Abstract
Curved membranes are a common and important attribute in cells. Protein and peptide curvature sensors are known to activate signaling pathways, initiate vesicle budding, trigger membrane fusion, and facilitate molecular transport across cell membranes. Nonetheless, there is little understanding how these proteins and peptides achieve preferential binding of different membrane curvatures. The current study is to elucidate specific factors required for curvature sensing. As a model system, we employed a recently identified peptide curvature sensor, MARCKS-ED, derived from the effector domain of the myristoylated alanine-rich C-kinase substrate protein, for these biophysical investigations. An atomistic molecular dynamics (MD) simulation suggested an important role played by the insertion of the Phe residues within MARCKS-ED. To test these observations from our computational simulations, we performed electron paramagnetic resonance (EPR) studies to determine the insertion depth of MARCKS-ED into differently curved membrane bilayers. Next, studies with varied lipid compositions revealed their influence on curvature sensing by MARCKS-ED, suggesting contributions from membrane fluidity, rigidity, as well as various lipid structures. Finally, we demonstrated that the curvature sensing by MARCKS-ED is configuration independent. In summary, our studies have shed further light to the understanding of how MARCKS-ED differentiates between membrane curvatures, which may be generally applicable to protein curvature sensing behavior.
Keywords: Curvature sensing, MARCKS-ED, peptide-lipid interactions, EPR
1. INTRODUCTION
Membrane curvature sensing is an important biological process. Nonetheless, the biophysical understanding of their molecular recognition is lacking. Many curvature sensing peptides and proteins share common structure and sequence characteristics [1]. For example, the Bin/Amphiphysin/Rvs (BAR) domains, found in various membrane proteins such as Amphiphysin and Endophilin-A1 [2, 3], insert an amphipathic helix into the bilayer, acting as a wedge to create more space in the outer leaflet thereby acting as an active membrane curvature inducer [4]. In another important study, Chapman and co-workers proposed that Synaptotagmin-1 passively senses positive membrane curvature upon membrane insertion [5]. Synaptotagmin-1 uses its flexible loops stemming from two C2 domains to penetrate into the membrane bilayer of loosely packed lipids, another common feature of positively curved membranes [5]. Lastly, electrostatic interactions have been suggested to be critical in stabilizing the interaction of protein domains on the membrane surface of highly curved regions [6]. For instance, the positively-charged regions of BAR domains have been shown to recognize highly curved membrane surfaces with exposed negatively charged lipids [6].
Recently, our group has reported a 25-amino acid peptide, MARCKS-ED (NH2-KKKKKRFSFKKSFKLSGFSFKKNKK-CONH2), derived from the effector domain of the myristoylated alanine-rich C kinase substrate (MARCKS) protein as a membrane curvature sensor [7]. In fact, the natural MARCKS protein has been observed to localize in areas of positive curvature in cells [8]. Nonetheless, the molecular mechanism of its curvature sensing behavior remains elusive. MARCKS is an 87-kDa cytoplasmic protein known to regulate Phospholipase C signaling via specific interactions with Phosphatidylinositol 4,5-bisphosphate (PIP2) along the inner cytosolic plasma membrane. Furthermore, MARCKS is a Protein Kinase C substrate that also binds at high affinity to Calcium-bound Calmodulin [7]. Membrane curvature (defined by the reciprocal of the particle radii) is known to regulate many important biological processes [9-11]. Curvature sensing by natural membrane proteins has drawn intense research interests recently [1, 12]. In our previous studies, we have shown that MARCKS's effector domain (MARCKS-ED) is a short sequence incorporated with both highly basic and hydrophobic residues that can sense highly curved membranes. In this current report, we employed a combination of various biophysical methods to investigate the basis of its curvature sensing behavior.
Based on simulation results, we hypothesized that the insertion of aromatic residue side chains significantly contribute to its curvature sensing, which is in agreement with previous reports that the Phe residues in the effector domain of MARCKS are buried into the membrane bilayer [13]. To further investigate the results of the simulation study, electron paramagnetic resonance (EPR) was used to determine the insertion depth of the Phe residues into lipid vesicles presenting different curvatures. Fluorescence enhancement experiments were performed using both L- and D-MARCKS-ED peptides to study whether the peptide recognizes curvature in a configuration-specific manner. To further expound on these fluorescence enhancement findings, we quantified the binding affinities of MARCKS-ED to differently sized lipid vesicles.
It has long been known that lipids have significant functions in membrane shape, mobility, and stiffness [14]. Lipid components give rise to membrane curvature primarily dictated by their shape and net charge [15, 16]. Importantly, lipid packing induces defects, which has been suggested to play a role in curvature sensing [12]. Therefore, we next studied the impact of various lipid components in biologically relevant membranes on the curvature sensing ability of MARCKS-ED, revealing their importance in the recognition of membrane curvature.
Taken together, we herein report that Phe insertion in conjunction with specific peptide-lipid interactions could contribute to the membrane curvature sensing by MARCKS-ED. These results could help to successfully design novel membrane curvature probes for studying biological processes.
2 MATERIAL AND METHODS
2.1 Atomistic Membrane Model
Molecular dynamics was employed to model two MARCKS-ED peptide molecules, with one of each located in the interfacial region of a hydrated, flat lipid bilayer system composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) and 1-palmitoyl-2-oleoyl-snglycero-3-phosphatidylserine (POPS). The Membrane Builder module in CHARMM-GUI [17] was used to construct the systems that are composed of a lipid bilayer, water and ions. The MARCKS-ED peptide was constructed from internal coordinates found in the CHARMM residue topology files. As part of this study aims to investigate the interactions of the MARCKS-ED peptide with the membrane interfacial region, the peptide was initially placed in the simulation system in such a way that the peptide lies flat along the interfacial region and thus, the xy-area of the system box includes the contribution of the peptide. This initial configuration of the peptide also orients the side chains to initially lie approximately parallel to the membrane surface. Each system was composed of two MARCKS-ED peptides (one in each leaflet), 250 lipids per leaflet (225 POPC and 25 POPS molecules) and ~20,000 water molecules. Based on the location of the MARCKS peptide where it is predicted to be constrained in one leaflet [18], two MARCKS-ED peptides were used for increased sampling of the peptide behavior in a membrane leaflet. This approach is similar to that employed by Maccallum et. al. [19]. Each simulation system contained on average ~130,000 atoms. A 0.15-M concentration of KCl was used for all simulations. Rectangular periodic boundary conditions were imposed with a variable box height of ~70 Å and xy-translation lengths of ~135 Å. The latter dimension was based on the area of the peptide and the 68.3-Å2 and 62-Å2 area/lipid ratios for POPC and POPS, respectively [17].
