstabilization, which in turn impairs cell function (Lin et al., 2009; Maekawa et al., 2012). Conversely, brains from Lrrk2−/− mice have increased free tubulin levels (Gillardon, 2009b). LRRK2 also plays a role in actin dynamics, as gene expression of encode cytoskeleton-related proteins are deregulated in blood mononuclear cells from patients with LRRK2 mutations (Mutez et al., 2011). Mutant LRRK2 leads to the accumulation of polymerized actin as well as phosphorylated ERM, both of which are reversed by LRRK2 knockout (Parisiadou et al., 2009). By applying QUICK (quantitative immunoprecipitation combined with knockdown) in NIH3T3 cells, Meixner and colleagues found that LRRK2 interacts with actin isoforms, and also with actin-associated proteins that contribute to filament assembly, organization, rearrangement, and maintenance (Meixner et al., 2011).
Further links between LRRK2 and microtubule polymerization are suggested by an observed interaction between LRRK2 and elongation factor 1α (EF1A) (Gillardon, 2009a). Besides its canonical role in mRNA translation, EF1A helps maintain the microtubule cytoskeleton. LRRK2 interacts with tau, another protein that contributes to microtubule stability, and may modulate microtubule dynamics through this interaction. LRRK2 can directly phosphorylate tubulin-associated tau directly (Kawakami et al., 2012), and G2019S and I2020T LRRK2 mutations increase tau-phosphorylation. This in turn reduces tau’s affinity for microtubules, which promotes its aggregation. Quantitative biochemical analysis shows the presence of unique phospho-tau species in G2019S mice (Melrose et al., 2010) and tau phosphorylation is reduced in brains from LRRK2 null mice (Parisiadou et al., 2009).
PD-associated LRRK2 mutations may directly influence the affinity of LRRK2 for microtubules or microtubule-bound proteins. Some studies suggest certain mutations increase affinity (Kett et al., 2012), while others (G2019S; R1441G, R1441H) reduce LRRK2-β-tubulin interactions (Law et al., 2013). Overall, in vivo and in vitro studies indicate tau is a substrate of LRRK2, and suggest LRRK2 may contribute to histology changes observed in tauopathy disorders (Bailey et al., 2013).
Both LRRK2 deficiency and overexpression alter neurite length and branching (Lin et al., 2010; MacLeod et al., 2006; Plowey et al., 2008). Neurons that express the G2019S mutation have shorter neurites, and shRNA-mediated LRRK2 suppression increases neurite length (MacLeod et al., 2006).
An emerging body of literature supports a role for LRRK2 function in microtubule dynamics and trafficking. LRRK2 may interact with and/or phosphorylate or regulate the phosphorylation of several structural and regulatory components of the actin cytoskeleton and microtubule network. Therefore understanding these cytoskeletal associations may provide insight into the PD pathogenesis.
2.5 Relation to protein quality control mechanisms
Protein inclusions in PD patients brains may reflect a failure in one of the two major intracellular protein breakdown pathways, the UPS and autophagy. In the UPS, E1, E2 and E3 enzymes tag proteins with ubiquitin. Ubiquination targets the substrate to the proteasome, a barrel-shaped multiprotein complex that as protease activity, and degrades targeted substrates into peptides. In autophagy, proteins and/or other cellular components are degraded by lysosomal hydrolases.
2.5.1 Lysosomal protein degradation
Changes in autophagy are consistently observed when mutant LRRK2 is overexpressed, or with knock-down of endogenous LRRK2. Nevertheless, delineating the precise mechanism(s) by which LRRK2 regulates autophagy has been difficult.
In C. elegans transgenic strains, with altered LRRK2 activity, bear changes autophagy-related gene expression (Ferree et al., 2012). LRRK2 interacts with CAMKK-β/AMPK, a Ca2+-dependent enzyme that induces the accumulation of autophagosomes (Gomez-Suaga et al., 2012). MAPKs, which positively regulate autophagy, are LRRK2 substrates (Gloeckner et al., 2009; Hsu et al., 2010).
In cultured cells, LRRK2 puncta co-localize with autophagic vacuoles (AVs) and multivesicular bodies (MVBs) patterns similar to those seen in human brains (Alegre-Abarrategui et al., 2009). In agreement with this, in rat brains punctate LRRK2 co-localizes with lysosomal and endosomal vesicles (Biskup et al., 2006). It is possible that alterations in the endocytic pathway are responsible for the deregulation of autophagy.
Reducing LRRK2 protein levels deregulates autophagy. For example, in the kidney, this manifests as an increase in LC3II, a protein marker of autophagy, and decreased levels of p62, an autophagic substrate. In addition, SNCA and protein carbonyls (a general marker of oxidative stress) levels also decrease whereas lysosomal proteins and proteases increase (Tong et al., 2012). However, neither autophagic nor lysosome-related structures accumulate in the brains of LRRK2 knock-out mice, which suggests LRRK2 may have different roles in different tissues, or that in the absence of LRRK2, homologs such as LRRK1 may compensate (Tong et al., 2010). Moreover the fact that LRRK2 expression levels in the central nervous system are decreased relatively to the renal tissue can signify that mutant LRRK2 or LRRK2 absence can cause subtle pathogenic effects throughout ageing which is in agreement with the late onset of the disease.
In view of that, the inhibition of LRRK2 kinase activity stimulates macroautophagy (Manzoni et al., 2013a). In HEK cells LRRK2 knockdown increased LC3II turnover and autophagy (Alegre-Abarrategui et al., 2009). The continuous induction of autophagy caused by the absence of LRRK2 in vivo can ultimately cause a deficiency in the clearance or recycling of autophagic components/autolysosomes. LRRK2, therefore may, in the long run, down-regulate autophagy.
Relative to cells transfected with WT LRRK2 or with the kinase-dead K1906M mutation, cells that express the G2019S LRRK2 mutation show striking increases in neuritic and somatic autophagic vacuoles, as well as decreased neurite length. (Plowey et al., 2008). Overexpression of G2019S LRRK2 causes autophagic and lysosomal structures to accumulate in primary cortical neurons and in neuronal cell lines (MacLeod et al., 2006). Similarly, in G2019S and, to a lesser degree, R1441C transgenic mice AV accumulation in the soma and processes of cortical and striatal neurons has been described (Ramonet et al., 2011). These studies suggest that in the presence of these mutations, either autophagy is induced or autophagosome-lysosomal clearance is impaired.
