Abstract
Despite their apparently good prognosis ~15% of high hyperdiploid (HD) childhood acute lymphoblastic leukemia (ALL) cases relapse. To search for responsible risk factors we determined copy number aberrations as well as copy neutral loss of heterozygosity (LOH) in 13 matched diagnosis and relapse samples and added the data of the only three available cases from the literature. Deletions and copy neutral LOH in 3 and 2 of the 16 cases directed us to the histone-modifying CREB-binding protein (CREBBP) gene, whose functional impairment is implicated in drug resistance. We therefore screened all samples for mutations in this gene and discovered 9 acquired sequence mutations in 7/16 cases, leading to an overall frequency of somatic CREBBP aberrations in HD ALL relapse cases of 63% that is considerably higher than that of the reported, mainly non-HD ALL (18.3%). Moreover, mutations in HD cases occur almost exclusively in the HAT domain (8/9; 89%). Hot spot mutations are present at diagnosis in 18.8% of relapsing HD ALL cases but in none of 40 respective cases remaining in long-term remission. Thus, the particular high incidence of CREBBP mutations in relapse-prone HD ALL cases could eventually be exploited for refined risk stratification and customized treatment in this genetic subgroup.
Keywords: childhood ALL, relapse leukemia, high hyperdiploid karyotype, CREBBP, SNP
INTRODUCTION
A high hyperdiploid karyotype (HD) defines the largest subgroup of childhood B-cell precursor acute lymphoblastic leukemia (ALL) and is characterized by a modal number of 51–68 gained chromosomes.1,2 Circumstantial evidence indicates that the mal-distribution of chromosomes occurs simultaneously during a single abnormal non-disjunction event. Alternative mechanisms, such as a duplication of an initial near-haploid chromosome set, the sequential gain of chromosomes during several cell divisions and the consecutive loss of chromosomes after a tetraploidization step, probably also exist but are much rarer.(reviewed in1) HD is already acquired in utero and, apart from non-random tri- and tetrasomies of specific chromosomes,3-5 contains hardly any recurrent structural abnormalities.6 Notwithstanding the fact that HD is an overall favorable prognostic feature, up to 15% of patients with this type of leukemia relapse and have a dismal outlook.1,7,8 Currently, no parameters exist that could help to predict which HD cases will eventually relapse. Although minimal residual disease (MRD) monitoring has significantly refined risk assessment in BFM-based treatment protocols, it is not sufficient to delineate relapse-prone HD cases, because most of them do not fall into the MRD high-risk group.9 The search for novel stratifying features aims to eventually refine treatment and is therefore highly warranted particularly in this specific subgroup. The most appropriate approaches to search for such prognostic markers include the genome-wide comparison of paired diagnostic and relapse samples with single nucleotide polymorphism (SNP) arrays to identify potentially relevant acquired copy number aberrations (CNA) as well as mutation screening of candidate genes.10,11
So far, SNP array analyses of HD ALL cases uncovered that microdeletions are more common at relapse and some, such as IKFZ1, PAX5, CDKN2A/B and AK3, are even recurrent; none of them, however, has so far been linked to a propensity to relapse.10–12 Mutation screening, on the other hand, revealed that the typical secondary mutations in receptor tyrosine kinase/RAS pathway genes do not seem to influence the disease course adversely.10 The only promising recent discovery, which eventually became also the focus of our analysis, concerns the potential role of the CREB-binding protein (CREBBP, also known as CBP). Germline mutations in this gene result in Rubinstein–Taybi Syndrome, a developmental disorder that is characterized by dysmorphic features, intellectual impairment and a susceptibility to developing solid tumors.13 Recently, somatic CREBBP mutations with potential prognostic relevance were identified in lymphoid malignancies.14–16 Mutations in this transcriptional coactivator/histone acetyl transferase/E3 ubiquitin ligase gene lead to haploinsufficiency and may render affected cells therapy resistant.14–16 The few data available to date in childhood ALL cases indicate that relapse-associated CREBBP mutations are present in about 18% of relapse cases with an apparent equal likelihood in all genetic subgroups. The analyzed cohorts, however, contained only three HD ALL cases.15