2.2 Molecular Dynamics Simulation
The Chemistry at Harvard Macromolecular Mechanics (CHARMM) program [20] was used for this study, making use of the CHARMM36 protein and lipid force fields [21-23]. The TIP3P model for water [24] was used with bonds to hydrogen atoms constrained with the SHAKE algorithm [25]. The particle-mesh Ewald (PME) method [26] was employed to calculate the electrostatics using a mesh size of ~1 Å for fast Fourier transformation, κ = 0.34/Å and a sixth-order B-spline interpolation. Non-bond pair lists were constructed using a 16-Å cutoff distance, and the Lennard-Jones potential was smoothly switched off at 10-12 Å using a force-switching function. Simulations were performed under constant temperature (330 K, to ensure bilayer fluidity) and normal pressure (1.0 atm) with a fixed lateral area using Nosé-Hoover methods [27] and the Langevin piston [28]. These conditions have been used in numerous previous simulations of biomolecular systems [29-32].
2.3 Solid Phase Peptide Synthesis
All peptides were synthesized using a CEM Liberty microwave-assisted peptide synthesizer using standard solid phase Fmoc chemistry. Peptides were conjugated to a 6-(N-(7- Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoic acid (NBD) fluorophore to the N-terminus via an aminohexanoic acid linker. For electron paramagnetic resonance (EPR) studies, the 11th amino acid residue, Lys, in MARCKS-ED was mutated to a Cys residue in order to conjugate the methanethiosulfonate (MTSL) label, generating the spin-labeled peptide, MTSL-K11C. Following NBD or MTSL conjugation, the resin beads were then washed, dried and cleaved from the peptide using a water/trifluoroacetic acid (TFA)/ triisopropylsilane (TIPS) cocktail of 2.5%/95%/2.5% for 1 hour under inert N2 conditions. Chilled Et2O was used to precipitate the peptides. Reverse phase high performance liquid chromatography was performed to purify each peptide using a semi-prep C8 column.
2.4 Lipid Vesicle Preparation
To generate homogeneous lipid vesicle solutions, a previously reported protocol was followed [33-35]. The following lipids were purchased from Avanti Polar Lipids: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), hydrogenated phosphatidylcholine (Hydro-PC), sphingomyelin and cholesterol. The wild type lipid composition was based on reported approximations of the outer leaflet of exosomes [36]. After mixing each lipid component, residual organic solvent was removed using a slow flow of N2 until a thin, white film was observed on the bottom of the glass vial. Each glass vial was placed in the dessicator in vacuo for at least 30 minutes. Phosphate buffer (pH = 7.40) was added to each glass vial containing the dry film and incubated overnight to hydrate at 4°C. A summary of the lipid compositions of the different vesicle systems studied is found in Table 1.
Table 1.
A list of different synthetic lipid vesicles prepared for biophysical assays.
| Lipid Models | Lipid Composition |
|---|---|
| WT (Wild type) | 28% POPC: 27% POPE: 17% POPS: 14% Chol: 14% Sph |
| NoPE | 55% POPC: 0% POPE: 17% POPS: 14% Chol: 14% Sph |
| NoPS | 45% POPC: 27% POPE: 0% POPS: 14% Chol: 14% Sph |
| NoSp | 42% POPC: 27% POPE: 17% POPS: 14% Chol: 0% Sph |
| NoCh | 42% POPC: 27% POPE: 17% POPS: 0% Chol: 14% Sph |
| DAG | 28% POPC: 27% DAG: 17% POPS: 14% Chol: 14% Sph |
| Hydro-PC | 28% POPC: 27% POPE: 17% POPS: 14% Hydro-PC: 14% Sph |
| LM1 | 70% POPC: 15% POPE: 0% POPS: 15% Chol |
| LM2 | 60% POPC: 15% POPE: 10% POPS: 15% Chol |
| LM3 | 50% POPC: 15% POPE: 20% POPS: 15% Chol |
The wild type model (WT) represents a composition closely resemble cell membranes [31].
The lipid vesicle solutions were prepared by an Avestin FL-50 pressurized extruder using polycarbonate membranes purchased by Avanti Polar Lipids following a previously established protocol [35]. The polycarbonate membrane filters used were of sizes 30, 100 and 400 nm. For each lipid vesicle size, the lipid vesicle solutions were extruded through the membrane filters at least 3 times to generate homogeneity. To characterize the actual vesicle diameter for each lipid vesicle solution, we used the nanoparticle tracking analysis (NTA) to produce a distribution curve displaying the vesicle size present in each solution. The LM10-HS instrument was used with a 638 nm laser at scatter mode and a 650 nm laser at fluorescence mode.
2.5 Fluorescence Enhancement Assay
The Fluorolog-3 fluorometer by Horiba Jobin Yvon was used to observe emission spectra for the NBD-labeled MARCKS-ED peptides. Excitation and emission wavelengths were set at γex = 480 nm and γem = 545 nm for the NBD fluorophore. Proper controls were performed to ensure that the observed fluorescence intensity was solely based on the peptide and not on the fluorophore [7]. The MARCKS-ED peptides (500 nM) were incubated with each lipid vesicle solution (500 μM) in PBS (pH = 7.40) prior to the experiment. The fluorescence emission spectrum was set from 500-650 nm.
2.6 Fluorescence Anisotropy Assay
Fluorescence anisotropy was performed using the Horiba Jobin Yvon Fluorolog-3 fluorometer. Various concentrations of lipid vesicles were titrated into the MARCKS-ED solution until the binding curve reached saturation. The excitation and emission wavelengths were set to γex = 480 nm and γem = 545 nm for the NBD fluorophore. As previously reported [37], the following equation was used to determine the apparent KD:
| (Eq 1) |
| (Eq 2) |
where Fb describes the fraction of peptide bound to the lipid vesicles, Kp describes the molar partition coefficient and [L] describes the lipid concentration. This equation was fitted to each anisotropy graph where the Kp is expressed as the inverse of the apparent dissociation constant, KD. Statistical analysis was performed using the Analysis of variance (ANOVA) test to produce a p-value between samples measured.
