Abstract
Background
Members of the thermophilic genus Geobacillus can grow at high temperatures and produce a battery of thermostable hemicellulose hydrolytic enzymes, making them ideal candidates for the bioconversion of biomass to value-added products. To date the molecular determinants for hemicellulose degradation and utilization have only been identified and partially characterized in one strain, namely Geobacillus stearothermophilus T-6, where they are clustered in a single genetic locus.
Results
Using the G. stearothermophilus T-6 hemicellulose utilization locus as genetic marker, orthologous hemicellulose utilization (HUS) loci were identified in the complete and partial genomes of 17/24 Geobacillus strains. These HUS loci are localized on a common genomic island. Comparative analyses of these loci revealed extensive variability among the Geobacillus hemicellulose utilization systems, with only seven out of 41–68 proteins encoded on these loci conserved among the HUS+ strains. This translates into extensive differences in the hydrolytic enzymes, transport systems and metabolic pathways employed by Geobacillus spp. to degrade and utilize hemicellulose polymers.
Conclusions
The genetic variability among the Geobacillus HUS loci implies that they have variable capacities to degrade hemicellulose polymers, or that they may degrade distinct polymers, as are found in different plant species and tissues. The data from this study can serve as a basis for the genetic engineering of a Geobacillus strain(s) with an improved capacity to degrade and utilize hemicellulose.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-836) contains supplementary material, which is available to authorized users.
Keywords: Geobacillus, Hemicellulose, Arabinoxylan, Arabinan, Xylanase, Acetylesterase, Arabinofuranose
Background
The plant cell wall consists of four major structural polymer components, namely cellulose, hemicellulose, pectin and lignin [1, 2]. In contrast to cellulose, which is a homopolymer of β-(1 → 4)-linked glucose monomers, hemicelluloses incorporate a wide range of heterogeneous polymers containing a backbone of xylose, arabinose, galactose or, mannose monomers [1, 3]. The most common hemicelluloses are the xylans, which have a backbone of β-(1 → 4)-linked xylose monomers, and are frequently decorated with side chains of arabinose, glucuronic acid or its 4-O-methyl derivative, and acetyl side groups [1]. The structure of the hemicellulose, its backbone monomers and degree of branching and type of side chains varies extensively between different plant species and even tissues within the same plant. For example, xyloglucans dominate the primary cell walls of dicotyledonous plants, while glucuronoarabinoxylans are prevalent in both the primary and secondary cell walls of commelinid monocots (e.g. sugarcane and maize). While the primary cell walls of conifers such as pine are dominated by xyloglucans, the hemicellulose fraction of their secondary cell walls is comprised mainly of galactoglucomannans [1]. Similarly, pectin represents a family of heterogeneous polymers exhibiting complex structures which are found in the plant primary cell wall [4].
In nature, plant biomass represents the most abundant carbohydrate source for microorganisms. As such, many bacteria, fungi and yeast can degrade hemicellulose polymers through the production of a battery of hydrolytic enzymes, including xylanases, xylosidases, arabinofuranosidases and glucuronidases [5]. In recent decades there has been increasing interest in the use of microorganisms and their enzymes for the hydrolysis of hemicelluloses, as a result of the various biotechnological and industrial applications in which the hydrolytic enzymes, the hemicelluloytic process and its products can be exploited. For example, xylanases can be applied in the bleaching of pulp and paper, increasing the digestibility of animal feeds, clarification of fruit juices and the bioconversion of plant bionass [5–7]. The latter can be exploited, in combination with the fermentative capacities of microorganisms in the conversion of lignocellulosic biomass to high yields of the renewable energy source bioethanol and other value-added products [8, 9]. The oligosaccharides generated from hemicellulose hydrolysis can also be applied as prebiotics that inhibit harmful microorganisms and stimulate the growth of bifidobacteria and lactobacilli in the human intestine [10]. There has been increasing interest in the use of thermophilic microorganisms for the hemicellulytic hydrolysis of plant biomass, as they display a number of advantages over mesophilic microorganisms [8, 11]. The rate of biomass conversion is enhanced at higher growth temperatures. Furthermore, as high temperatures are required for the pre-treatment and delignification of plant biomass in many industrial processes, the ability of thermophilic microorganisms to tolerate higher temperatures, as well as producing thermostable hydrolytic enzymes, can potentially reduce process costs [8, 11].