Human iPSC dopaminergic neurons derived from either idiopathic or LRRK2 PD keratinocytes and fibroblasts show a decreased autophagic flux. This occurs in conjunction with increases in p62 protein, autophagosome, and lipid droplets and is consistent with impaired autophagic clearance (Sanchez-Danes et al., 2012). Compared to control fibroblasts, fibroblasts that express the G2019S mutation exhibit increased autophagy (Manzoni et al., 2013b). This is increase is mediated by the MAPK1/3 (Bravo-San Pedro et al., 2013). In SH-SY5Y cells, LRRK2 also appears to activate ERK1/2 (Liou et al., 2008). More interestingly, it is hypothesized that when excessive mitochondrial damage or excessive mitophagy is induced by neurotoxins the regenerative capacity of nigral neurons can be affected promoting PD-related pathogenic mechanisms (Dagda et al., 2008)
Calcium contributes to autophagy regulation, and links exist between calcium and LRRK2. One link is likely mediated by NAADP/Ca2+, a Ca2+ mobilizing messenger that targets acidic (lysosome-like) Ca2+ stores and endoplasmic reticular stores (Guse and Lee, 2008). Recently, it was reported that the Ca2+ chelator BAPTA prevented an LRRK2-induced increase in autophagosomes, thereby suggesting LRRK2’s autophagy effects are Ca2+ dependent. Further, exposing cells to a cell-permeate NAADP analogue restored the number of autophagosomes, lysosomal pH, and lipid droplet content to those that were observed when LRRK2 was overexpressed (Gomez-Suaga et al., 2012). One interpretation of this study is that LRRK2 localizes to lysosomes, where it interacts with NAADP receptors such as TPC2 to cause lysosomal calcium release. Calcium from the lysosomes then induces an endoplasmic reticulum calcium release, which activates the CaMKK/AMPK pathway. This, in turn, leads to lysosomal alkalinization and increases autophagosomes.
Rab7, a small GTP binding protein involved in late endosomal transport, lysosomal biogenesis, autophagosome-lysosome fusion, and endosome-lysosome fusion interacts with LRRK2. In morphologically altered neurons from subjects with neurodegenerative diseases, and especially those that show LBs accumulation, LRRK2 localizes to the endosomallysosomal compartment and co-localizes with Rab7B (Higashi et al., 2009). The drosophila LRRK2 homologue was also shown to localize late endosome and lysosome membranes, where it physically interacts with Rab7 and reduces Rab7-dependent targeting of lysosomes to the perinuclear region (Dodson et al., 2012). This suggests LRRK2-Rab7 interactions may regulate autophagosome-lysosome and endosome-lysosome fusion, which ultimately would perturb autophagic-lysosomal clearance pathways. Whether this happens in human neurons, though, is currently unclear.
LRRK2 itself appears to be removed via chaperone-mediated autophagy (CMA). While WT LRRK2 undergoes efficient CMA-mediated lysosomal degradation, G2019S LRRK2 does not. This raises the question of whether lysosomal G2019S LRRK2 accumulation might secondarily compromise the CMA-mediated removal of SNCA from neurons (Orenstein et al., 2013).
Overall there is still some controversy regarding how LRRK2 regulates autophagy. We can speculate that different LRRK2 mutations may cause impairment in the autophagic balance due to improper autophagic-lysosomal clearance or by an increase in the autophagic flux. Both paradigms can lead to the accumulation of autophagic components and substrates.
2.5.2 Relationship to the ubiquitin-proteasome system
In addition to being degraded by CMA, LRRK2 is also removed by the UPS. LRRK2 forms a complex with heat shock protein 90 (Hsp90) in vivo, and preventing this association via Hsp90 inhibition induces LRRK2 proteasomal degradation. Interestingly, a reduction in axon growth that occurs when G2019S LRRK2 is over-expressed can be reversed by Hsp90 inhibition (Wang et al., 2008). Indeed, a complex formed between LRRK2, CHIP, and Hsp90 appears to regulate cell LRRK2 levels. Hsp90 overexpression reduces CHIP-mediated LRRK2 degradation and geldanamycin, an Hsp90 inhibitor, increases CHIP-mediated LRRK2 degradation (Ding and Goldberg, 2009; Ko et al., 2009). The effect of whether or not LRRK2 mutations interfere with its Hsp90/CHIP interactions, though, is unknown. LRRK2 overexpression, both in vitro and in vivo, impairs UPS function. This leads to an accumulation of diverse substrates, including SNCA and ubiquitin (Lichtenberg et al., 2011).
3. LRRK2 and other PD-linked proteins: common pathways
Several reports suggest LRRK2 interacts with Parkin, DJ-1 and PINK1 (Samann et al., 2009; Smith et al., 2005; Venderova et al., 2009). It has further been postulated that autophagy may represent a point of functional convergence between these proteins and the pathways they contribute to (Cuervo et al., 2004; Dagda et al., 2009; Geisler et al., 2010; Narendra et al., 2008; Plowey et al., 2008).
Co-expression of human parkin in LRRK2 G2019S-expressing flies protects against DA neurodegeneration that arises with advancing age or following rotenone treatment (Ng et al., 2009). DJ-1 can also rescue neurons from LRRK2 G2019S-induced toxicity (Heo et al., 2010b). Moreover, neural cells generated from iPSCs derived from PD patients fibroblasts carrying mutations in the PINK1 and LRRK2 genes share mitochondrial abnormalities providing insight into convergence of cellular disease mechanisms between different familial forms of PD (Cooper et al., 2012).
4. LRRK2 and the Immune System
LRRK2 is expressed in circulating immune cells, and this expression is enhanced in the presence of microbial structures and viral particles. During bacterial phagocytosis, LRRK2 localizes near bacterial membranes. LRRK2 also increases NF-kB-dependent transcription and represses Nuclear Factor of activated T-cells (NFAT)-dependent transcription (Gardet et al., 2010; Hakimi et al., 2011; Liu et al., 2011b).
In monocytes, interferon-γ induces LRRK2 expression. This action is mediated through a novel ERK-dependent, interferon-γ signal transduction pathway (Kuss et al., 2014). LRRK2 may also contribute to monocyte maturation (Thevenet et al., 2011).
Microglial activation is observed in post-mortem PD brain tissue (Imamura et al., 2003), and PD patients also show other evidence of increased inflammation. Other inflammation-related changes are observed in PD subject lymphocytes, monocytes, and natural killer cells. Pro-inflammatory cytokine levels are elevated in PD subject blood, cerebrospinal fluid, and brain tissue (Collins et al., 2012). Peripheral blood mononuclear cells from PD patients with LRRK2 mutations have changes similar to those seen in mononuclear cells from sporadic PD patients (Mutez et al., 2014). Consequently, a role for LRRK2 has been implicated in microglial pro-inflammatory responses in the brains of PD subjects (Moehle et al., 2012).