2.7 Electron Paramagnetic Resonance (EPR)
EPR was performed to study the interactions of the mutated MARCKS-ED peptide, MTSL-K11C, with different sized vesicles. Methanethiosulfonate (MTSL) provides the unpaired electron required for an EPR response. The EPR data were collected using a Bruker Elexsys E500 spectrometer and a loop gap resonator. To determine the depth parameter values of the MTSL-K11C peptide within the membrane bilayer, EPR power saturation experiments were performed by incrementally increasing the incident microwave power while recording the changes of the amplitude of the central resonance peak until signal saturation is reached. The equations shown below describe how the depth parameter was measured. The power at which the system was at half saturation, P1/2, was calculated based on Eq. 3. The P1/2 values describe the frequency of collisions between the nitroxide label, MTSL, and the O2 or NiEDDA probes. The P1/2 value is then used to solve for the solvent accessibility, π, for both paramagnetic probes, O2 and NiEDDA using Eq. 4, respectively. A higher π value points to more collision frequencies between the paramagnetic probe with the nitroxide-labeled K11C peptide, MTSL-K11C. The solvent accessibility values are required to calculate the depth parameter values (Φ) as shown in Eq. 5 below.
| (Eq 3) |
| (Eq 4) |
| (Eq 5) |
Where A desribes the amplitude, c desribes the scaling, P describes the power, P1/2 describes the power at half saturation and ΔH describes the width of the central resonance peak.
3 RESULTS
3.1 Computational Simulations of MARCKS-ED
We have previously reported that L-MARCKS-ED binds to biologically relevant vesicles while its Phe and Lys residues are both essential [7]. As structural analyses of peptides/proteins in the membranes are difficult to conduct, computational simulations provide a useful alternative. For a microscopic view of this peptide-lipid interaction, the MARCKS-ED peptide was simulated with an atomistic membrane bilayer. Figure 1 shows a snapshot of the system after 40 ns of dynamics simulation. As mentioned in the Methods section, the initial placement of the peptide placed it in a configuration that has the side chains sticking out along the sides of the protein backbone where they lie approximately parallel to the membrane surface. From Fig. S1, it can be seen that within the first 2 ns, initial rearrangements were already taking place, particularly with the depth of the Phe side chains. From their initial positions at ~ ±20 Å, centers of mass of the Phe side chains are already starting to move 7-8 Å deeper towards the center of the membrane. The positions of the Lys side chains also change during the early stages and have started to move around 3-4 Å towards the bulk solvent. The cationic terminal regions of MARCKS-ED are more exposed to water, due to the interactions of the Lys side chains with the bulk solvent. By contrast, the central stretch of the peptide, where the Phe residues (represented in gray) are located, is buried into the interface and appears to hold the MARCKS-ED peptide to the bilayer.
Figure 1.
Atomistic model of MARCKS-ED interacting with membrane bilayers. a) Snapshot showing MARCKS-ED binding to the membrane bilayers after 40 ns of simulations using the CHARMM forcefield. Lipid tails are shown in cyan. Water molecules are shown in stick and ball representation with the oxygen atoms shown in red and the hydrogen atoms in white. Lysine residues of MARCKS-ED are shown in blue, phenylalanine in gray and serine in red. The residue in yellow is the Lys residue that was mutated into Cys for the sequential EPR experiments. The lipid head groups are shown as: choline nitrogen (light violet), phosphate phosphorus (light yellow) and one of the sn-2 carbonyl oxygen (pink). For clarity, atoms in the foreground are not shown and the size of the head group atoms have been decreased. b) A different view with the lipid tails and water molecules removed. Colors and representations are the same as that in a).
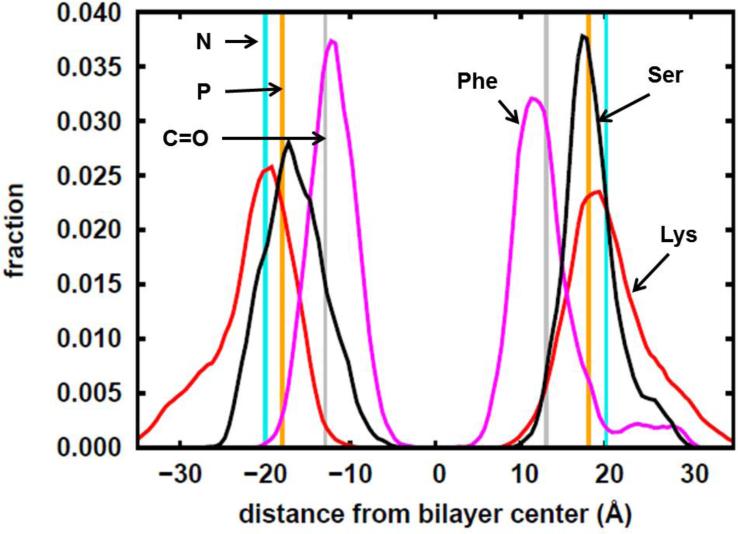
To quantify the positions of the main residues comprising the peptide, the coordinates of the centers of mass for each side chain of the Lys, Phe and Ser residues were averaged after 10 ns of equilibration. Then the histograms of the z-coordinates relative to the bilayer center were constructed. Figure S1 in the Supplementary Information shows that at the 10 ns point, the z-coordinates of the centers of masses of the side chains of the Lys, Ser and Phe residues are fluctuating within a constant range. This indicates that an equilibrium state had been established. In the simulation, the average distances from the bilayer center to the lipid head group components are as follows: choline N= ~±20 Å, phosphate P= ~18 Å, and carbonyl C= ~± 13 Å. As shown in Figure 2, the Lys residues (red curve) are found furthest away from the bilayer center, with some found past the choline region. Ser residues (black curve) are also found closer to the bulk solvent owing to the fact that its side chain is a hydrogen bond donor and thus can interact with the bulk solvent and the head groups. Phe residues, due to their elevated hydrophobicity, are found closer to the bilayer center (magenta curve) at ~± 12 Å. Implicit membrane simulations by Zhang, Yetiraj and Cui [38] likewise report this trend for the MARCKS-ED peptide where the charged and polar residues are found in the high dielectric constant region and the Phe residues are found close to the dielectric boundary. In comparison to Ellena et al [18] ~5-10 Å below the lipid phosphate was where they observed the Phe residues in the presence of Phosphatidylinositol 4,5-bisphosphate (PIP2). Overall, MARCKS-ED adopts a “boat” conformation upon membrane association. This observation is in agreement with another previous literature report [13], supporting our computational simulation as a valid platform to study the peptide-lipid interactions by the MARCKS-ED peptide.