Obligately thermophilic species within the genus Bacillus have been accommodated in the genus Geobacillus, which currently encompasses nineteen species [12, 13]. Members of this genus are rod-shaped, aerobic endo-spore formers and have optimum growth temperatures of between 45 and 75°C [13]. Geobacillus spp. serve as an attractive resource for production of a wide range of thermostable enzymes for a variety of biotechnological applications [8, 14]. Geobacillus strains have been shown to be highly effective in the degradation of hemicellulose polymers, and considerable attention has focused on the thermostable extra- and intra-cellular xylanases and β-xylosidases [15–18]. The molecular determinants underlying the capacity of this genus to utilize hemicellulose have, however, been largely elucidated in only one strain, Geobacillus stearothermophilus T-6, and have been shown to reside on a single genetic locus [19–21]. On the basis of characterization of some of the gene clusters within this locus, a model for the hemicellulolytic capacity of this strain has been developed [19–21]. The model hemicellulose substrate, acetylglucuronoarabinoxylan (GAX), is cleaved by the extracellular GH10 xylanase XynA1 into large xylooligomers, including xylooligosaccharides and aldotetraouronic acid, which are subsequently transport across the cellular membrane by specific oligosaccharide transport systems [19, 20]. The glucuronic acid is cleaved from the aldotetrauronic acid (2-O-α-(4-O-methyl-D-glucuronosyl)-xylotriose or MeGlcAXyl3) polymer by the action of a GH67 α-glucuronidase and is metabolized via the uronate metabolic pathway, also encoded in the hemicellulose utilization locus [19]. The xylooligosaccharides and xylotriose derived from MeGlcAXyl3 polymers are further cleaved into D-xylose monomers through the concerted action of a second intracellular GH10 xylanase, XynA2, and three β-xylosidases XynB1, XynB2 and XynB3, which are then metabolized via the pentose phosphate and glycolysis pathways [19, 20]. Pathways for the hydrolysis and utilization of arabinofuranosyl substituents and for the main chain deacetylation have also been described [20, 22, 23]. A gene cluster within the hemicellulose utilization locus of G. stearothermophilus T-6 has also been shown to play a role in the degradation of the pectin-associated L-arabinan polymer [21], further highlighting the capacity of Geobacillus spp. to degrade and utilize polymers in plant biomass.
To date, the genomes of twenty-four Geobacillus spp. have been sequenced. Here, using the G. stearothermophilus T-6 hemicellulose utilization system (HUS) as a model, we have identified and characterized loci coding for the degradation and utilization of hemicellulose in most of the sequenced strains. Our analyses reveal the extensive variability in the Geobacillus hemicellulose utilization loci, which could be exploited to generate recombinant strains with improved hemicellulolytic capacities.
Results and discussion
A hemicellulose utilization system is a common feature among most Geobacillusspp
The capacity of G. stearothermophilus T-6 to degrade and utilize hemicellulose is dependent on a single complex gene locus, which has recently been sequenced (NCBI Acc. # DQ868502). This locus is ~76 kb in size, has a G + C content of 46.93% and encodes 60 proteins. The HUS locus of T-6 can be roughly divided into thirteen distinct gene clusters (Figure 1 – clusters A-M). Twelve of the gene clusters have been characterized functionally and/or in silico and have been shown to play roles in the concerted degradation and utilization of the hemicellulose polymer glucuronoarabinoxylan in this strain [19, 20]. Of these, seven can be considered as central to hemicellulose degradation, with the proteins encoded in these clusters driving the extracellular degradation of arabinoglucuronoxylan (cluster K), transport of the cleaved oligosaccharide products into the cell (clusters F and H) and further intracellular degradation of these intermediates into metabolizable monosaccharides (clusters D, G, I and L). Four additional gene clusters (clusters B, E, J and M) are not essential for the degradation of the hemicellulose polymer, but rather encode pathways for the metabolism of the end-product pentose sugars arabinose and xylose and for uronic acids, as well as a putative alternative pathway for pentose sugar metabolism [19, 20]. An additional cluster which is not essential for hemicellulose degradation, encodes a transporter of L-arabinose monosaccharides (cluster A). A gene cluster with a role in the degradation and utilization of the pectin-associated polymer L-arabinan has also been described in G. stearothermophilus T-6 [21] and is positioned in the hemicellulose utilization cluster of this strain (Figure 1 - cluster C).