In our pursuit to identify novel risk factors in HD ALL cases we originally compared SNP array patterns of 13 matched diagnosis and relapse HD ALL samples. Amongst the observed CNA and uniparental isodisomies/trisomies (UPID/T), those involving chromosome 16 caught our special attention, because such abnormalities are rare in HD cases and the only potentially relevant gene present in the respective minimal overlapping region is CREBBP. We therefore screened all samples for CREBBP mutations as well and included the only three available corresponding HD cases from the literature in our analysis.12,15
MATERIALS AND METHODS
Patient samples
This study comprises 16 paired diagnosis and relapse samples of HD B-cell precursor-ALL (Table 1). Eleven cases were from Austria and two from the Czech Republic. The SNP array data of three further patients with available remission samples and CREBBP sequence data were kindly provided by CG Mullighan, St Jude Children’s Research Hospital (Hyperdip50-SNP-#27, #53, #54). Cases from AT and CZ were selected on the basis of a chromosome number between 51 and 67 according to cytogenetic data at diagnosis and a blast count of >85% in diagnosis and relapse leukemic samples. Written informed consent from parents was obtained according to the declaration of Helsinki for inclusion of patients into this study. The ethical committees of the CCRI and St Anna Kinderspital approved this study. The Austrian patients were enrolled in the ALL-BFM 95 and ALL-BFM 2000 and the Czech patients in the ALL IC BFM 2002 and ALL interim 2007 treatment protocol for initial disease. All patients experienced a bone marrow relapse except for one patient (#4) who had two CNS relapses before the bone marrow relapse that was analyzed here. Median age of patients #1–13 was 6.7 years (range 3–13 years) and median remission duration was 44 months (range 12–97 months). Early relapse was defined by first remission duration of up to 30 months. For the determination of the constitutional genotype we used early remission bone marrow samples, which were available in all cases.
Table 1. Clinical characteristics of HD ALL relapse cases.
| Pt ID | Gender | Age at Dx (years) | MRD | Rem (months) | Rel site | MRD Rel | Outcome after Rel (months) | Source |
|---|---|---|---|---|---|---|---|---|
| 1 | m | 13 | IR | 12 | BM | 10−1 | SCTa | Austria |
| 2 | f | 6, 1 | IRb | 17 | BM | 10−1 | SCT, rem 18 | Czech Republic |
| 3 | m | 8, 9 | IR | 18 | BM | 10−1 | SCT, rela | Austria |
| 4 | m | 3 | IRb | 25 | BM | 3×10−3 | SCTa | Austria |
| 5 | f | 5, 7 | IR | 36 | BM | 10−2 | SCTa | Austria |
| 6 | f | 3, 9 | IR | 42 | BM | 10−1 | SCT, rem 52 | Czech Republic |
| 7 | f | 7, 3 | na | 44 | BM | <10−4 | rel, SCTa | Austria |
| 8 | f | 6, 9 | IRb | 49 | BM | 10−2 | SCTa | Austria |
| 9 | m | 5, 8 | na | 55 | BM | 10−2 | SCT, rem 98 | Austria |
| 10 | m | 4, 7 | LR | 55 | BM | <10−4 | SCT, rem 22 | Austria |
| 11c | f | 9, 8 | na | 61 | BM | <10−4 | rem 46 | Austria |
| 12 | m | 10, 7 | na | 88 | BM | 5×10−3 | SCT, rem 68 | Austria |
| 13 | m | 6, 7 | IR | 97 | BM | <10−4 | SCT, rem 34 | Austria |
| 14 | m | na | na | na | BM | na | na | St Judes #27 |
| 15 | m | na | na | na | BM | na | na | St Judes #53 |
| 16 | m | na | na | na | BM | na | na | St Judes #54 |
Abbreviations: ALL, acute lymphoblastic leukemia; BM, bone marrow; Dx, initial diagnosis; HD, hyperdiploid; IR, intermediate risk; LR, low risk; m/f, male/female; MRD, minimal residual disease risk classification by Ig/TCR rearrangements and risk assignment as previously reported;17,24 MRD rel, tumor load at day 36 of treatment, where a tumor load of equal or more than 10−3 has been established as a parameter for SCT;25 na, not available; Pt, patient; Rel, relapse; Rem, remission; SCT, hematopoietic stem cell transplantation.
Death.
Indicates patients with a slow early response and poor outcome.
Down’s syndrome.
To determine the frequency of CREBBP mutations in non-relapse HD ALL we screened 40 diagnostic samples from cases with a median remission duration of 114 months (range 84–144) for mutations in exons 25–27 of the HAT domain.