In dermal fibroblasts prepared from PD subjects with LRRK2 mutations, LRRK2 silencing reduces basal and provoked cyclooxygenase (COX)-2 RNA levels (Lopez de Maturana et al., 2013). Reducing LRRK2 in murine brain microglia attenuates a lipopolysaccharide (LPS)-induced increase in cytokine mRNA and protein expression (Kim et al., 2012). Compared to isolated murine microglia obtained from mice that express WT LRRKs, microglia from mice that express the LRRK2 R1441G mutation produce more pro-inflammatory cytokines. When added to neuron cultures, conditioned medium from LPS-stimulated microglia that have the LRRK2 R1414G mutation triggers cell death (Gillardon et al., 2012). Both central and peripheral LPS administration was shown to only affect dopaminergic neurons, with no damage to either GABAergic or serotoninergic neurons and this damage was permanent in tyrosine hydroxylase-immunoreactive neurons in the substantia nigra (Herrera et al., 2000; Qin et al., 2007).
Altogether these reports pinpoint that LRRK2 dysfunction in PD may also involve immune response pathways.
5. Outlook
A better understanding of LRRK2 biology will provide insight into PD neurodegeneration. Accumulated evidence suggests LRRK2’s kinase activity represents a reasonable therapeutic target for both autosomal dominant PD due to LRRK2 mutations, as well as for those with sporadic PD. It must be kept in mind, however, that LRRK2 plays a role in multiple cell pathways, including cytoskeleton maintenance and on various signalling cascades. This could limit the clinical applicability of LRRK2-directed interventions. A better molecular-level understanding of how LRRK2 mutations change LRRK2 function, as well as its cellular role, may help overcome this practical limitation.
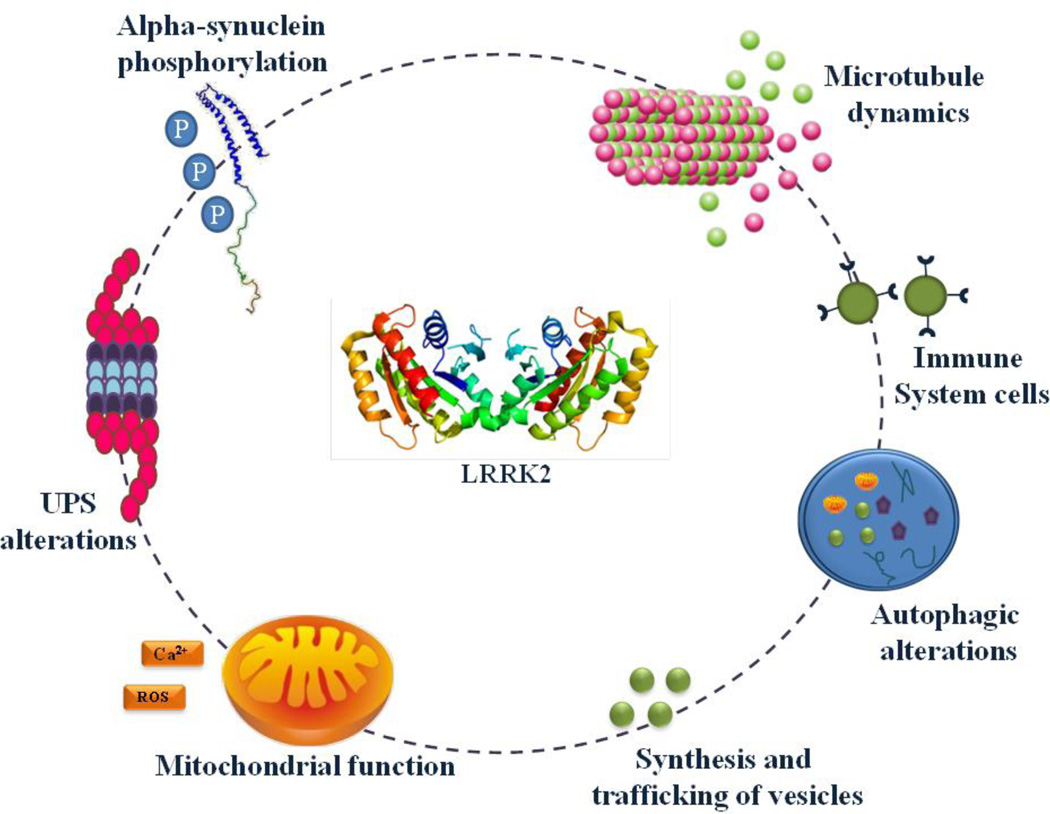
Figure 2.
LRRK2 involvement in cellular mechanisms. Several data posit that LRRK2 through its kinase and/or GTPase domain can affect mitochondrial function, ubiquitin-proteasome system, autophagy-lysosomal pathway, microtubule dynamics as well as trafficking of vesicles and proteins, alpha-synuclein phosphorylation content and immune system cells.
Acknowledgements
This work was supported by the project PEst-C/SAU/LA0001/2011 from Portuguese Foundation for Science and Technology (FCT-MCTES, Portugal). AR Esteves is supported by Post-Doctoral Fellowship (SFRH/BPD/75044/2010) from Portuguese Foundation for Science and Technology (FCT-MCTES, Portugal). RHS is supported by the University of Kansas Alzheimer’s Center (NIH P30AG035982).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alegre-Abarrategui J, Ansorge O, Esiri M, Wade-Martins R. LRRK2 is a component of granular alpha-synuclein pathology in the brainstem of Parkinson's disease. Neuropathol Appl Neurobiol. 2008;34:272–283. doi: 10.1111/j.1365-2990.2007.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, Liu G, Beal MF, Huso DL, Dawson TM, Dawson VL. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles DC, Gan BH, Onstead L, Zhao Y, Lim KL, Dachsel J, Melrose H, Farrer M, Wszolek ZK, Dickson DW, Tan EK. Mutations in LRRK2 increase phosphorylation of peroxiredoxin 3 exacerbating oxidative stress-induced neuronal death. Hum Mutat. 2011;32:1390–1397. doi: 10.1002/humu.21582. [DOI] [PubMed] [Google Scholar]
- Bailey RM, Covy JP, Melrose HL, Rousseau L, Watkinson R, Knight J, Miles S, Farrer MJ, Dickson DW, Giasson BI, Lewis J. LRRK2 phosphorylates novel tau epitopes and promotes tauopathy. Acta neuropathologica. 2013;126:809–827. doi: 10.1007/s00401-013-1188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z, Smith KA, Lavoie MJ. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49:5511–5523. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosa A, Trancikova A, Civiero L, Glauser L, Bubacco L, Greggio E, Moore DJ. GTPase activity regulates kinase activity and cellular phenotypes of Parkinson's disease-associated LRRK2. Hum Mol Genet. 2013;22:1140–1156. doi: 10.1093/hmg/dds522. [DOI] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, Faull RL, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Biskup S, West AB. Zeroing in on LRRK2-linked pathogenic mechanisms in Parkinson's disease. Biochim Biophys Acta. 2009;1792:625–633. doi: 10.1016/j.bbadis.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Bravo-San Pedro JM, Niso-Santano M, Gomez-Sanchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, Climent V, Lopez de Maturana R, Sanchez-Pernaute R, Lopez de Munain A, Fuentes JM, Gonzalez-Polo RA. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell Mol Life Sci. 2013;70:121–136. doi: 10.1007/s00018-012-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar M, Zach S, Carlson CB, Brockmann K, Gasser T, Gillardon F. Leucine-rich repeat kinase 2 functionally interacts with microtubules and kinase-dependently modulates cell migration. Neurobiol Dis. 2013 doi: 10.1016/j.nbd.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Carballo-Carbajal I, Weber-Endress S, Rovelli G, Chan D, Wolozin B, Klein CL, Patenge N, Gasser T, Kahle PJ. Leucine-rich repeat kinase 2 induces alpha-synuclein expression via the extracellular signal-regulated kinase pathway. Cell Signal. 2010;22:821–827. doi: 10.1016/j.cellsig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Weng YH, Chien KY, Lin KJ, Yeh TH, Cheng YP, Lu CS, Wang HL. (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ. 2012;19:1623–1633. doi: 10.1038/cdd.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ, 3rd, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182:474–484. doi: 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LM, Toulouse A, Connor TJ, Nolan YM. Contributions of central and systemic inflammation to the pathophysiology of Parkinson's disease. Neuropharmacology. 2012;62:2154–2168. doi: 10.1016/j.neuropharm.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, Hargus G, Deleidi M, Lawson T, Bogetofte H, Perez-Torres E, Clark L, Moskowitz C, Mazzulli J, Chen L, Volpicelli-Daley L, Romero N, Jiang H, Uitti RJ, Huang Z, Opala G, Scarffe LA, Dawson VL, Klein C, Feng J, Ross OA, Trojanowski JQ, Lee VM, Marder K, Surmeier DJ, Wszolek ZK, Przedborski S, Krainc D, Dawson TM, Isacson O. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci Transl Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci U S A. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Goldberg MS. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One. 2009;4:e5949. doi: 10.1371/journal.pone.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum Mol Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett EA, Zhao J, Mork CN, Hu D, Nichols RJ. Phosphorylation of LRRK2 serines 955 and 973 is disrupted by Parkinson's disease mutations and LRRK2 pharmacological inhibition. J Neurochem. 2012;120:37–45. doi: 10.1111/j.1471-4159.2011.07537.x. [DOI] [PubMed] [Google Scholar]
- Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ. Inhibition of LRRK2 kinase activity leads to dephosphorylation of SeR910)/SeR935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada AA, Liu X, Baker-Glenn C, Beresford A, Burdick DJ, Chambers M, Chan BK, Chen H, Ding X, DiPasquale AG, Dominguez SL, Dotson J, Drummond J, Flagella M, Flynn S, Fuji R, Gill A, Gunzner-Toste J, Harris SF, Heffron TP, Kleinheinz T, Lee DW, Le Pichon CE, Lyssikatos JP, Medhurst AD, Moffat JG, Mukund S, Nash K, Scearce-Levie K, Sheng Z, Shore DG, Tran T, Trivedi N, Wang S, Zhang S, Zhang X, Zhao G, Zhu H, Sweeney ZK. Discovery of highly potent, selective, and brain-penetrable leucine-rich repeat kinase 2 (LRRK2) small molecule inhibitors. J Med Chem. 2012;55:9416–9433. doi: 10.1021/jm301020q. [DOI] [PubMed] [Google Scholar]
- Ferree A, Guillily M, Li H, Smith K, Takashima A, Squillace R, Weigele M, Collins JJ, Wolozin B. Regulation of physiologic actions of LRRK2: focus on autophagy. Neurodegener Dis. 2012;10:238–241. doi: 10.1159/000332599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Gandhi PN, Chen SG, Wilson-Delfosse AL. Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson's disease. J Neurosci Res. 2009;87:1283–1295. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ, Podolsky DK. LRRK2 is involved in the IFN-gamma response and host response to pathogens. Journal of immunology. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gillardon F. Interaction of elongation factor 1-alpha with leucine-rich repeat kinase 2 impairs kinase activity and microtubule bundling in vitro. Neuroscience. 2009a;163:533–539. doi: 10.1016/j.neuroscience.2009.06.051. [DOI] [PubMed] [Google Scholar]
- Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability--a point of convergence in parkinsonian neurodegeneration? J Neurochem. 2009b;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- Gillardon F, Schmid R, Draheim H. Parkinson's disease-linked leucine-rich repeat kinase 2(R1441G) mutation increases proinflammatory cytokine release from activated primary microglial cells and resultant neurotoxicity. Neuroscience. 2012;208:41–48. doi: 10.1016/j.neuroscience.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Boldt K, von Zweydorf F, Helm S, Wiesent L, Sarioglu H, Ueffing M. Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. J Proteome Res. 2010;9:1738–1745. doi: 10.1021/pr9008578. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Suaga P, Luzon-Toro B, Churamani D, Zhang L, Bloor-Young D, Patel S, Woodman PG, Churchill GC, Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2012;21:511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Wey M, Kortholt A, Van Haastert PJ, Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 2008;27:2352. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E. Role of LRRK2 kinase activity in the pathogenesis of Parkinson's disease. Biochem Soc Trans. 2012;40:1058–1062. doi: 10.1042/BST20120054. [DOI] [PubMed] [Google Scholar]
- Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Greggio E, Singleton A. Kinase signaling pathways as potential targets in the treatment of Parkinson's disease. Expert Rev Proteomics. 2007;4:783–792. doi: 10.1586/14789450.4.6.783. [DOI] [PubMed] [Google Scholar]
- Greggio E, Taymans JM, Zhen EY, Ryder J, Vancraenenbroeck R, Beilina A, Sun P, Deng J, Jaffe H, Baekelandt V, Merchant K, Cookson MR. The Parkinson's disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochem Biophys Res Commun. 2009;389:449–454. doi: 10.1016/j.bbrc.2009.08.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro PS, Huang Y, Gysbers A, Cheng D, Gai WP, Outeiro TF, Halliday GM. LRRK2 interactions with alpha-synuclein in Parkinson's disease brains and in cell models. J Mol Med (Berl) 2012 doi: 10.1007/s00109-012-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse AH, Lee HC. NAADP: a universal Ca2+ trigger. Sci Signal. 2008;1:re10. doi: 10.1126/scisignal.144re10. [DOI] [PubMed] [Google Scholar]
- Hakimi M, Selvanantham T, Swinton E, Padmore RF, Tong Y, Kabbach G, Venderova K, Girardin SE, Bulman DE, Scherzer CR, LaVoie MJ, Gris D, Park DS, Angel JB, Shen J, Philpott DJ, Schlossmacher MG. Parkinson's disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. Journal of neural transmission. 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BS, Iacovitti L, Katano T, Hattori N, Seol W, Kim KS. Expression of the LRRK2 gene in the midbrain dopaminergic neurons of the substantia nigra. Neurosci Lett. 2008;442:190–194. doi: 10.1016/j.neulet.2008.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugarvoll K, Wszolek ZK. Clinical features of LRRK2 parkinsonism. Parkinsonism Relat Disord. 2009;15(Suppl 3):S205–S208. doi: 10.1016/S1353-8020(09)70815-6. [DOI] [PubMed] [Google Scholar]
- Heo HY, Kim KS, Seol W. Coordinate Regulation of Neurite Outgrowth by LRRK2 and Its Interactor, Rab5. Experimental neurobiology. 2010a;19:97–105. doi: 10.5607/en.2010.19.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp Cell Res. 2010b;316:649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Herrera AJ, Castano A, Venero JL, Cano J, Machado A. The single intranigral injection of LPS as a new model for studying the selective effects of inflammatory reactions on dopaminergic system. Neurobiol Dis. 2000;7:429–447. doi: 10.1006/nbdi.2000.0289. [DOI] [PubMed] [Google Scholar]
