Abstract
The FDA Adverse Event Reporting System (FAERS) is a database for post-marketing drug safety monitoring and influences FDA safety guidance documents, such as changes in drug labels. The number of cases in the FAERS has rapidly increased with the improvement of submission methods and data standard and thus has become an important resource for regulatory science. While the FAERS has been predominantly used for safety signal detection, this study explored its utility for disease monitoring.
Publicly available health-care information has grown dramatically with recent advances in computing, internet, and database technologies. With the large amounts of newly available medical data from diverse sources, we can now approach public-health issues in ways not previously possible. Electronic medical records (EMRs), clinical studies, and epidemiological studies remain the fundamental sources of information for disease monitoring. Intelligently integrating the wealth of health-related data to address current biomedical challenges has gained momentum to improve service delivery and public health. In addition, mining public data from PubMed, FAERS, and FDA drug labels represents a new venue for health surveillance. Applying data-mining approaches to these public-health databases provides unique information that will 1) improve health-service delivery by identifying new trends in the prevalence of diseases and adverse events (AEs), 2) guide the development of expensive epidemiological studies, and 3) identify new opportunities in translational medicine and regulatory science.
The FAERS is a database that supports the FDA's post-marketing drug-safety monitoring efforts [1]. The database contains valuable information about AEs, medication errors, patient demographics, and more. Since its inception, millions’ cases have been reported to the FAERS by manufacturers, health-care professionals, and consumers. Most data-mining efforts to date have used information from the FAERS for pharmacovigilance, such as drug-safety signal detection, drug-drug interaction identification, and idiosyncratic adverse drug-reaction. However, the potential for the database to be used as a disease surveillance tool had not yet been explored. We hypothesize that the disease information embedded in the FAERS can be translated into signals indicating the disease prevalence in a population. This was demonstrated by analyzing >4 million cases in the FAERS between 1997 and 2011 to assess diseases showing sex difference (Figure 1). We identified 115 diseases exhibiting a significantly biased prevalence between sexes. Almost half of these sex-biased diseases can be confirmed with literature data. By examining eight diseases using the patient data from Marshfield Clinic’s EMRs, we found that the sex-biased prevalence for each disease was consistent across all three sources (i.e., the FAERS, literature report, and EMRs) (Figure 2), implying that the FAERS could be a potential resource for disease monitoring.
Figure 1.
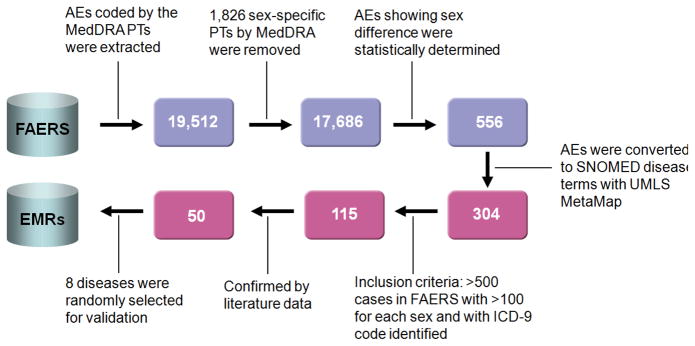
Flowchart of study on FDA Adverse Event Reporting System (FAERS) for disease monitoring. Blue boxes show the number of AEs (top row) and red boxes (bottom row) show the number of diseases.