MRD analysis
MRD using immunoglobulin/T-cell receptor (TCR) rearrangements was performed as described previously.17
DNA extraction and sample preparation
DNA was extracted from leukemic and remission bone marrow MNCs according to the standard protocols as described previously.18
SNP array analyses
Affymetrix (Santa Clara, CA, USA) GeneChip Human mapping 500k arrays (#13), Genome-Wide Human SNP Array 5.0 (#5, 8, 9, 11, 12) and 6.0 (#1, 2, 3, 4, 6, 7, 10) arrays were used for the analyses. Array hybridization, quality control and genotyping were performed by Atlas Biolabs (Berlin, Germany). Files are available upon request.
Partek Genomic Suite software version 6.5 (Partek, St Louis, MO, USA) was used to analyze signal intensities and genotype calls. Diagnosis, remission and relapse samples of all cases were matched according to genotype data. We applied the genomic segmentation algorithm in paired analysis for the detection of copy number alterations in diagnosis and relapse compared with the respective remission sample essentially as described recently.18 Segmentation parameters were chosen as follows: 10 minimum genomic markers per segment (defining how small a region can be), a P-value threshold of 0.001 (controlling for outlying probes) and a signal-to-noise ratio of 0.8. Copy number changes detected by genomic segmentation were verified by comparison with the log2 ratio visualization in the Partek Genome Browser.
Deletions and duplications associated with immunoglobulin/TCR rearrangements at 2p11.2 (IGK), 7p14.1 (TCRG), 7q34 (TCRB), 14q11.2 (TCRD/A), 14q32.33 (IGH) and 22q11.22 (IGL) were excluded from the analyses. Microdeletions and focal deletions were defined by a size smaller than 10 Mb and 300 kb, respectively. Regions of UPID/T were analyzed by unpaired loss of heterozygosity (LOH) analysis with the HapMap270 reference file provided by Affymetrix in Partek, which uses a Hidden Markov Model (to find regions that are most likely to be loss events based on the genotype error and the expected heterozygous frequency at each SNP). To define copy neutral LOH, LOH data were compared with the copy number data in the specific regions. Regions of LOH in diagnosis and relapse samples were compared with remission samples to verify their somatic origin. Allele specific copy number analysis in Partek with genotypes from the remission sample (paired analysis) was used to confirm LOH and copy number data. Genomic positions of CNA are indicated according to the University of California, Santa Cruz Genome Browser (NCBI Build, Version 36.1; http://genome.ucsc.edu/cgi-bin/hgTracks).
Mutational analysis of CREBBP and JAK2
DNA from diagnosis, relapse and remission samples was used to analyze all exons and exon/intron junctions of the CREBBP gene by PCR amplification. Primers used for CREBBP screening are listed in Supplementary Table S1. Sequencing was performed by VBC Biotech Service GmbH (Vienna, Austria). Mutations were sequenced twice from different PCR reactions to verify the aberration. Remission samples were used to assess the origin (somatic versus constitutional) of the mutations.
All samples were screened for JAK2 mutations in exon 16 using published primers19 but did not show any mutation.
To test for potential functional consequences of CREBBP mutations, we used an in silico analysis based on a method that combines Grantham variation (which measures the degree of biochemical variation among amino acids found at a given position in the multiple sequence alignment) and Grantham deviation scores (which reflects the ‘biochemical distance’ of the mutant amino acid from the observed amino acid at a particular position), and Align-Grantham variation–Grantham deviation, to predict whether our non-synonymous CREBBP mutations affect protein function. For the alignment 11 orthologs were taken.
Statistical analysis
Statistical significance of frequencies of microdeletions between diagnosis and relapse was calculated using the paired t-test and the frequency of CREBBP mutations in relapse versus non-relapse cases was determined by the Fisher’s exact test. Probability values (P < 0.05) were considered statistically significant.