Figure 2.
Histograms calculated from the simulation trajectory showing the distribution of the z-coordinates of the side-chain centers of mass of the Lys, Phe and Ser residues of the MARCKS-ED peptide. N, P and C=O respectively refer to the average positions of the nitrogen, phosphorus and carbonyl carbon atoms of the lipid molecules. The z-coordinates are relative to the bilayer center where z=0.
3.2 Peptide configuration and MARCKS-ED curvature sensing
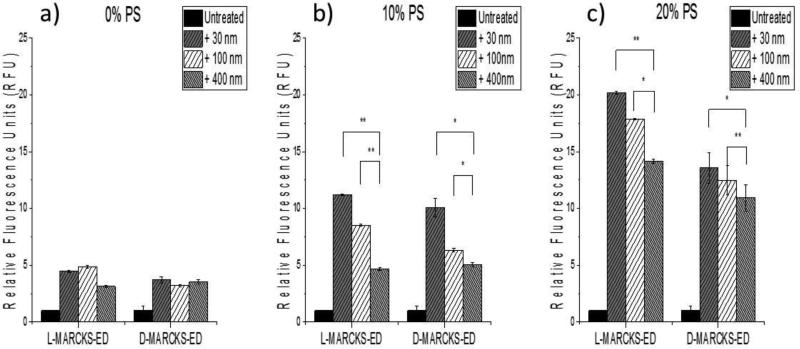
Previous literature has shown that the natural MARCKS-ED regions do not adopt any secondary structures [37, 39]. To further explore the molecular recognition of MARCKS-ED to differently curved membranes, we next carried out fluorescence enhancement assays with MARCKS-ED along with its mirror-image peptide D-MARCKS-ED, composed of unnatural D-amino acids. Fluorescence enhancement describes a change in fluorescence intensity brought about by the environmental change surrounding the fluorophore (conjugated to the N-terminus of each peptide) from a polar, aqueous solvent to a hydrophobic bilayer. An increase in fluorescence intensity indicates peptide-vesicle binding. Vesicles with various percentages of the anionic lipid, phosphatidylserine (PS), were used in these experiments as previous studies showed that PS contributes to most of the binding affinity of MARCKS-ED [7]. As can be observed from Figure 3, D-MARCKS-ED behaves similarly to L-MARCKS-ED. For bilayers with 10% and 20% PS, both D- and L-MARCKS are able to distinguish between the different membrane curvatures. However, curvature sensing ceases when there is no PS present in the bilayer. To better quantify the curvature-sensing ability of D-MARCKS-ED, the binding affinity was measured by fluorescence anisotropy assays. Lipid vesicles with different diameters (smaller vesicles present surfaces with more curvature) were titrated to the NBD-labeled D-MARCKS-ED, which results in an increase in polarization upon peptide-vesicle association. The dissociation constants (KD) were calculated according to Eq 1 and 2. Table 2 shows the KD values of D-MARCKS-ED for 30-, 100- and 400-nm pore size vesicles of lipid models 1, 2 and 3 and Figure S2 shows their titration curves. We could not obtain the KD values for the lipid model 1 that contains 0% PS because the titration curve never reached the saturation. Table 2 also shows KD values of NBD-labeled L-MARCKS-ED to the models 1, 2 and 3 that we have previously reported as a comparison [7]. Model 2 showed the most curvature sensing behaviors for both D- and L-MARCKS-ED. KD values of D-MARCKS-ED to 30-, 100- and 400-nm diameters vesicles were determined to be 69±9 μM, 303±65 μM and 394±133 μM, respectively and this trend is similar to L-MARCKS-ED whose KD values of 30-, 100- and 400-nm diameters were 24±3 μM, 42±13 μM and 86±20 μM, respectively. Our results also indicated that DMARCKS-ED binds more tightly to highly curved vesicles that contain more PS. Taken together, these results demonstrate that the curvature sensing behavior does not depend on the configuration of amino acids in MARCKS-ED, consistent with that MARCKS-ED binds in an unstructured conformation and the binding is primarily driving by electrostatic interactions.
Figure 3.
Fluorescence enhancement results comparing L-MARCKS-ED and D-MARCKS-ED peptides. Maximum fluorescence emission by the NBD label attached to the N-terminus of L-and D-MARCKS-ED in the presence of vesicles of different diameters. [Peptide] = 500 nM. [Lipid vesicle] = 500 μM. Fluorescence (in relative fluorescence units, RFU) was normalized to the untreated MARCKS-ED peptides. *P value < 0.01 compared to 400 nm polycarbonate membrane pores. **P value < 0.05 compared to vesicles generated by 400 nm polycarbonate membrane pores. Statistical analysis was performed using the Analysis of variance (ANOVA) test to produce a p-value between samples measured.
Table 2.
The dissociation constants, KD, of D-MARCKS-ED and L-MARCKS-ED for 30-, 100- and 400-nm pore size vesicles of lipid models 1, 2, 3 and wild type (WT).
| D-MARCKS-ED | ||||
|---|---|---|---|---|
| Vesicle Size (nm) | Model 1: 0% PS | Model 2: 10% PS | Model 3: 20% PS | WT: 17%PS |
| 30 | n.d. | 69±9 μM | 14±12 μM | 12±46 μM |
| 100 | n.d. | 303±65 μM | 15±50 μM | 29±45 μM |
| 400 | n.d. | 394±133 μM | 23±6 μM | 30±6 μM |
| L-MARCKS-ED | ||||
|---|---|---|---|---|
| Vesicle Size (nm) | Model 1: 0% PS | Model 2: 10% PS | Model 3: 20% PS | WT: 17%PS |
| 30 | 141±17 μM | 24±3 μM | 16±3 μM | 14±1 μM |
| 100 | 64±5 μM | 42±13 μM | 18±1 μM | 17±2 μM |
| 400 | 146±18 μM | 86±20 μM | 26±6 μM | 33±2 μM |
n.d. = not determined.