Figure 1.
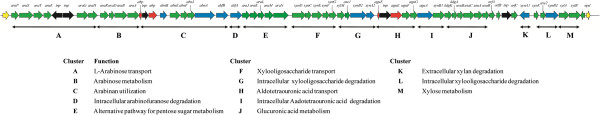
Schematic diagram of the G. stearothermophilus T-6 hemicellulose utilization locus. Each arrow represents a gene in the locus, with genes encoding predicted transposons colored in black, while open reading frames interrupted by transposons are colored in red. Genes encoding glycosyl hydrolases are colored in blue. The G. stearothermophilus HUS locus was subdivided into thirteen gene clusters on the basis of their predicted function.
The genomes of 24 Geobacillus strains, thirteen complete and eleven draft genomes, have been sequenced (Table 1) [24–34]. The twelve gene clusters of T-6 with a reported role in hemicellulose utilization were used as units for the identification of orthologous clusters in the sequenced Geobacillus genomes, by localized BlastN analysis. Between three and twelve partial or complete orthologous clusters were found in twenty of the sequenced strains, while no orthologous clusters could be found in the genomes of the remaining four strains (Table 2; Figure 2). A further three strains, namely G. thermoglucosidasius CCB_US3_UF5 and B23, as well as G. kaustophilus HTA426 only carry three orthologous gene clusters, namely those for xylose metabolism, arabinose transport and L-arabinose metabolism. The lack of orthologs of all T-6 clusters required for hemicellulose degradation suggests that these strains do not have this capacity, and rather make use of L-arabinose and D-xylose monomers that may be present in the environment. The remaining seventeen sequenced Geobacillus strains contain between eight and twelve gene clusters sharing orthology with the thirteen clusters that make up the hemicellulose utilization locus of G. stearothermophilus T-6 (Table 2), including between five and seven of the clusters with a role in hemicellulose degradation. The genomic locations of the orthologous clusters indicate that, as is the case of T-6, they are arranged into a single chromosomal hemicellulose utilization locus. Thus, including G. stearothermophilus T-6, eighteen Geobacillus strains (72% of the twenty-five strains analyzed) can be considered to contain a Hemicellulose Utilization System (HUS). In addition to T-6, the genomes of three further sequenced Geobacillus strains, namely Y412MC52, Y412MC61 and WSUCF1, were observed to encode an L-Arabinan Utilization System (AUS) incorporated within the hemicellulose utilization locus.
Table 1.