RESULTS
Patterns of chromosomal imbalances, CNA and UPID/T in paired diagnosis and relapse HD ALL samples
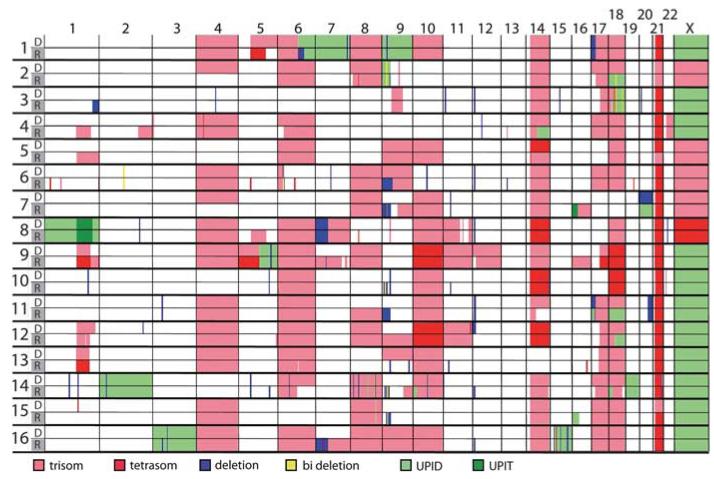
The overall CNA and UPID patterns obtained from our HD cases were similar to those described in previous studies (Figure 1; Supplementary Figure S1 and Table S2).6 The vast majority of tri- and tetrasomies were conserved at relapse, the partial or complete loss of originally trisomic chromosomes produced whole or partial chromosomal UPIDs in 10 cases (#1, 2, 4, 5, 6, 7, 11, 12, 13, 14; 62%) and whole or partial chromosomal gains affected primarily initial disomic chromosomes in 9 cases (#4, 5, 7, 8, 9, 11, 12, 14, 16; 56%; Figure 1; Supplementary Figure S1 and Table S2).
Figure 1. Genome-wide CNAs and LOH in HD ALL relapse cases.
CNAs and UPID/Ts in diagnosis and relapse samples. Copy number changes and UPID/Ts are mapped according to their chromosomal position and indicated by color for each sample (row) and each chromosome from 1 to X (column). The numbers at the beginning of each row refer to the patient’s ID. Sample types are indicated by the color bars: white for diagnosis (D) and gray for relapse (R). Color code indicating losses, gains and UPID/T at the bottom of the graph; bi deletion, biallelic deletion.
Acquired wUPID/T and pUPID/T were seen in 14 cases (78%). Four cases (#1, 8, 14, 16; 25%) had wUPIDs of chromosomes 1, 2, 3, 7, 9, 15 and 19, which do not belong to the group of non-random tri- and tetrasomies that usually constitute the HD karyotype (Figure 1). This pattern contradicts the notion that in these instances the HD karyotype evolved from a single non-disjunction event.1 The only conceivable alternative explanation is that these UPIDs are relicts of an originally tetraploid karyotype.1,6 Taking into account that 20% of unbalanced structural abnormalities in HD ALL are already present in a subclonal form at diagnosis, the higher rate of deletions and pUPIDs in our cases at relapse is most probably because of clonal selection rather than clonal evolution.6,20
In line with previous studies,10–12,21,22 microdeletions were generally rare at diagnosis with a mean frequency of 4 (range 0–13) per case. Their frequency became only slightly higher at relapse (mean 5; range 0–20; P<0.007; Supplementary Table S3). In addition to the deletions affecting the known ALL target genes CDKN2A/B, ETV6, PAX5, IKZF1 and NR3C16,18,21,22 (Table 2; Supplementary Table S3), we also found three 16p deletions (cases #6, 12, 16; Figure 2a; Supplementary Figures S1–3). This region contains the CREBBP gene, which was only recently identified to be recurrently mutated in lymphoid malignancies.14–16 As two other cases from our cohort also had a 16p pUPIT/D at relapse (#7, 15; Figure 1; Supplementary Figures S1 and S2), we considered this gene a valid target for our further investigations.
Table 2. Recurrent CNAs in 16 HD ALL cases at initial diagnosis and relapse.