- Higashi S, Biskup S, West AB, Trinkaus D, Dawson VL, Faull RL, Waldvogel HJ, Arai H, Dawson TM, Moore DJ, Emson PC. Localization of Parkinson's disease-associated LRRK2 in normal and pathological human brain. Brain Res. 2007a;1155:208–219. doi: 10.1016/j.brainres.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Higashi S, Moore DJ, Colebrooke RE, Biskup S, Dawson VL, Arai H, Dawson TM, Emson PC. Expression and localization of Parkinson's disease-associated leucine-rich repeat kinase 2 in the mouse brain. J Neurochem. 2007b;100:368–381. doi: 10.1111/j.1471-4159.2006.04246.x. [DOI] [PubMed] [Google Scholar]
- Higashi S, Moore DJ, Yamamoto R, Minegishi M, Sato K, Togo T, Katsuse O, Uchikado H, Furukawa Y, Hino H, Kosaka K, Emson PC, Wada K, Dawson VL, Dawson TM, Arai H, Iseki E. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. J Neuropathol Exp Neurol. 2009;68:994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkle KM, Yue M, Behrouz B, Dachsel JC, Lincoln SJ, Bowles EE, Beevers JE, Dugger B, Winner B, Prots I, Kent CB, Nishioka K, Lin WL, Dickson DW, Janus CJ, Farrer MJ, Melrose HL. LRRK2 knockout mice have an intact dopaminergic system but display alterations in exploratory and motor co-ordination behaviors. Mol Neurodegener. 2012;7:25. doi: 10.1186/1750-1326-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, Ferree A, Raghavan K, Shen GC, Segal L, Ryu H, Cookson MR, Wolozin B. MKK6 binds and regulates expression of Parkinson's disease-related protein LRRK2. J Neurochem. 2010;112:1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulihan MM, Ishihara-Paul L, Kachergus J, Warren L, Amouri R, Elango R, Prinjha RK, Upmanyu R, Kefi M, Zouari M, Sassi SB, Yahmed SB, El Euch-Fayeche G, Matthews PM, Middleton LT, Gibson RA, Hentati F, Farrer MJ. LRRK2 Gly2019Ser penetrance in Arab-Berber patients from Tunisia: a case-control genetic study. Lancet Neurol. 2008;7:591–594. doi: 10.1016/S1474-4422(08)70116-9. [DOI] [PubMed] [Google Scholar]
- Hurley MJ, Patel PH, Jackson MJ, Smith LA, Rose S, Jenner P. Striatal leucine-rich repeat kinase 2 mRNA is increased in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-lesioned common marmosets (Callithrix jacchus) with L-3, 4-dihydroxyphenylalanine methyl ester-induced dyskinesia. Eur J Neurosci. 2007;26:171–177. doi: 10.1111/j.1460-9568.2007.05638.x. [DOI] [PubMed] [Google Scholar]
- Iaccarino C, Crosio C, Vitale C, Sanna G, Carri MT, Barone P. Apoptotic mechanisms in mutant LRRK2-mediated cell death. Hum Mol Genet. 2007;16:1319–1326. doi: 10.1093/hmg/ddm080. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Hishikawa N, Sawada M, Nagatsu T, Yoshida M, Hashizume Y. Distribution of major histocompatibility complex class II-positive microglia and cytokine profile of Parkinson's disease brains. Acta neuropathologica. 2003;106:518–526. doi: 10.1007/s00401-003-0766-2. [DOI] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen ND, Peng Y, Ho CC, Rideout HJ, Petrey D, Liu P, Dauer WT. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One. 2009;4:e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawaji S, Ito G, Iwatsubo T. Identification of the autophosphorylation sites of LRRK2. Biochemistry. 2009;48:10963–10975. doi: 10.1021/bi9011379. [DOI] [PubMed] [Google Scholar]
- Kawakami F, Yabata T, Ohta E, Maekawa T, Shimada N, Suzuki M, Maruyama H, Ichikawa T, Obata F. LRRK2 phosphorylates tubulin-associated tau but not the free molecule: LRRK2-mediated regulation of the tau-tubulin association and neurite outgrowth. PLoS One. 2012;7:e30834. doi: 10.1371/journal.pone.0030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay DM, Kramer P, Higgins D, Zabetian CP, Payami H. Escaping Parkinson's disease: a neurologically healthy octogenarian with the LRRK2 G2019S mutation. Mov Disord. 2005;20:1077–1078. doi: 10.1002/mds.20618. [DOI] [PubMed] [Google Scholar]
- Kett LR, Boassa D, Ho CC, Rideout HJ, Hu J, Terada M, Ellisman M, Dauer WT. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum Mol Genet. 2012;21:890–899. doi: 10.1093/hmg/ddr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Yang MS, Choi D, Kim JH, Kim HS, Seol W, Choi S, Jou I, Kim EY, Joe EH. Impaired inflammatory responses in murine Lrrk2-knockdown brain microglia. PLoS One. 2012;7:e34693. doi: 10.1371/journal.pone.0034693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, Zhang YJ, Jiang H, Ross CA, Moore DJ, Patterson C, Petrucelli L, Dawson TM, Dawson VL. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Obitsu S, Teshima R. alpha-Synuclein aggregation and transmission are enhanced by leucine-rich repeat kinase 2 in human neuroblastoma SH-SY5Y cells. Biol Pharm Bull. 2011;34:1078–1083. doi: 10.1248/bpb.34.1078. [DOI] [PubMed] [Google Scholar]
- Kubo M, Kamiya Y, Nagashima R, Maekawa T, Eshima K, Azuma S, Ohta E, Obata F. LRRK2 is expressed in B-2 but not in B-1 B cells, and downregulated by cellular activation. J Neuroimmunol. 2010;229:123–128. doi: 10.1016/j.jneuroim.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Kumar A, Cookson MR. Role of LRRK2 kinase dysfunction in Parkinson disease. Expert Rev Mol Med. 2011;13:e20. doi: 10.1017/S146239941100192X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss M, Adamopoulou E, Kahle PJ. Interferon-gamma induces leucine-rich repeat kinase LRRK2 via extracellular signal-regulated kinase ERK5 in macrophages. J Neurochem. 2014 doi: 10.1111/jnc.12668. [DOI] [PubMed] [Google Scholar]