Genome characteristics of twenty-four sequenced Geobacillus strains
| Species | Strain | Isolation source | Sequencing status | NCBI Acc # | Replicons/contigs | Size (Mb) | G + C% | Ref |
|---|---|---|---|---|---|---|---|---|
| G. thermoleovorans | B23 | Subterraneon oil reservoir, Japan | Draft | BATY00000000 | 209 | 3.35 | 52.29 | [24] |
| G. thermoleovorans | CCB_US3_UF5 | Ulu Slim hot spring, Malaysia | Complete | NC_016593.1 | 1 | 3.60 | 52.28 | [25] |
| G. kaustophilus | HTA426 | Deep sea sediment, Marian Trench | Complete | BA000043.1 | 2 | 3.59 | 51.98 | [26] |
| Geobacillus sp. | CAMR5420 | - | Draft | JHUS01000000 | 96 | 3.49 | 52.00 | [27] |
| G. kaustophilus | GBlys | Pasteurized milk | Draft | BASG00000000 | 216 | 3.54 | 52.05 | [28] |
| Geobacillus sp. | MAS1 | Hot spring, Pakistan | Draft | AYSF00000000 | 121 | 3.50 | 52.21 | - |
| Geobacillus sp. | A8 | Deep mine water, South Africa | Draft | AUXP01000000 | 173 | 3.35 | 52.41 | - |
| Geobacillus sp. | CAMR12739 | - | Draft | JHUR01000000 | 74 | 3.38 | 52.19 | [27] |
| Geobacillus sp. | C56-T3 | Sandy’s Spring W, Nevada, USA | Complete | CP002050.1 | 1 | 3.65 | 52.49 | - |
| Geobacillus sp. | Y412MC61 | Obsidian Hot Spring, YNP, USA | Complete | NC_013411.1 | 2 | 3.67 | 52.33 | - |
| Geobacillus sp. | Y412MC52 | Obsidian Hot Spring, YNP, USA | Complete | NC_014915.1 | 2 | 3.67 | 52.33 | - |
| Geobacillus sp. | WSUCF1 | Compost, USA | Draft | ATCO00000000 | 346 | 3.40 | 52.21 | [29] |
| Geobacillus sp. | GHH01 | Soil, Germany | Complete | NC_020210.1 | 1 | 3.58 | 52.28 | [30 |
| Geobacillus sp. | C56-T2 | Double Hot Springs, Nevada | Draft | SAMN0017395 | 3 | 3.55 | 52.39 | - |
| Geobacillus sp. | JF8 | Compost, Japan | Complete | NC_022080.4 | 2 | 3.49 | 52.79 | [31] |
| Geobacillus sp. | G11MC16 | - | Draft | ABVH00000000 | 31 | 3.55 | 48.80 | - |
| G. thermodenitrificans | NG80-2 | Subterraneon oil reservoir, China | Complete | NC_009328.1 | 2 | 3.61 | 48.86 | [32] |
| G. thermodenitrificans | DSM465T | Sugar beet juice, Austrua | Partial | AYKT00000000 | 76 | 3.40 | 49.05 | [33] |
| G. caldoxylolyticus | CIC9 | Hot Spring, Indonesia | Partial | AMRO0000000 | 82 | 3.83 | 44.17 | - |
| G. thermoglucosidasius | C56YS93 | Obsidian Hot Spring, YNP, USA | Complete | NC_015660.1 | 3 | 4.00 | 43.93 | - |
| G. thermoglucosidasius | TNO-09.020 | Dairy processing plant, Netherlands | Complete | NZ_CM001483 | 1 | 3.74 | 43.82 | [34] |
| G. thermoglucosidasius | M10EXG | Waste-composting unit, Australia | Complete | 2501416905a | 1 | 3.67 | 43.74 | - |
| Geobacillus sp. | Y4.1MC1 | Bath Hot Spring, YNP, USA | Complete | NC_014650.1 | 2 | 3.84 | 44.02 | - |
| Geobacillus sp. | WCH70 | Middleton, USA | Complete | NC_012793.1 | 3 | 3.51 | 42.80 | - |
NCBI Acc # refers to the NCBI accession numbers of the genomes for the specified organisms. arefers to the Integrated Microbial Genomes (IMG) object identifier for the genome of G. thermoglucosidasius M10EXG ( http://img.jgi.doe.gov).
Table 2.