| Patient ID | 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletions | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | |
| Cytoband | Genes in region | ||||||||||||||||||||||||||||||||
| 5q31.3 | NR3C1 | x | x | x | |||||||||||||||||||||||||||||
| 7p12.2 | IKZF1 | x | x | x | x | ||||||||||||||||||||||||||||
| 6p22.2 | Histone cluster | x | b | ||||||||||||||||||||||||||||||
| 6q21 | FYN | x | |||||||||||||||||||||||||||||||
| 9p13.3 | BAG1 | x | x | x | x | ||||||||||||||||||||||||||||
| 9p13.2 | PAX5 | x | x | x | x | x | x | x | |||||||||||||||||||||||||
| 9p21.3 | MLLT3 | b | b | x | x | x | x | x | |||||||||||||||||||||||||
| 9p21.3 | PTPLAD2 | b | b | x | x | x | x | x | b | ||||||||||||||||||||||||
| 9p21.3 | IFNA | b | b | x | x | x | x | x | b | ||||||||||||||||||||||||
| 9p21.3 | KLHL9 | b | b | x | x | b | x | x | x | ||||||||||||||||||||||||
| 9p21.3 | MTAP | b | b | x | x | b | x | b | b | ||||||||||||||||||||||||
| 9p21.3 | C9orf53 | b | b | x | b | b | x | b | b | ||||||||||||||||||||||||
| 9p21.3 | CDKN2A/B | b | b | x | b | b | x | b | b | ||||||||||||||||||||||||
| 9p23 | PTPRD | x | x | ||||||||||||||||||||||||||||||
| 9p24.1 | AK3 | x | x | x | |||||||||||||||||||||||||||||
| 10q21.2 | ARID5B | x | x | ||||||||||||||||||||||||||||||
| 11p12 | RAG1 | x | x | ||||||||||||||||||||||||||||||
| 11p12 | RAG2 | x | x | x | |||||||||||||||||||||||||||||
| 12q12 | ARID2 | x | x | ||||||||||||||||||||||||||||||
| 12p13.1 | ATF7IP | x | x | x | |||||||||||||||||||||||||||||
| 12p13.2 | BCL2L14 | x | x | x | |||||||||||||||||||||||||||||
| 12p13.2 | ETV6 | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||
| 13q12.2 | PAN3 | x | |||||||||||||||||||||||||||||||
| 20p12.2 | C20orf94 | x | x | ||||||||||||||||||||||||||||||
| 20q13.3 | BIRC7 | x | x | x | |||||||||||||||||||||||||||||
| 16p13.3 | CREBBP | x | x | x | |||||||||||||||||||||||||||||
Abbreviations: ALL, acute lymphoblastic leukemia; b, biallelic deletions; D, diagnosis; HD, hyperdiploid; R, relapse; x, monoallelic deletions. Genes listed in the table were selected based on their recurrences either in our analysis or in recently published articles including SNP array analyses.6,12,26
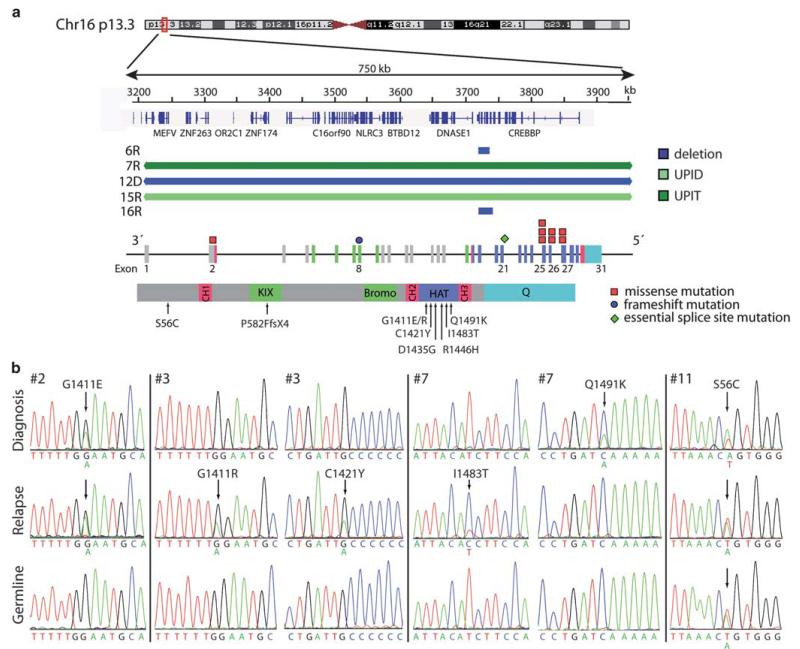
Figure 2. CREBBP aberrations in HD ALL relapse cases.
(a) Extent and localization of CREBBP aberrations. Top: genomic deletions including the CREBBP gene at 16p13.3. At the top a partial ideogram of the 16p13 region is shown. The scale indicates genomic positions of CREBBP and neighboring genes and the deletions and UPID/T in kilobases according to the University of California Santa Cruz Genome Browser (NCBI Build, Version 36.1). Each blue or green bar represents a heterozygous deletion or region of LOH in a single patient indicated as inferred by SNP array. Bottom: schematic diagram of the CREBBP gene and protein. The colors of the exons refer to the corresponding functional protein domains. The mutations in various exons are indicated by a rectangle for non-synonymous SNPs, circle for frameshift mutations and a diamond shape for essential splice site mutations. All somatic non-synonymous SNPs occur in the HAT domain. In the protein view the amino-acid changes are listed and the arrows indicate the location of the mutations (Bromo, bromodomain; CH, cysteine–histidine rich; HAT, histone acetyl transferase domain; KIX, CREB binding; Q, polyglutamine stretch). (b) Sequence traces labelled with nucleotides displaying the non-synonymous mutations in diagnosis and relapse samples compared with matched remission sample (constitutional control). Patient #3 harbored two mutations at relapse. The amino-acid substitution Q1491K found at diagnosis in #7 was lost at relapse. Instead, three uni-parental alleles together with an I1483T occurred. Patient #11 had a constitutional sequence change of unknown significance.