- Latourelle JC, Sun M, Lew MF, Suchowersky O, Klein C, Golbe LI, Mark MH, Growdon JH, Wooten GF, Watts RL, Guttman M, Racette BA, Perlmutter JS, Ahmed A, Shill HA, Singer C, Goldwurm S, Pezzoli G, Zini M, Saint-Hilaire MH, Hendricks AE, Williamson S, Nagle MW, Wilk JB, Massood T, Huskey KW, Laramie JM, DeStefano AL, Baker KB, Itin I, Litvan I, Nicholson G, Corbett A, Nance M, Drasby E, Isaacson S, Burn DJ, Chinnery PF, Pramstaller PP, Al-hinti J, Moller AT, Ostergaard K, Sherman SJ, Roxburgh R, Snow B, Slevin JT, Cambi F, Gusella JF, Myers RH. The Gly2019Ser mutation in LRRK2 is not fully penetrant in familial Parkinson's disease: the GenePD study. BMC Med. 2008;6:32. doi: 10.1186/1741-7015-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law BM, Spain VA, Leinster VH, Chia R, Beilina A, Cho HJ, Taymans JM, Urban MK, Sancho RM, Blanca Ramirez M, Biskup S, Baekelandt V, Cai H, Cookson MR, Berwick DC, Harvey K. A direct interaction between Leucine-rich Repeat Kinase 2 and specific beta-tubulin isoforms regulates tubulin acetylation. J Biol Chem. 2013 doi: 10.1074/jbc.M113.507913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Dawson VL, Dawson TM. Leucine-rich repeat kinase 2 (LRRK2) as a potential therapeutic target in Parkinson's disease. Trends Pharmacol Sci. 2012;33:365–373. doi: 10.1016/j.tips.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the Parkinson disease-associated leucine-rich repeat kinase 2. J Biol Chem. 2010a;285:29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Patel JC, Wang J, Avshalumov MV, Nicholson C, Buxbaum JD, Elder GA, Rice ME, Yue Z. Enhanced striatal dopamine transmission and motor performance with LRRK2 overexpression in mice is eliminated by familial Parkinson's disease mutation G2019S. J Neurosci. 2010b;30:1788–1797. doi: 10.1523/JNEUROSCI.5604-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang QJ, Pan N, Lee S, Zhao Y, Chait BT, Yue Z. Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson's disease. PLoS One. 2011;6:e17153. doi: 10.1371/journal.pone.0017153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, Beal MF, Burke RE, Li C. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg M, Mansilla A, Zecchini VR, Fleming A, Rubinsztein DC. The Parkinson's disease protein LRRK2 impairs proteasome substrate clearance without affecting proteasome catalytic activity. Cell Death Dis. 2011;2:e196. doi: 10.1038/cddis.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tsai PI, Wu RM, Chien CT. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ss. J Neurosci. 2010;30:13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, Chen ZZ, Gallant PE, Tao-Cheng JH, Rudow G, Troncoso JC, Liu Z, Li Z, Cai H. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AK, Leak RK, Li L, Zigmond MJ. Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiol Dis. 2008;32:116–124. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Dobson B, Glicksman MA, Yue Z, Stein RL. Kinetic mechanistic studies of wild-type leucine-rich repeat kinase 2: characterization of the kinase and GTPase activities. Biochemistry. 2010;49:2008–2017. doi: 10.1021/bi901851y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hamamichi S, Lee BD, Yang D, Ray A, Caldwell GA, Caldwell KA, Dawson TM, Smith WW, Dawson VL. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson's disease models. Hum Mol Genet. 2011a;20:3933–3942. doi: 10.1093/hmg/ddr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nature immunology. 2011b;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Maturana R, Aguila JC, Sousa A, Vazquez N, Del Rio P, Aiastui A, Gorostidi A, Lopez de Munain A, Sanchez-Pernaute R. Leucine-rich repeat kinase 2 modulates cyclooxygenase 2 and the inflammatory response in idiopathic and genetic Parkinson's disease. Neurobiology of aging. 2013 doi: 10.1016/j.neurobiolaging.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Luerman GC, Nguyen C, Samaroo H, Loos P, Xi H, Hurtado-Lorenzo A, Needle E, Stephen Noell G, Galatsis P, Dunlop J, Geoghegan KF, Hirst WD. Phosphoproteomic evaluation of pharmacological inhibition of leucine-rich repeat kinase 2 reveals significant off-target effects of LRRK-2-IN-1. J Neurochem. 2013 doi: 10.1111/jnc.12483. [DOI] [PubMed] [Google Scholar]
- Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kubo M, Yokoyama I, Ohta E, Obata F. Age-dependent and cell-population-restricted LRRK2 expression in normal mouse spleen. Biochem Biophys Res Commun. 2010;392:431–435. doi: 10.1016/j.bbrc.2010.01.041. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Mori S, Sasaki Y, Miyajima T, Azuma S, Ohta E, Obata F. The I2020T Leucine-rich repeat kinase 2 transgenic mouse exhibits impaired locomotive ability accompanied by dopaminergic neuron abnormalities. Mol Neurodegener. 2012;7:15. doi: 10.1186/1750-1326-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamais A, Raja M, Manzoni C, Dihanich S, Lees A, Moore D, Lewis PA, Bandopadhyay R. Divergent alpha-synuclein solubility and aggregation properties in G2019S LRRK2 Parkinson's disease brains with Lewy Body pathology compared to idiopathic cases. Neurobiol Dis. 2013;58:183–190. doi: 10.1016/j.nbd.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics: Authors' correction. Trends in cell biology. 1999;9:289. doi: 10.1016/s0962-8924(99)01607-4. [DOI] [PubMed] [Google Scholar]
- Manzoni C, Mamais A, Dihanich S, Abeti R, Soutar MP, Plun-Favreau H, Giunti P, Tooze SA, Bandopadhyay R, Lewis PA. Inhibition of LRRK2 kinase activity stimulates macroautophagy. Biochim Biophys Acta. 2013a;1833:2900–2910. doi: 10.1016/j.bbamcr.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni C, Mamais A, Dihanich S, McGoldrick P, Devine MJ, Zerle J, Kara E, Taanman JW, Healy DG, Marti-Masso JF, Schapira AH, Plun-Favreau H, Tooze S, Hardy J, Bandopadhyay R, Lewis PA. Pathogenic Parkinson's disease mutations across the functional domains of LRRK2 alter the autophagic/lysosomal response to starvation. Biochemical and biophysical research communications. 2013b;441:862–866. doi: 10.1016/j.bbrc.2013.10.