The number (#) of orthologous gene clusters from the G . stearothermophilus T-6 HUS loci in sequenced Geobacillus strains
| Strain number | Geobacillusstrain | # orthologous clusters |
|---|---|---|
| 1 | G. thermoleovorans B23 | 3 |
| 2 | G. thermoleovorans CCB_US3_UF5 | 3 |
| 3 | G. kaustophilus HTA426 | 3 |
| 4 | Geobacillus sp. CAMR5420 | 12 |
| 5 | G. kaustophilus GBlys | 12 |
| 6 | G. thermopakistaniensis MAS1 | 11 |
| 7 | Geobacillus sp. A8 | 12 |
| 8 | Geobacillus sp. CAMR12739 | 12 |
| 9 | Geobacillus sp. C56-T3 | 12 |
| 10 | Geobacillus sp. Y412MC61 | 12 |
| 11 | Geobacillus sp. Y412MC52 | 12 |
| 12 | Geobacillus sp. WSUCF1 | 12 |
| 13 | Geobacillus sp. GHH01 | 11 |
| 14 | Geobacillus sp. C56-T2 | 10 |
| 15 | Geobacillus sp. T-6 | 12 |
| 16 | Geobacillus sp. JF8 | 8 |
| 17 | Geobacillus sp. G11MC16 | 12 |
| 18 | G. thermodenitrificans NG80-2 | 12 |
| 19 | G. thermodenitrificans DSM465T | 12 |
| 20 | G. caldoxylolyticus CIC9 | 11 |
| 21 | G. thermoglucosidasius C56YS93 | 8 |
| 22 | G. thermoglucosidasius TNO-09.020 | 0 |
| 23 | G. thermoglucosidasius M10EXG | 0 |
| 24 | Geobacillus sp. Y4.1MC1 | 0 |
| 25 | Geobacillus sp. WCH70 | 0 |
Figure 2.
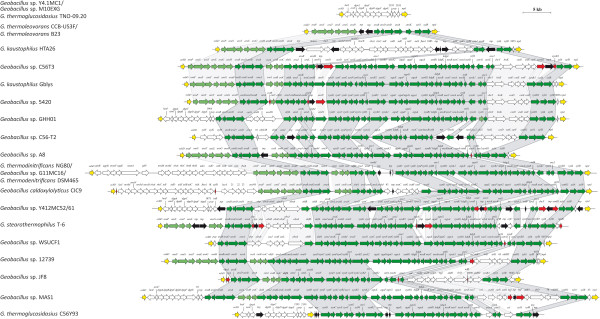
Comparative diagram of the Geobacillus hemicellulose loci. The aligned echD-npd islands of the twenty-four sequenced HUS+ and HUS- Geobacillus strains and that of G. stearothermophilus T-6 are shown. The flanking echD and npd genes are colored in yellow, genes coding for transposons in black, and genes in which the reading frames are transposon-disrupted are in red. Those genes conserved among >70% of the HUS+ strains are colored in green, those conserved among >50% and <70% in light green, while those conserved among <50% of the HUS+ strains are colored in white.
A phylogenetic tree including all sequenced Geobacillus spp. and twelve Geobacillus spp. type strains was constructed using the recN gene. Phylogeny using the recN gene has been demonstrated to have great resolving power at the species and subs-species level for the genus Geobacillus [35]. The topology (Figure 3) showed that the hemicellulose utilization system is encoded across all major branches of the Geobacillus genus phylogeny, although clusters of HUS- strains can be observed in the G. thermoleovorans-kaustophilus and G. thermoglucosidasius-thermantarcticus-toebii branches.
Figure 3.
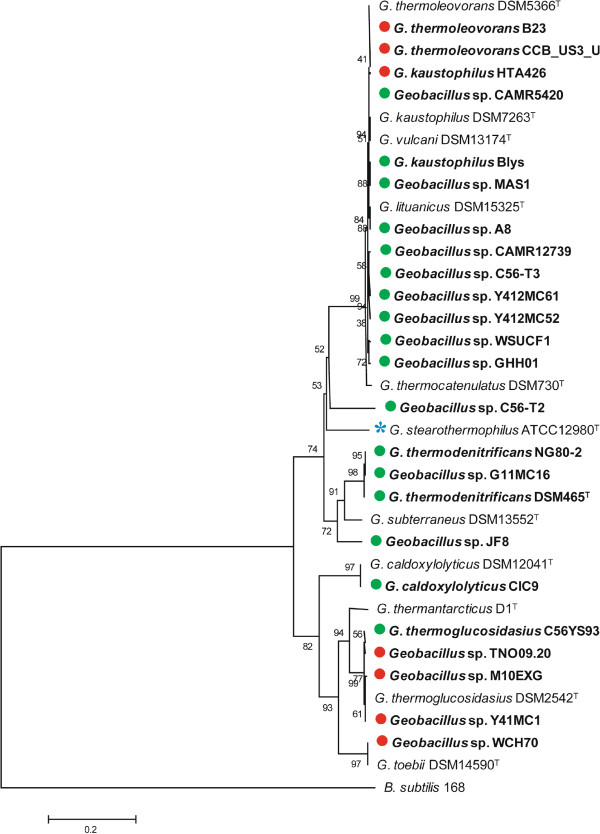
Phylogeny of the sequenced Geobacillus strains showing the distribution of hemicellulose loci. A neighbour-joining phylogeny was constructed on the basis of the recN gene for the twenty-four sequenced Geobacillus strains as well as twelve Geobacillus spp. type strains. The presence or absence of HUS loci in the sequenced strains is indicated by green and red dots, respectively. The blue star next to G. stearothermophilus ATCC 12980T indicates the predicted phylogenetic position of G. stearothermophilus T-6 for which a recN sequence is not available. The recN gene sequence of Bacillus subtilis 168 was used as outgroup. Bootstrap values (n = 1,000) are shown.