Sequence and deletion mutations of CREBBP at diagnosis and relapse
Mutation screening revealed altogether 9 somatically acquired CREBBP sequence mutations in 7 of 16 HD ALL cases. They comprised seven non-synonymous point mutations, one nonsense as well as one essential splice site mutation (Figures 1b and c). The latter mutation affects a splice site in exon 21 and is predicted to alter splicing in the HAT domain (Table 3). Of note, 8/9 (89%) mutations occurred in the HAT domain. Our in silico analyses indicate that eight mutations will alter the protein function but one is less likely to do so (Table 3). The only additional mutation outside the HAT domain most likely induces a truncating frame shift mutation in the KIX domain, which is involved in CREB binding. Consultation of the catalog of somatic mutations in cancer (http://www.sanger.ac.uk/genetics/CGP/cosmic/) revealed that the mutations we identified at positions D1435 and I1483 were recurrent but except for one, not identical to the ones described earlier.15,16,23 The constitutional exon 2 missense mutation in the interdomain region of the CREBBP gene in the patient with the Down’s Syndrome (#11), on the other hand, is neither predicted to cause a functional change (Table 3) nor did it cause any apparent Rubinstein–Taybi syndrome-associated phenotypic effects.13
Table 3. Sequence mutations of CREBBP in 16 HD ALL relapse cases.
| Sample ID | Exon | Nucleotide change | AA change | Affected domain |
|---|---|---|---|---|
| Non-synonymous SNPs | ||||
| 2D+R | 25 | c.4232G>A | G1411E | HAT |
| 3R | 25 | c.4231G>A | G1411R | HAT |
| 3R | 25 | c.4262G>A | C1421Ya | HAT |
| 7D | 27 | c.4471C>A | Q1491K | HAT |
| 7R | 27 | c.4448T>C | I1483T | HAT |
| 10R | 26 | c.4304A>G | D1435G | HAT |
| 11D+R+C | 2 | c.166A>T | S56Ca | Interdomain region |
| 15D+R | 26 | c.4337G>A | R1446Hb | HAT |
| Frameshift/splice site mutation | ||||
| 1D+R | 8 | c.1744_1745delCCinsTTT | P582PhefsX4 | KIX |
| 4R | 21 | c.3836+1G>A | splicing | HAT |
Abbreviations: AA, amino acid; ALL, acute lymphoblastic leukemia; D, diagnosis; HD, hyperdiploid; R, relapse. Reference sequence: NM_004380.2. Accession numbers of CREBBP mutations (NCBI dbSNP database http://www.ncbi.nlm.nih.gov/projects/SNP/): ID1, ss 479152796; ID2, 479152787; ID3 G1411R, 479152788; ID3 C1421Y, 479152789; ID4, 479152795; ID7 Q1491K, 479152790; ID7 I1483T, 479152791; ID10, 479152792; ID11, 479152793.
Mutation predicted to be less likely to affect protein function by Align-GVGD analysis.
Functionally relevant mutation reported by Mullighan et al.15
Overall, 12 distinct somatic CREBBP deletion and sequence mutations were found in 10/16 cases (63%; Table 4). Five of them were already present at diagnosis and five only at relapse. Two cases (#7, 12) lost the original mutation and one of them (#7) acquired a new one instead (Figure 2b). A UPIT/D at 16p was observed in two cases (#7, 15) at relapse (Supplementary Figures S1 and S2). Whether, in analogy to the UPID case (#15),15 the mutation-carrying allele was also duplicated or even triplicated in the case with the partial trisomy remains open, as it is not known whether the small wild-type sequence peak derives from the main mutation-carrying clone, a smaller variant subclone or from contaminating normal cells (Figures 1b and c; Supplementary Figure S2). Furthermore, case #3 had two distinct mutations at relapse (Figure 2b, Table 3).