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata IF, Checkoway H, Hutter CM, Samii A, Roberts JW, Kim HM, Agarwal P, Alvarez V, Ribacoba R, Pastor P, Lorenzo-Betancor O, Infante J, Sierra M, Gomez-Garre P, Mir P, Ritz B, Rhodes SL, Colcher A, Van Deerlin V, Chung KA, Quinn JF, Yearout D, Martinez E, Farin FM, Wan JY, Edwards KL, Zabetian CP. Common variation in the LRRK2 gene is a risk factor for Parkinson's disease. Mov Disord. 2012;27:1823–1826. doi: 10.1002/mds.25226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, Delbroek L, Swerts J, Chavez-Gutierrez L, Esposito G, Daneels G, Karran E, Holt M, Gevaert K, Moechars DW, De Strooper B, Verstreken P. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Meixner A, Boldt K, Van Troys M, Askenazi M, Gloeckner CJ, Bauer M, Marto JA, Ampe C, Kinkl N, Ueffing M. A QUICK screen for Lrrk2 interaction partners--leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol Cell Proteomics. 2011;10:M110 001172. doi: 10.1074/mcp.M110.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose H. Update on the functional biology of Lrrk2. Future Neurol. 2008;3:669–681. doi: 10.2217/14796708.3.6.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose HL, Dachsel JC, Behrouz B, Lincoln SJ, Yue M, Hinkle KM, Kent CB, Korvatska E, Taylor JP, Witten L, Liang YQ, Beevers JE, Boules M, Dugger BN, Serna VA, Gaukhman A, Yu X, Castanedes-Casey M, Braithwaite AT, Ogholikhan S, Yu N, Bass D, Tyndall G, Schellenberg GD, Dickson DW, Janus C, Farrer MJ. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol Dis. 2010;40:503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle MS, Webber PJ, Tse T, Sukar N, Standaert DG, DeSilva TM, Cowell RM, West AB. LRRK2 inhibition attenuates microglial inflammatory responses. J Neurosci. 2012;32:1602–1611. doi: 10.1523/JNEUROSCI.5601-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–2020. doi: 10.1212/WNL.0b013e3181ff9685. [DOI] [PubMed] [Google Scholar]
- Mutez E, Larvor L, Lepretre F, Mouroux V, Hamalek D, Kerckaert JP, Perez-Tur J, Waucquier N, Vanbesien-Mailliot C, Duflot A, Devos D, Defebvre L, Kreisler A, Frigard B, Destee A, Chartier-Harlin MC. Transcriptional profile of Parkinson blood mononuclear cells with LRRK2 mutation. Neurobiol Aging. 2011;32:1839–1848. doi: 10.1016/j.neurobiolaging.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Mutez E, Nkiliza A, Belarbi K, de Broucker A, Vanbesien-Mailliot C, Bleuse S, Duflot A, Comptdaer T, Semaille P, Blervaque R, Hot D, Lepretre F, Figeac M, Destee A, Chartier-Harlin MC. Involvement of the immune system, endocytosis and EIF2 signaling in both genetically determined and sporadic forms of Parkinson's disease. Neurobiol Dis. 2014;63:165–170. doi: 10.1016/j.nbd.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, Alessi DR. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Yu M, Wang C, Xu Z. Leucine-rich repeat kinase 2 disturbs mitochondrial dynamics via Dynamin-like protein. J Neurochem. 2012;122:650–658. doi: 10.1111/j.1471-4159.2012.07809.x. [DOI] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, Raya A, Sulzer D, Cuervo AM. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013 doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Nath P, Washecka N, Gibbs JR, Singleton AB. Comprehensive analysis of LRRK2 in publicly available Parkinson's disease cases and neurologically normal controls. Hum Mutat. 2008;29:485–490. doi: 10.1002/humu.20668. [DOI] [PubMed] [Google Scholar]
- Papkovskaia TD, Chau KY, Inesta-Vaquera F, Papkovsky DB, Healy DG, Nishio K, Staddon J, Duchen MR, Hardy J, Schapira AH, Cooper JM. G2019S leucine-rich repeat kinase 2 causes uncoupling protein-mediated mitochondrial depolarization. Hum Mol Genet. 2012;21:4201–4213. doi: 10.1093/hmg/dds244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisiadou L, Cai H. LRRK2 function on actin and microtubule dynamics in Parkinson disease. Commun Integr Biol. 2010;3:396–400. doi: 10.4161/cib.3.5.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G, Zhu X, Babar AK, Siedlak SL, Yang Q, Ito G, Iwatsubo T, Smith MA, Chen SG. Leucine-rich repeat kinase 2 colocalizes with alpha-synuclein in Parkinson's disease, but not tau-containing deposits in tauopathies. Neurodegener Dis. 2008;5:222–224. doi: 10.1159/000113708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli G, Condliffe SB, Bauer M, Giesert F, Boldt K, De Astis S, Meixner A, Sarioglu H, Vogt-Weisenhorn DM, Wurst W, Gloeckner CJ, Matteoli M, Sala C, Ueffing M. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J Neurosci. 2011;31:2225–2237. doi: 10.1523/JNEUROSCI.3730-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos M, Cortes E, Vonsattel JP, Fahn S, Waters C, Cote LJ, Moskowitz C, Honig LS, Clark LN, Marder KS, Alcalay RN. Clinical and pathological characteristics of LRRK2 G2019S patients with PD. J Mol Neurosci. 2012;47:139–143. doi: 10.1007/s12031-011-9696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Wu X, Block ML, Liu Y, Breese GR, Hong JS, Knapp DJ, Crews FT. Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia. 2007;55:453–462. doi: 10.1002/glia.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing H, Wong W, McGeer EG, McGeer PL. Lrrk2 phosphorylates alpha synuclein at serine 129: Parkinson disease implications. Biochem Biophys Res Commun. 2009a;387:149–152. doi: 10.1016/j.bbrc.2009.06.142. [DOI] [PubMed] [Google Scholar]
- Qing H, Zhang Y, Deng Y, McGeer EG, McGeer PL. Lrrk2 interaction with alpha-synuclein in diffuse Lewy body disease. Biochem Biophys Res Commun. 2009b;390:1229–1234. doi: 10.1016/j.bbrc.2009.10.126. [DOI] [PubMed] [Google Scholar]
- Ramonet D, Daher JP, Lin BM, Stafa K, Kim J, Banerjee R, Westerlund M, Pletnikova O, Glauser L, Yang L, Liu Y, Swing DA, Beal MF, Troncoso JC, McCaffery JM, Jenkins NA, Copel NG, Galter D, Thomas B, Lee MK, Dawson TM, Dawson VL, Moore DJ. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Liu M. Current understanding of LRRK2 in Parkinson's disease: biochemical and structural features and inhibitor design. Future Med Chem. 2012;4:1701–1713. doi: 10.4155/fmc.12.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko IN, Chia R, Cookson MR. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson's disease? BMC medicine. 2012;10:20. doi: 10.1186/1741-7015-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko IN, Cookson MR. 14-3-3 proteins are promising LRRK2 interactors. Biochem J. 2010;430:e5–e6. doi: 10.1042/BJ20101200. [DOI] [PubMed] [Google Scholar]