In addition to the lipid models 1, 2 and 3, we also measured the KD values of both D- and L-MARCKS-ED with the “wild type” lipid vesicles, whose lipid composition closely mimic biological exosome membranes [36], of 30-, 100- and 400-nm diameters: they were determined to be 12±46 μM, 29±45 μM and 30±6 μM for D-MARCKS-ED and 14±1 μM, 17±2 μM and 33±2 μM for L-MARCKS-ED, respectively. This preferential binding is comparable with literature reports that the curvature sensing protein, Synaptotagmin-1 (Syt-1), shows a 1.9-fold increase in binding affinity between small (d=105 nm) and large (d=252 nm) vesicles [5].
3.3 Characterizing the Contribution of Different Insertion Depths in Curvature Sensing
To explore the second contributing factor of different insertion depths, electron paramagnetic resonance (EPR) spectroscopy was performed. Previous studies with single sized vesicles (d=50 nm) showed that MARCKS-ED penetrated into membrane bilayers [13]. Herein, the interaction of MARCKS-ED with vesicles of different sizes was explored. Supplementary Figure S3 shows spectra for the methanethiosulfonate (MTSL, a commonly used paramagnetic probe)-labeled MARCKS-ED, MTSL-K11C, in the presence (red) and absence (blue) of lipid vesicles. Consistent with a previous report [13], EPR spectra with lipid vesicles all showed significant peak broadening, an indicative of the reduced tumbling rate of MTSL-K11C caused by its membrane association.
To measure the depth of MARCKS-ED insertion into different membrane curvatures, power saturation experiments were performed for each lipid vesicle size. Figure S4 shows the power saturation curves for each vesicle size in the absence (red curve) and presence of the paramagnetic probes O2 and NiEDDA (blue and green curves, respectively). Measurements of the probe-free and the NiEDDA-containing vesicles were performed under N2. Using the equations summarized in Materials and Methods, the depth parameter (Φ) was then calculated.
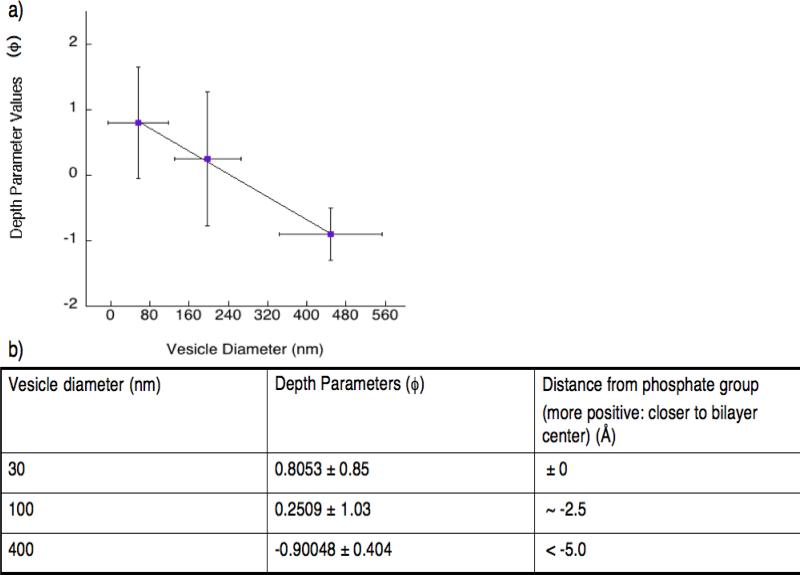
Figure 4a shows how the depth parameter values Φ (larger values indicate deeper insertion) correlate to different vesicle diameters. These Φ values were then calibrated using five commercially available, spin-labeled nitroxide phosphatidylcholine (PC) lipids. The head group labeled PC was given a value of −5 Å (distance from the phosphate group, increasing positive values indicate greater insertion) based on a previous report [13]. These lipids are labeled with doxyl nitroxides at positions C5, C7, C10 and C12 on the acyl chain as well as on the head group, respectively [13]. Calibration plots display a linear correlation between the distance of the nitroxide label on the lipid chain relative to the membrane bilayer and their depth parameters (Figure S5). The distance of the MTSL label in MTSL-K11C from the phosphate group was calculated as shown in Figure 4b (more positive values indicate deeper membrane insertion into the bilayer). From these results, we conclude that MARCKS-ED inserts more deeply into highly curved membrane bilayers.
Figure 4.
Plot showing the correlation between the depth parameters (φ) and different vesicle sizes. a) These results demonstrate that MARCKS-ED inserts more deeply in the membrane bilayer with more curvatures. b) A list of each synthetic lipid vesicle size describing its depth parameters (Φ) and distance from the membrane bilayer (Å).
3.4 Does Lipid Composition Influence Curvature Sensing?
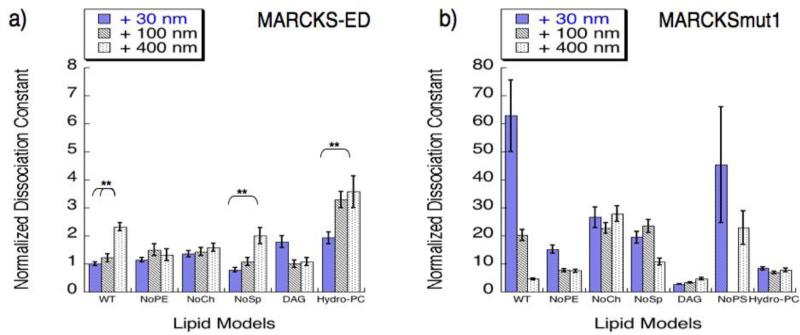
To explore the role of lipids in MARCKS-ED curvature sensing behavior, we measured its binding affinity towards vesicles with different lipid composition and sizes (Table 1). A mutant peptide, MARCKSmut1 (NH2-KKKKKRASAKKSAKLSGASAKKNKK-CONH2), in which the Phe residues in the original sequence of MARCKS-ED were replaced with Ala (underlined) was used as a control. The bolded amino acids are positively charged, so MARCKSmut1 can only use the electrostatic interactions for the binding. Figure 5 shows normalized dissociation constants (KD) in relation to vesicle size. In vesicles consisting of the wild type membrane model, MARCKS-ED binds to smaller vesicles with significantly higher affinity. We observed similar results in the vesicles lacking sphingomyelin, suggesting that sphingomyelin has no apparent contribution in MARCKS-ED's curvature sensing behavior. We previously showed that in the absence of phosphatidylserine (PS), the preferential binding of MARCKS-ED was compromised [7]. Herein, we find that phosphatidylethanolamine (PE), or cholesterol (Figure 5a, NoPE and NoCh, respectively) are also required for MARCKS-ED's curvature sensing. Across all these lipid systems, MARCKSmut1 does not show any size discrimination (Figure 5b), further confirming the critical role of the Phe residues.