The Geobacillushemicellulose system forms part of a genomic island
Genomic islands were identified in the genomes of the thirteen complete Geobacillus genomes using the Islandviewer server [36]. Between thirteen and thirty genomic islands were predicted for each strain. One predicted island, flanked by genes encoding an enoyl-CoA hydratase (echD) and a nitropropane dioxygenases (npd), is present on the chromosome of twelve out of thirteen complete sequenced Geobacillus strains. BlastN analysis with these genes identified similar islands in all eleven draft genomes (Figure 2), suggesting that an echD-npd island is present in twenty-three of the twenty-four sequenced strains. While orthologs of the echD and npd genes were detected in the genome sequence of Geobacillus sp. WCH70, these are separated by ~540 kb of sequence and were not identified as an island. The echD-npd islands range in size from 10.5 to 87.6 kilobases and carry between eleven and sixty-eight protein coding sequences (Table 3). The G + C content of the echD-npd island in all Geobacillus strains is on average 4.90 ± 1.15% (average G + C content ± standard deviation) lower than the genomic G + C content, indicative of horizontal acquisition of this island.
Table 3.
Characteristics of the echD - npd islands of twenty-five Geobacillus strains
| Strain # | Species | Strain | Island size (kb) | # CDS | G + C% | G + C% island vs genome |
|---|---|---|---|---|---|---|
| 1 | G. thermoleovorans | B23 | 22.0 | 17 | 48.24 | -4.05 |
| 2 | G. thermoleovorans | CCB_US3_UF5 | 22.0 | 16 | 48.27 | -4.01 |
| 3 | G. kaustophilus | HTA426 | 54.9 | 41 | 48.04 | -3.94 |
| 4 | Geobacillus sp. | CAMR5420 | 66.4 | 53 | 46.54 | -5.46 |
| 5 | G. kaustophilus | GBlys | 68.1 | 53 | 46.20 | -5.85 |
| 6 | Geobacillus sp. | MAS1 | 74.7 | 62 | 46.12 | -6.09 |
| 7 | Geobacillus sp. | A8 | 60.4 | 48 | 46.75 | -5.66 |
| 8 | Geobacillus sp. | CAMR12739 | 67.9 | 54 | 46.65 | -5.54 |
| 9 | Geobacillus sp. | C56-T3 | 69.7 | 55 | 46.49 | -6.00 |
| 10 | Geobacillus sp. | Y412MC61 | 79.4 | 64 | 47.28 | -5.05 |
| 11 | Geobacillus sp. | Y412MC52 | 79.4 | 64 | 47.28 | -5.05 |
| 12 | Geobacillus sp. | WSUCF1 | 58.3 | 47 | 46.86 | -5.35 |
| 13 | Geobacillus sp. | GHH01 | 71.5 | 57 | 46.29 | -5.99 |
| 14 | Geobacillus sp. | C56-T2 | 60.8 | 44 | 47.00 | -5.39 |
| 15 | Geobacillus stearothermophilus | T-6 | 76.1 | 60 | 46.92 | - |
| 16 | Geobacillus sp. | JF8 | 50.7 | 41 | 46.16 | -6.63 |
| 17 | Geobacillus sp. | G11MC16 | 86.3 | 68 | 43.73 | -5.07 |
| 18 | G. thermodenitrificans | NG80-2 | 86.4 | 68 | 43.75 | -5.11 |
| 19 | G. thermodenitrificans | DSM465T | 86.4 | 68 | 43.76 | -5.29 |
| 20 | G. caldoxylolyticus | CIC9 | 87.6 | 68 | 38.83 | -5.34 |
| 21 | G. thermoglucosidasius | C56YS93 | 12.4 | 12 | 41.89 | -2.04 |
| 22 | G. thermoglucosidasius | TNO-09.020 | 10.6 | 11 | 40.69 | -3.13 |
| 23 | G. thermoglucosidasius | M10EXG | 10.5 | 11 | 40.65 | -3.09 |
| 24 | Geobacillus sp. | Y4.1MC1 | 10.6 | 11 | 40.56 | -3.46 |
| 25 | Geobacillus sp. | WCH70 | - | - | - | - |