Table 4. Summary of all genomic CREBBP aberrations in 16 HD ALL relapse cases.
| Patient ID | 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aberration | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R | D | R |
| Deletion | x | x | x | |||||||||||||||||||||||||||||
| UPID/UPIT | x | x | ||||||||||||||||||||||||||||||
| Mutation | f | f | x | x | a | s | b | b | x | c | c | x | d | |||||||||||||||||||
Abbreviations: a, two distinct mutations; ALL, acute lymphoblastic leukemia; b, different mutations at diagnosis and relapse; c, constitutional; CREBBP, CREB-binding protein; d, biallelic; D, diagnosis; f, frame shift; HD, hyperdiploid; R, relapse; s, splice site; UPID/UPIT, uniparental isodisomies/trisomies; x, presence of aberration.
Response to treatment and time to relapse of CREBBP mutated HD ALL cases
In all, 4 of 13 cases had an early relapse and all of them harbored a CREBBP mutation (Table 1) whereas only 4 of 9 cases with a late relapse carried mutations (including the case that lost the initial mutation).
Eight of nine cases with available MRD information were in the intermediate risk group and one (#10) in the low risk group (Table 1). Interestingly, 3/8 intermediate risk cases had a slow early molecular response (MRD at time point 1: equal or more than 10E-3, and MRD at time point 2: positive, but less than 10E-3), a response pattern that had recently been associated with an increased relapse propensity in the AIEOP/BFM protocols.9 Consequently, such patients are currently switched to the high-risk arm of BFM protocols. At relapse, the overall treatment response was rather poor, which precludes any further interpretation of the potential role of CREBBP mutations in this context.
CREBBP HAT domain mutations predominate in HD ALL relapse cases
To corroborate the potential relevance of CREBBP mutations in relapse-prone HD ALL relapse cases, we sequenced exons 25–27 of 40 HD ALL cases in long-term remission and found only one constitutional non-synonymous point mutation in exon 27 (c.4480C>A leading to AA change P1494T; NCBI dbSNP database accession number ss479152794) in a patient with an originally unclear syndrome. Thus, none of the 40 non-relapse cases versus 18.8% (3/16) of relapse HD ALL cases already have a somatic mutation in these 3 CREBBP exons at initial diagnosis (P = 0.02).
DISCUSSION
To search for genetic alterations that could eventually be exploited as predictive and prognostic markers, we compared genome-wide CNA and LOH of 16 matched diagnosis and relapse samples from childhood HD ALL cases. Of particular interest was the involvement of 16p in the form of copy number alterations as well as UPID in 5/16 cases. A similar SNP array study uncovered only two 16p deletions and no UPID, albeit in 74 mainly non-relapse HD ALL cases.6 This finding eventually led to the identification of CREBBP mutations as the potential culprit in altogether 10/16 cases (63%) in this cohort. The CREBBP mutations identified in our HD ALL cases are predicted to alter protein function and primarily affect the HAT domain (89%). In half of the cases these mutations were already present at diagnosis whereas in the other half they emerged only in relapse. All four early but only four of the remaining nine late relapses carried CREBBP mutations.
Another rather unique characteristic of HD ALL cases is that CREBBP mutations cluster especially in exons 25–27 of the HAT domain. This pattern differs considerably from their distribution in non-HD ALL (3/13; 23%) and is more similar to that in B-cell non-Hodgkin’s lymphoma cases where the majority of mutations occur in the HAT domain.15,16 In accordance with our findings, CREBBP sequence mutations outnumbered deletions by far and, as in the two former studies, did generally also not overlap. Moreover, the predominant monoallelic manifestation of the respective lesions further supports their proposed haploinsufficient tumor suppressor role in the leukemic process.16
Initial observations suggested that CREBBP mutations are a particular feature of high-risk ALL and that they occur in 18.3% of relapse cases.15 This notion also derived from the fact that only one CREBBP mutation–but no deletion–was identified in 170 non-relapsed ALL cases.15 Moreover, such mutations seem to occur with an equally low frequency in all genetic subgroups of high-risk childhood ALL.15 The results of our analyses, on the other hand, imply that CREBBP mutations are highly enriched in relapse-prone HD ALL cases. As virtually all relapses take place in HD ALL cases within the MRD intermediate risk group, we suggest that CREBBP mutations represent the first relevant delineating parameter that allows an early prediction of potential relapses in HD ALL cases.