Figure 5.
Membrane curvature sensing by a) MARCKS-ED and b) MARCKSmut1 in vesicles with different lipid composition. KD values were determined by fluorescence anisotropy assays for each lipid vesicle size and composition (Table S2). For direct comparison, all KD values were normalized to that for wild type 30 nm vesicle by MARCKS-ED. *P value < 0.01 compared to vesicles generated by 400 nm polycarbonate membrane pores. **P value < 0.05 compared to vesicles generated by 400 nm polycarbonate membrane pores, indicating curvature sensing behavior. Statistical analysis was performed using the Analysis of variance (ANOVA) test to produce a p-value between samples measured.
To further understand why both PE and cholesterol are critical to MARCKS-ED's curvature sensing behavior, we chose to replace each of these lipids with another lipid component having similar structural characteristics to determine whether any rescue of curvature sensing behavior could be observed. First, we replaced PE with diacyl glycerol (DAG), a lipid sharing a common conical shape with PE [14]. The removal of PE from wild type vesicles eliminated vesicle size distinction (Figure 5a, NoPE). Upon replacement of the PE with DAG (Figure 5a, DAG), no significant rescue of curvature sensing was observed. MARCKSmut1 measured stronger binding affinities for the DAG mutant model relative to all the other lipid models. Although smaller KD values were observed, there is only a slight difference between the 30- and 400-nm vesicle solutions.
Cholesterol is known to increase membrane rigidity in fluid phases [40, 41]. Figure 5a shows that the curvature sensing behavior by MARCKS-ED is abolished in the absence of cholesterol. To explore cholesterol's contribution in the context of MARCKS-ED targeting highly curved vesicles, we replaced cholesterol with hydrogenated-phosphatidylcholine (Hydro-PC) to probe the rigidity-enhancing role. With fully saturated lipid tails, Hydro-PC can enhance lipid packing, thereby increasing membrane rigidity [42]. As shown in Figure 5a (Hydro-PC), a rescue of curvature sensing was observed with the replacement of cholesterol by Hydro-PC, indicating that membrane rigidity indeed contributes to MARCKS-ED's differential binding to vesicles of different sizes. In comparison to the wild type MARCKS-ED peptide, MARCKSmut1 showed no curvature sensing for either mutant model.
4 DISCUSSION
In this current report, the roles of both the peptide and the lipids were explored to understand MARCKS-ED's curvature sensing behavior. Molecular modeling shows the central stretch of MARCKS-ED inserted into the membrane despite Lys residues that pull both termini towards the bulk solvent forming a ‘boat’ conformation, which was previously hinted by Cafiso and co-workers [18]. The preferred position we observed for Phe (~± 12 Å) is also in agreement with previous simulations of individual side-chain analogs by MacCallum and co-workers [19]. However, in the same cited work, a position of ~14 Å for Ser, was obtained. This position is closer to the bilayer compared to our simulation results presented in this study (~± 18 Å). This difference may be attributed to the fact that the presence of a number of Lys residues in the MARCKS-ED pulls the whole peptide towards the bulk solvent. Nonetheless, the observation that the Phe residues stay buried in the membrane despite the pull lends further support to the essential role of these Phe residues in membrane curvature sensing by MARCKS-ED.
MARCKS-ED adopts an extended, unstructured conformation in solution and its interaction with the membrane is not expected to be dependent on the L- or D-conformation of the comprising amino acid residues. D-MARCKS-ED behaves similarly to L-MARCKS-ED as expected in being able to distinguish between different curvatures of liposomes with 10% and 20% PS. To look more closely at the insertion of MARCKS-ED into the bilayer, we employed EPR, a well-established method to study membrane protein insertion [13]. Consistent with our speculation that the insertion of Phe residues are critical to the preferential binding of MARCKS-ED to highly curved membranes, we observed a trend that shows a deeper insertion of Phe as the lipid vesicle gets smaller. The EPR results are comparable to our observations from simulations. For the largest vesicle, it can be seen from Figure 4 that the depth of the peptide is ~5 Å away from the phosphate region. Figure 2 shows that there is still a considerable population of Lysine residues at that distance from the phosphate region.
Our results suggest that different lipid shapes and packing have major impacts on the interaction of the bilayer with MARCKS-ED. PE, with its small head group, can create surface defects and lipid gaps on the surface for larger vesicle sizes. However, to produce surface defects in smaller vesicles, the presence of defect-inducing lipids is not as critical as they are in larger vesicles. This is due to the fact that, for smaller vesicles, the higher surface curvature itself induces defects. As shown in Figure 5a, the removal of POPE did result to a significant change in MARCKS-ED binding to the 30- and 100-nm. The presence of POPE can affect the distribution of POPS on the surface of bilayers [43], which could be a reason for the stronger binding of MARCKS-ED to the 400-nm POPE-deficient vesicles. More specifically, this perhaps suggests that POPS membrane surface localization could be disrupted in larger vesicles in the presence of POPE, thus interfering with electrostatic interactions with a result of weaker binding. Without POPE, it could be inferred that POPS is more localized thus the result of stronger binding between MARCKS-ED and larger vesicles. Next, we tested whether DAG could rescue the curvature sensing by MARCKS-ED in the absence of POPE due to their similar shapes. However, the replacement of POPE with DAG shows no significant rescue of curvature sensing, which reveals that shape alone could not fully explain the essential role of POPE in MARCKS ED's curvature sensing. This could be due to the lack of negatively charged phosphate group in the DAG head group that makes its behavior markedly different from that of POPE. Taken together, curvature sensing behavior was not observed for either peptide with this mutant model, indicating the essential role that POPE plays in MARCKS-ED's curvature sensing.