BlastN analysis with the echD and npd genes from the complete genomes against the G. stearothermophilus T-6 HUS locus revealed that a complete copy of echD and a partial npd gene were present at the 5′ and 3′ ends of this locus, respectively. A search up- and downstream of the hemicellulose degradation and utilization gene clusters in sixteen of the sequenced HUS+ strains showed that, similarly, all of the hemicellulose utilization loci in these strains were maintained within the echD-npd genomic islands, The three Geobacillus strains which contained orthologs of only three of the thirteen clusters in the T-6 hemicellulose utilization locus also carry these clusters within the echD-npd island, indicating that the echD-npd genomic islands serve as a centralized repository for the hemicellulose degradation and utilization genes within Geobacillus spp. One exception is the HUS+ strain G. thermoglucosidasius C56YS93, where the orthologous clusters for hemicellulose degradation and utilization are localized downstream of the npd-echD island and are flanked by transposon genes (Figure 2).
The average difference in G + C content between the echD-npd island and the genome is 3.61 ± 0.41% for HUS- strains, while the mean difference for HUS+ is somewhat larger (5.55 ± 0.44%). An exception is again observed for G. thermoglucosidasius C56YS93, where the G + C content of the echD-npd island is only 2.04% below that of the genome, while that of the island plus the HUS locus is 0.06% above that the genome. Of note is the great variability in genomic G + C contents observed for the different sequenced Geobacillus strains. For example, the mean G + C content of the G. caldoxylolyticus CIC9 genome is 44.17%, and that of Geobacillus sp. A8 is 52.41%, while the mean G + C contents of their echD-npd islands are 46.75% and 38.83%, respectively. We suggest that these differences, given that the islands are highly variable, might be due to the non-conserved content of the island. The mean G + C contents for 41 conserved genes and those of the non-conserved genes in the echD-npd islands of these two strains were determined. This showed that G + C content differences existed between the islands of the A8 and CIC9 strains for both conserved and non-conserved genes, but that there was negligible variation in the G + C contents of both the conserved and non-conserved gene complement in the two strains (Additional file 1: Table S1). The concept of G + C amelioration [37] implies that while the G + C contents of genomic islands that have recently been horizontally acquired reflect those of the donor strain(s), the genes in these islands are subject to the same mutational processes as all other genes in the recipient strain and will become more similar to that of the recipient genomes over time. We suggest that the observed differences of genomic and island G + C contents among Geobacillus strains is due to the fact that the echD-npd islands may have derived from a common ancestor with a much lower genomic G + C content, and that these islands show G + C contents which are more, or less, ameliorated, depending on the time since acquisition.