CREBBP mutations are already present at diagnosis in half of our cases that experienced a relapse. This finding is in accordance with Mullighan et al.15 who showed that merely 7/15 ALL cases have the mutation in the major clone at initial presentation. The order of mutation appearance in two of our cases stresses the difficulty of predicting to which extent, if at all, a particular CREBBP mutation-carrying clone will eventually succeed to evolve into a major relapse clone. It seems that this selective process is significantly influenced and codetermined by several other intrinsic genetic and extrinsic (micro) environmental factors.15 A particular contribution of other common genetic lesions, such as CDKN2A/B, IKZF1, NR3C1, ETV6 and PAX5 in the evolution of such clones is conceivable, but far from clear. Given the heterogeneous appearance patterns of such mutations during the course of the disease and the profound overall drug resistance encountered in the respective disease recurrences, one cannot rely only on the presence of CREBBP mutation carrying clones to distinguish drug-resistant from drug-sensitive relapse cases. The hitherto already established wide-ranging deregulating effects of CREBBP mutations on gene expression and their likely contribution to the development of glucocorticoid resistance can nevertheless pave the way for a more targeted and individualized form of treatment of affected cases, for instance with already available HDAC inhibitors.15,16
In conclusion, the results of our investigation provide robust evidence that CREBBP gene alterations are particularly common in HD ALL relapse cases. Consequently, CREBBP mutations could serve as the first important parameter in this specific genetic subgroup to predict their likelihood to relapse. The profound clustering of CREBBP mutations in the HAT domain provides a comparatively simple, accessible screening target. Most importantly, however, the identification of such CREBBP mutations may eventually be exploited to adapt the treatment of affected cases accordingly.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their parents as well as physicians involved in this study, Andishe Attarbaschi for providing clinical data and Marion Zavadil for proofreading. This study is supported in part by Grants from the Austrian National Bank (ÖNB 12519 and 13881) to GM, Grant P18046-B12 of the Austrian Science Fund (FWF P 22073-B19) and the St Anna Kinderkrebsforschung to RP-G.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- 1.Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 3.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 4.Maia AT, van der Velden VH, Harrison CJ, Szczepanski T, Williams MD, Griffiths MJ, et al. Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia. 2003;17:2202–2206. doi: 10.1038/sj.leu.2403101. [DOI] [PubMed] [Google Scholar]
- 5.Panzer-Grumayer ER, Fasching K, Panzer S, Hettinger K, Schmitt K, Stockler-Ipsiroglu S, et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood. 2002;100:347–349. doi: 10.1182/blood-2002-01-0144. [DOI] [PubMed] [Google Scholar]
- 6.Paulsson K, Forestier E, Lilljebjorn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107:21719–21724. doi: 10.1073/pnas.1006981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 8.Tallen G, Ratei R, Mann G, Kaspers G, Niggli F, Karachunsky A, et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: results of trial ALL-REZ BFM 90. J Clin Oncol. 2010;28:2339–2347. doi: 10.1200/JCO.2009.25.1983. [DOI] [PubMed] [Google Scholar]
- 9.Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 10.Davidsson J, Paulsson K, Lindgren D, Lilljebjorn H, Chaplin T, Forestier E, et al. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- 11.Kawamata N, Ogawa S, Seeger K, Kirschner-Schwabe R, Huynh T, Chen J, et al. Molecular allelokaryotyping of relapsed pediatric acute lymphoblastic leukemia. Int J Oncol. 2009;34:1603–1612. doi: 10.3892/ijo_00000290. [DOI] [PubMed] [Google Scholar]
- 12.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roelfsema JH, Peters DJ. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med. 2007;9:1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, et al. Key pathways are frequently mutated in high-risk childhood acute lympho-blastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flohr T, Schrauder A, Cazzaniga G, Panzer-Grumayer R, van der Velden V, Fischer S, et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22:771–782. doi: 10.1038/leu.2008.5. [DOI] [PubMed] [Google Scholar]
- 18.Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–2667. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- 19.Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talamo A, Chalandon Y, Marazzi A, Jotterand M. Clonal heterogeneity and chromosomal instability at disease presentation in high hyperdiploid acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2010;203:209–214. doi: 10.1016/j.cancergencyto.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 23.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Velden VH, Panzer-Grumayer ER, Cazzaniga G, Flohr T, Sutton R, Schrauder A, et al. Optimization of PCR-based minimal residual disease diagnostics for childhood acute lymphoblastic leukemia in a multi-center setting. Leukemia. 2007;21:706–713. doi: 10.1038/sj.leu.2404535. [DOI] [PubMed] [Google Scholar]
- 25.Eckert C, Biondi A, Seeger K, Cazzaniga G, Hartmann R, Beyermann B, et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. Lancet. 2001;358:1239–1241. doi: 10.1016/S0140-6736(01)06355-3. [DOI] [PubMed] [Google Scholar]
- 26.Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.