- Saha S, Guillily MD, Ferree A, Lanceta J, Chan D, Ghosh J, Hsu CH, Segal L, Raghavan K, Matsumoto K, Hisamoto N, Kuwahara T, Iwatsubo T, Moore L, Goldstein L, Cookson M, Wolozin B. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J Biol Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A, Canals JM, Memo M, Alberch J, Lopez-Barneo J, Vila M, Cuervo AM, Tolosa E, Consiglio A, Raya A. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J Biol Chem. 2009;284:36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol W. Biochemical and molecular features of LRRK2 and its pathophysiological roles in Parkinson's disease. BMB reports. 2010;43:233–244. doi: 10.5483/bmbrep.2010.43.4.233. [DOI] [PubMed] [Google Scholar]
- Sheng Z, Zhang S, Bustos D, Kleinheinz T, Le Pichon CE, Dominguez SL, Solanoy HO, Drummond J, Zhang X, Ding X, Cai F, Song Q, Li X, Yue Z, van der Brug MP, Burdick DJ, Gunzner-Toste J, Chen H, Liu X, Estrada AA, Sweeney ZK, Scearce-Levie K, Moffat JG, Kirkpatrick DS, Zhu H. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med. 2012;4:164ra161. doi: 10.1126/scitranslmed.3004485. [DOI] [PubMed] [Google Scholar]
- Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. Mutant LRRK2 Toxicity in Neurons Depends on LRRK2 Levels and Synuclein But Not Kinase Activity or Inclusion Bodies. J Neurosci. 2014;34:418–433. doi: 10.1523/JNEUROSCI.2712-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Francis SH, Corbin JD. Autophosphorylation: a salient feature of protein kinases. Mol Cell Biochem. 1993;127–128:51–70. doi: 10.1007/BF01076757. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EK, Jankovic J. Genetic testing in Parkinson disease: promises and pitfalls. Arch Neurol. 2006;63:1232–1237. doi: 10.1001/archneur.63.9.1232. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Vancraenenbroeck R, Ollikainen P, Beilina A, Lobbestael E, De Maeyer M, Baekelandt V, Cookson MR. LRRK2 kinase activity is dependent on LRRK2 GTP binding capacity but independent of LRRK2 GTP binding. PLoS One. 2011;6:e23207. doi: 10.1371/journal.pone.0023207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet J, Pescini Gobert R, Hooft van Huijsduijnen R, Wiessner C, Sagot YJ. Regulation of LRRK2 expression points to a functional role in human monocyte maturation. PLoS One. 2011;6:e21519. doi: 10.1371/journal.pone.0021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener. 2012;7:2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc Natl Acad Sci U S A. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ, 3rd, Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci U S A. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsika E, Moore DJ. Mechanisms of LRRK2-mediated neurodegeneration. Curr Neurol Neurosci Rep. 2012;12:251–260. doi: 10.1007/s11910-012-0265-8. [DOI] [PubMed] [Google Scholar]
- Venderova K, Kabbach G, Abdel-Messih E, Zhang Y, Parks RJ, Imai Y, Gehrke S, Ngsee J, Lavoie MJ, Slack RS, Rao Y, Zhang Z, Lu B, Haque ME, Park DS. Leucine-Rich Repeat Kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Hum Mol Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- Vitte J, Traver S, Maues De Paula A, Lesage S, Rovelli G, Corti O, Duyckaerts C, Brice A. Leucine-rich repeat kinase 2 is associated with the endoplasmic reticulum in dopaminergic neurons and accumulates in the core of Lewy bodies in Parkinson disease. J Neuropathol Exp Neurol. 2010;69:959–972. doi: 10.1097/NEN.0b013e3181efc01c. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, Moore DJ, Dawson TM, Dawson VL, Chiosis G, Cookson MR, Cai H. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, Perry G, Casadesus G, Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber PJ, Smith AD, Sen S, Renfrow MB, Mobley JA, West AB. Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J Mol Biol. 2011;412:94–110. doi: 10.1016/j.jmb.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Westerlund M, Ran C, Borgkvist A, Sterky FH, Lindqvist E, Lundstromer K, Pernold K, Brene S, Kallunki P, Fisone G, Olson L, Galter D. Lrrk2 and alpha-synuclein are co-regulated in rodent striatum. Mol Cell Neurosci. 2008;39:586–591. doi: 10.1016/j.mcn.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Yuan C, Chen R, Dawson TM, Dawson VL. ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J Neurosci. 2012;32:3877–3886. doi: 10.1523/JNEUROSCI.4566-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YL, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase Activity Plays a Key Role in the Pathobiology of LRRK2. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoubian TA, Slone SR, Harrington AJ, Hamamichi S, Schieltz JM, Caldwell KA, Caldwell GA, Standaert DG. Differential neuroprotective effects of 14-3-3 proteins in models of Parkinson's disease. Cell Death Dis. 2010;1:e2. doi: 10.1038/cddis.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Johnson WM, Gao Y, Wang W, Zhang J, Deak M, Alessi DR, Zhu X, Mieyal JJ, Roder H, Wilson-Delfosse AL, Chen SG. Kinase inhibitors arrest neurodegeneration in cell C. elegans models of LRRK2 toxicity. Hum Mol Genet. 2013;22:328–344. doi: 10.1093/hmg/dds431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hermanson SB, Carlson CB, Riddle SM, Vogel KW, Bi K, Nichols RJ. Pharmacological inhibition of LRRK2 cellular phosphorylation sites provides insight into LRRK2 biology. Biochem Soc Trans. 2012;40:1158–1162. doi: 10.1042/BST20120137. [DOI] [PubMed] [Google Scholar]
- Zhou H, Huang C, Tong J, Hong WC, Liu YJ, Xia XG. Temporal expression of mutant LRRK2 in adult rats impairs dopamine reuptake. Int J Biol Sci. 2011;7:753–761. doi: 10.7150/ijbs.7.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]