There are many factors that could affect lipid packing such as temperature, lipid composition and the compression brought upon by inherent bilayer curvature; therefore, the specific role of cholesterol as a modulator of lipid packing should be investigated. Indeed we acknowledge cholesterol's dynamic behavior in generating ordered lipid domains in membrane environments, which may influence curvature sensing. We can only propose this suggestion taking in consideration the multiple functions of cholesterol in membranes. In fact, its depletion has been suggested to disrupt these domains, currently recognized as lipid rafts [44]. Furthermore, in the context of highly curved vesicles, cholesterol appears to contribute to MARCKS-ED's curvature sensing behavior, proposing how lipid mobility and structure may influence this behavior.
The generation and maintenance of surface defects, which arise from lipid packing, appear to influence curvature sensing by MARCKS-ED. We explored the membrane rigidity effects following our observation that curvature sensing behavior was eliminated upon the replacement of cholesterol by POPC. Likewise, the binding of MARCKS-ED to the 30- and 100-nm vesicles did not show an appreciable response to the removal of cholesterol but resulted in tighter binding to the 400-nm. These results suggest that any change that cholesterol brings to membrane rigidity has more effect on the largest vesicle than on the two smaller ones. In larger vesicles, cholesterol may add more rigidity to both stabilize surface defects and increase lipid constraints to allow for better bilayer insertion. On the other hand, the high degree of curvature present in the smaller vesicles can bring about such a degree of lipid packing that the effect of cholesterol becomes unnoticeable. That is, even with the removal of cholesterol, the rigidity of the smaller bilayers is not impacted greatly because of the lipid packing that occurs inherently due to curvature. This may also suggest why the wild type model shows weaker binding for larger vesicles. In contrast to their deletion models, cholesterol and PE could be influencing membrane rigidity and lipid surface defects, which may interfere with electrostatic interactions of larger vesicles. Given previous studies showing PE's influence on PS membrane exposure [43], this may explain how MARCKS-ED selectively targets highly curved vesicles with stronger binding affinity in their absence.
The binding affinities revealed by the replacement of cholesterol with Hydro-PC (rigidity-enhancing lipid) points to the role of lipid packing in membrane bilayers on the curvature sensing behavior of MARCKS-ED. The replacement of cholesterol with Hydro-PC successfully rescued the curvature sensing ability MARCKS-ED. These studies suggest that rigidity-altering lipid components may be a significant factor in influencing curvature sensing. Furthermore, lipid packing may also be a contributing factor. As previous curvature sensing studies have shown, we acknowledge that curvature sensing observed by MARCKS-ED may indeed be influenced by the inherent behavior of highly curved membranes [45]. Our data indeed reveals the importance of the insertion-inducing residue Phe as well as lipid packing on curvature sensing; however, these observations only help support an additional influence: the impact of surface defects, highlighting the significant role in lipid-lipid interactions which may be influencing the peptide-lipid interactions we have observed.
Results observed for the mutant peptide MARCKSmut1 displayed weaker binding compared to the wild type MARCKS-ED peptide, and importantly without membrane curvature distinction. These results further confirmed the importance of Phe insertion in both MARCKS-ED membrane binding and curvature sensing.
5 CONCLUSIONS
Previous and current membrane curvature studies have introduced a variety of mechanisms to illustrate dynamic protein-membrane interactions; however, there are still unresolved questions to fully characterize how proteins are promoted to highly curved membranes. Our current studies have presented new insights to further progress the field of membrane curvature. Previously, certain studies have thoroughly recognized specific protein domains (e.g. BAR, scaffolding etc) as being a contributor to membrane curvature sensing based on Baumgart et al [46]. Hatzakis and coworkers have acknowledged how highly curved membranes themselves recruit proteins with insertion motifs [45]. Furthermore, lipid composition has also been shown to influence how proteins target highly curved vesicles based on Antonny et al [47]; however, similar to our own findings, they suggested how insertion motifs in conjunction to lipid defects initiate curvature sensing. Taken together, our work reveals multiple influences observed by those in the field, all contributing to membrane curvature sensing, suggesting the complexity of this behavior. These include peptide specificity, the presence of defect-inducing lipids as well as lipid rigidity effects stemming from cholesterol.
Using both computational and experimental biophysical techniques, we have probed the mechanism underlying the curvature sensing ability of MARCKS-ED. This was done by examining alterations in curvature sensing brought about by changes in both the peptide and the bilayers. The results of this study contribute to the molecular mechanism that governs the interaction between proteins and curved membranes. This understanding will positively impact the identification and rational design of novel proteins and peptides that, in their ability to target highly curved vesicles, can be used for various biological applications.
Supplementary Material
Highlights.
We investigated the molecular mechanism of the curvature sensing behaviors of MARCKS ED.
The curvature sensing by MARCKS-ED is independent of the L/D-configuration of the amino acids.
Phe residues insert more deeply into the highly curved membrane, driving its preferential binding.
The curvature sensing behaviors are affected by the membrane rigidity, fluidity and the lipid components.
ACKNOWLEDGMENTS
We thank Dr. Annette Erbse and Dr. Joseph Falke for thoughtful discussion of the EPR analysis. This work utilized the Janus supercomputer, which is supported by the National Science Foundation (award number CNS-0821794), the University of Colorado Boulder, the University of Colorado Denver, and the National Center for Atmospheric Research. The Janus supercomputer is operated by the University of Colorado Boulder. We also acknowledge the use of the Vieques cluster at the Biofrontiers Institute of the University of Colorado Boulder.
Funding Sources
We thank the National Institutes of Health (R01 GM103843) for financial support. L.A.M. is supported by a National Institute of Health Ruth L. Kirschtein National Research Service Award (F31 CA165349).
ABBREVIATIONS
- EPR
electron paramagnetic resonance
- EVs
extracellular vesicles
- MARCKS
myristoylated alanine-rich C-kinase subtrate
- MTSL
methanethiosulfonate
- NBD
6-(N- (7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoic acid
- NTA
nanoparticle tracking analysis
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information. EPR spectra of MARCKS-ED with lipid vesicles, EPR calibration plots, Table of anisotropy values for each lipid model, Time course experiments of lipid vesicles with and without MARCKS-ED peptide, Anisotropy assays of D-MARCKS-ED. This material is available free of charge via the internet at http://www.sciencedirect.com/science/journal/00052736
The authors declare no competing financial interest.