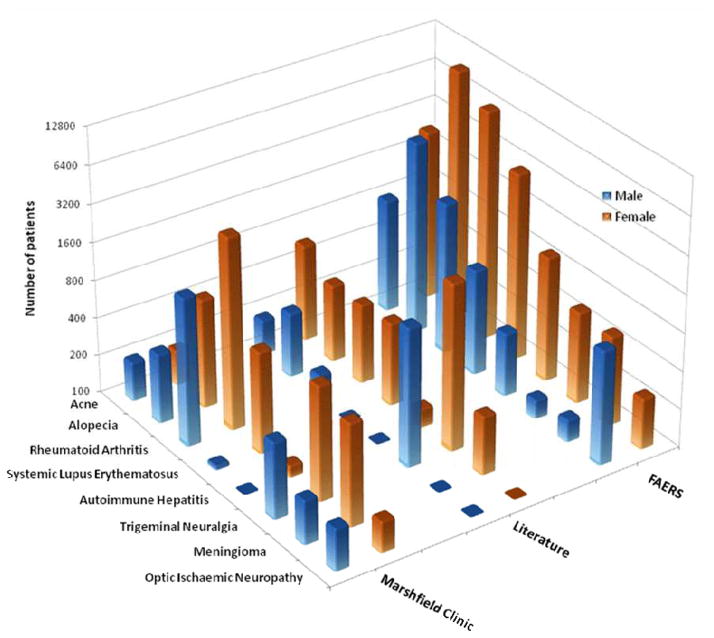
Figure 2.
Comparison of eight sex-based diseases (acne, alopecia, rheumatoid arthritis, lupus, autoimmune hepatitis, optic ischemic neuropathy, trigeminal neuralgia, and meningioma) found in FAERS, Marshfield Clinic, and population studies from publications.
Study Design and Results
As depicted in Figure 1, the study can be divided into two parts: AEs-centric (top row) and disease-centric (bottom row) analysis. Four ontology-based standards and tools were applied for data manipulation and conversion, which include Medical Dictionary for Regulatory Activities (MedDRA) for AEs, Systematized Nomenclature Of Medicine (SNOMED) for clinical terms, International Classification of Diseases book 9 (ICD-9) for diseases, and Unified Medical Language System (UMLS) for ontology mapping. Specifically, a total of 19,512 AE terms coded by MedDRA preferred terms (PT) in the FAERS was identified. We excluded these terms (1,826 PTs) specified by MedDRA as sex-related PTs. Of the 17,686 AEs that remained, 556 exhibited statistically significant differences between sexes with p-value < 10−10 and at least a two-fold difference between sexes. To interpret the context of terms with respect to clinical application, the 556 AE terms were mapped to the SNOMED clinical term using the UMLS MetaMap [2]. This resulted in the identification of 304 sex-biased clinical terms (Supplementary Table 1 and Supplementary Materials and Methods). Using ICD-9 code, 115 sex-biased diseases (Supplementary Table 2 and Supplementary Materials and Methods) met the inclusion criteria (at least 500 total FAERS case reports and >100 cases for each sex).
Of the 115 diseases, 53 had literature reports (Supplementary Table 3 and Supplementary Materials and Methods) and 50 of those showed the sex-biased effect consistent in fold changes and direction with a 94.34% concordance with the FAERS data. To further confirm the literature reports, we selected eight diseases to be investigated using Marshfield Clinic EMRs (Supplementary Table 4 and Supplementary Materials and Methods). The results confirmed the findings for seven of the eight diseases (alopecia, rheumatoid arthritis, lupus, autoimmune hepatitis, optic ischemic neuropathy, trigeminal neuralgia, and meningioma). The eighth disease, acne, has a controversial report regarding sex-differences [3] (Figure 2). Two important observations were made from this analysis: (1) three known sex-biased autoimmune diseases (rheumatoid arthritis, lupus, and autoimmune hepatitis) were successfully identified by our FAERS-based approach and confirmed by both publication and the EMRs, demonstrating the potential utility of the FAERS for disease monitoring [4]; and (2) it appears that sex-biased diseases are organ-independent as evident that these diseases are associate with various organs (i.e., alopecia for hair, trigeminal neuralgia for nerve, acne for skin, optic ischemic neuropathy for eye, and meningioma for brain).