REFERENCES
- 1.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 2.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 3.Gerlach H, Laumann V, Martens S, Becker CF, Goody RS, Geyer M. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol. 2010;6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 4.Arkhipov A, Yin Y, Schulten K. Membrane-bending mechanism of amphiphysin N-BAR domains. Biophys J. 2009;97:2727–2735. doi: 10.1016/j.bpj.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 7.Morton LA, Yang H, Saludes JP, Fiorini Z, Beninson L, Chapman ER, Fleshner M, Xue D, Yin H. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem Biol. 2013;8:218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J Biol Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- 9.Ouimet CC, Wang JK, Walaas SI, Albert KA, Greengard P. Localization of the MARCKS (87 kDa) protein, a major specific substrate for protein kinase C, in rat brain. J Neurosci. 1990;10:1683–1698. doi: 10.1523/JNEUROSCI.10-05-01683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Blackshear PJ, Johnson JD, McLaughlin S. Phosphorylation reverses the membrane association of peptides that correspond to the basic domains of MARCKS and neuromodulin. Biophys J. 1994;67:227–237. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JY, Shishido T, Jiang XL, Aderem A, Mclaughlin S. Phosphorylation, High Ionic-Strength, and Calmodulin Reverse the Binding of Marcks to Phospholipid-Vesicles. J Biol Chem. 1994;269:28214–28219. [PubMed] [Google Scholar]
- 12.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin Z, Cafiso DS. Membrane structure of protein kinase C and calmodulin binding domain of myristoylated alanine rich C kinase substrate determined by site-directed spin labeling. Biochemistry. 1996;35:2917–2925. doi: 10.1021/bi9521452. [DOI] [PubMed] [Google Scholar]
- 14.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 16.Shemesh T, Luini A, Malhotra V, Burger KN, Kozlov MM. Prefission constriction of Golgi tubular carriers driven by local lipid metabolism: a theoretical model. Biophys J. 2003;85:3813–3827. doi: 10.1016/S0006-3495(03)74796-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellena JF, Burnitz MC, Cafiso DS. Location of the myristoylated alanine-rich C-kinase substrate (MARCKS) effector domain in negatively charged phospholipid bicelles. Biophys J. 2003;85:2442–2448. doi: 10.1016/s0006-3495(03)74667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacCallum JL, Bennett WFD, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. Journal of Computational Chemistry. 1983;4:187–217. [Google Scholar]
- 21.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr., Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackerell AD, Jr., Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 23.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 25.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 26.Darden TA, Pedersen LG. Molecular modeling: an experimental tool. Environ Health Perspect. 1993;101:410–412. doi: 10.1289/ehp.93101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 28.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. Journal of Chemical Physics. 1995;103:4613–4621. [Google Scholar]
- 29.Li LB, Vorobyov I, Allen TW. Potential of mean force and pK(a) profile calculation for a lipid membrane-exposed arginine side chain. J Phys Chem B. 2008;112:9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 30.Dorairaj S, Allen TW. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. P Natl Acad Sci USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen TW, Andersen OS, Roux B. Structure of gramicidin A in a lipid bilayer environment determined using molecular dynamics simulations and solid-state NMR data. J Am Chem Soc. 2003;125:9868–9877. doi: 10.1021/ja029317k. [DOI] [PubMed] [Google Scholar]
- 32.de Jesus AJ, Allen TW. The determinants of hydrophobic mismatch response for transmembrane helices. Biochimica et biophysica acta. 2013;1828:851–863. doi: 10.1016/j.bbamem.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Connor J, Bucana C, Fidler IJ, Schroit AJ. Differentiation-dependent expression of phosphatidylserine in mammalian plasma membranes: quantitative assessment of outer-leaflet lipid by prothrombinase complex formation. Proc Natl Acad Sci U S A. 1989;86:3184–3188. doi: 10.1073/pnas.86.9.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith SA, Morrissey JH. Rapid and efficient incorporation of tissue factor into liposomes. J Thromb Haemost. 2004;2:1155–1162. doi: 10.1111/j.1538-7836.2004.00772.x. [DOI] [PubMed] [Google Scholar]
- 35.Morton LA, Saludes JP, Yin H. Constant Pressure-controlled Extrusion Method for the Preparation of Nano-sized Lipid Vesicles. J Vis Exp. 2012 doi: 10.3791/4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arbuzova A, Wang L, Wang J, Hangyas-Mihalyne G, Murray D, Honig B, McLaughlin S. Membrane binding of peptides containing both basic and aromatic residues. Experimental studies with peptides corresponding to the scaffolding region of caveolin and the effector region of MARCKS. Biochemistry. 2000;39:10330–10339. doi: 10.1021/bi001039j. [DOI] [PubMed] [Google Scholar]
- 38.Zhang LL, Yethiraj A, Cui Q. Free Energy Calculations for the Peripheral Binding of Proteins/Peptides to an Anionic Membrane. 1. Implicit Membrane Models. J Chem Theory Comput. 2014;10:2845–2859. doi: 10.1021/ct500218p. [DOI] [PubMed] [Google Scholar]
- 39.Arbuzova A, Schmitz AAP, Vergeres G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubenstein JL, Smith BA, McConnell HM. Lateral diffusion in binary mixtures of cholesterol and phosphatidylcholines. Proc Natl Acad Sci U S A. 1979;76:15–18. doi: 10.1073/pnas.76.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofsass C, Lindahl E, Edholm O. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys J. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J, Pipe SW, Rasmussen JT, Heegaard CW, Gilbert GE. Lactadherin blocks thrombosis and hemostasis in vivo: correlation with platelet phosphatidylserine exposure. J Thromb Haemost. 2008;6:1167–1174. doi: 10.1111/j.1538-7836.2008.03010.x. [DOI] [PubMed] [Google Scholar]
- 44.Kabouridis PS, Janzen J, Magee AL, Ley SC. Cholesterol depletion disrupts lipid rafts and modulates the activity of multiple signaling pathways in T lymphocytes. Eur J Immunol. 2000;30:954–963. doi: 10.1002/1521-4141(200003)30:3<954::AID-IMMU954>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 45.Nikos VKB, Hatzakis S, Larsen Jannik, Madsen Kenneth L, Bolinger Pierre-Yves, Kunding Andreas H, Castillo John, Gether Ulrik, Hedegard Per, Stamou Dimitrios. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature Chemical Biology. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 46.Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.