Closing Thoughts and Perspective
Our study demonstrated that the FAERS has the utility to identify sex-biased diseases previously unstudied or unreported, and the methodology could be extended to investigate other risk factors, such as age, geographical location, and ethnicity. This optimistic view is encouraged by the following observations/facts. Firstly, the FAERS-based results were consistent with the literature reports and EMRs data although the cases studied were limited. Secondly, using the FAERS data before 2011, we were able to identify optic ischemic neuropathy as a potential sex-biased disease which was only reported in 2012 [5]. Thirdly, we noticed that there was a much larger patient count in the FAERS reports compared to published reports for the diseases investigated, indicating that the FAERS could be a better and rich resource for disease monitoring. Lastly, unlike most EMRs, the FAERS are publicly available which could create a better environment than EMRs for developing innovative methodologies, thus improving the disease monitoring strategies.
The number of cases in the FAERS has rapidly increased in the past 15 years and will grow even more rapidly with the improvement of submission methods and data standards, and thus become a “Big Data” challenge. Some of these challenges involve data manipulation, association and conversion, which could be minimized with standardization and ontology. Here, we applied several ontology tools to investigate disease terms in FAERS coded in MedDRA terminologies. Our results demonstrate the power of using existing standard vocabularies to achieve information exchange and integration.
Several aspects of our study require further consideration. The current FAERS data may not reflect the ‘true’ prevalence of disease because the population observed for drug exposure and disease prevalence is restricted to patients who report AEs. Under-reporting and incomplete information in FAERS contributed to a lesser extent. These shortcomings might be minimized as standards for data submission improve, the number of case reports increases, statistical approaches advance, and other relevant databases for cross-referencing and validation are integrated into the analysis. It is worthwhile to point out that the indication field in the FAERS database could also be used for the source of disease prevalence information. Though potentially valuable, approximately 36% of the data in the indication field is missing —primarily data from older reports. Furthermore, one may argue that using the AE field might result in analysis of drug-induced effects rather than the specified disease. Nevertheless, as a proof-of-concept study, we explore the FDA FAERS to generate hypotheses for disease monitoring.
The data-integration example presented herein may serve as an approach for utilizing health information and accelerating global disease monitoring. A variety of publicly available databases exist for disease surveillance. Some of these data sources include 1) CDC’s National Institute for Occupational Safety and Health, 2) CDC’s National Notifiable Diseases Surveillance Systems, 3) WHO’s Global Atlas of the Health Workforce, 4) WHO’s Global Health Observatory, 5) VigiBase™, 6) National Organization for Rare Disease, 7) eHealthMe, and others. In addition, private medical records at clinics, hospitals, health-care providers, and insurances are potential resources for data mining as well. Integrating these data sources is the future of disease monitoring, and may have the power to uncover hidden relationships and knowledge at minimal cost. In summary, we aim to integrate the rich information in FAERS, EMRs, publications, and other public-health databases, and translate that information to advance medical research and regulatory science.
Supplementary Material
Acknowledgments
The project described was supported, in part, by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR000427 (AM and SML). The authors thank Crystal Jacobson on data query using the Marshfield Clinic Research Data Warehouse. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Disclaimer: The views presented in this article do not necessarily reflect current or future opinion or policy of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification and not intended as endorsement.
References
- 1.Pratt LA, Danese PN. More eyeballs on AERS. Nature Biotechnology. 2009;27:601–602. doi: 10.1038/nbt0709-601. [DOI] [PubMed] [Google Scholar]
- 2.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schafer T, Nienhaus A, Vieluf D, Berger J, Ring J. Epidemiology of acne in the general population: the risk of smoking. British Journal of Dermatology. 2001;145:100–104. doi: 10.1046/j.1365-2133.2001.04290.x. [DOI] [PubMed] [Google Scholar]
- 4.Whitacre CC. Sex differences in autoimmune disease. Nature Immunology. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 5.The Postoperative Visual Loss Study, G. Risk Factors Associated with Ischemic Optic Neuropathy after Spinal Fusion Surgery. Anesthesiology. 2012;116:15–24. doi: 10.1097/ALN.1090b1013e31823d31012a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.