Comparison of the hemicellulose utilization loci reveals extensive variation among Geobacillusstrains
The protein coding sequences (CDSs) on the echD-npd islands of all twenty-five Geobacillus strains were predicted using FgenesB [38]. The translated CDS sets from both HUS- and HUS+ strains were compared by localized BlastP analyses and the proteins were annotated on the basis of orthologous proteins identified by comparison against the NCBI non-redundant protein database (Additional file 2: Table S2). No proteins were conserved among the echD-npd islands of all twenty-five analyzed strains, while only seven proteins are conserved among the HUS+ strains. As between 41 and 68 proteins are encoded on the echD-npd islands of HUS+ strains, the low number of conserved proteins (10.3-17.1% of the total proteins encoded in the islands) highlights the extensive variability among the Geobacillus HUS loci. CDSs conserved among the eighteen HUS+ strains include two coding for a two component system (xynDC), three coding for a xylotriose transporter (xynEFG) and two genes (xylAB) required for xylose metabolism. The translated proteins products share an average amino acid identity of 87.35% between pair-wise compared strains, although lower orthology is observed for the XynDCEFG-XylAB sets of G. caldoxylolyticus CIC9 and Geobacillus sp. C56-T2, which share 60.5% and 67.4% average amino acid identity with each of the other sixteen HUS+ strains, respectively, and 80.7% amino acid identity between them. A neighbour-joining phylogeny was constructed on the basis of the concatenated XynDCEFG-XylAB amino acid sequences of the eighteen HUS+Geobacillus strains, as well as sixteen phylogenetic relatives for which whole genome sequences are available and in which orthologous proteins are present. The resultant tree (Figure 4) shows congruence in the clustering of most HUS+Geobacillus strains with the associated RecN tree, with the exception of Geobacillus sp. C56-T2 and G. caldoxylolyticus CIC9, suggesting that the hemicellulose utilization loci of sixteen strains have been derived from a common ancestor and have subsequently diverged in parallel with their genomes, while those of the other two strains are derived from a separate, but possibly common, ancestor.
Figure 4.
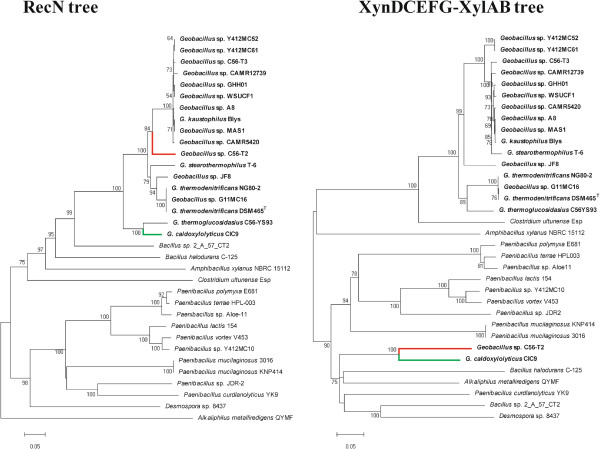
Phylogenetic comparison of the HUS + Geobacillus strains on the basis of RecN and XynDCEFG-XylAB. A neighbour-joining tree was constructed on the basis of the RecN amino acid sequences of the HUS+ Geobacillus strains as well as sixteen representatives of related genera in which orthologs of XynDCEFG-XylAB are present. Similarly a neighbour-joining tree was constructed based on alignment of the concatenated XynDCEFG-XylAB amino acid sequences. Desmospora sp. 8437 was used as outgroup and bootstrap values are shown (n = 1,000). The branches in red and in green, represent the phylogenetic positions of Geobacillus sp. C56-T2 and G. caldoxylolyticus CIC9, respectively, in both trees.
Variability in the extracellular hydrolytic enzyme complement encoded on the Geobacillus HUS loci
The subcellular localizations of all the HUS-encoded CDSs were predicted by comparison of their translated protein products against the PsortB 3.0 server [39] and the detection of signal peptides by submission to the SignalP 4.1 server [40]. SignalP analysis predicted seventeen distinct HDI-encoded proteins with typical Gram-positive signal peptide/cleavage sites required for the Sec-dependent transport of proteins across the cellular membrane. PsortB analysis showed that the majority of these (fifteen out of seventeen proteins) are integrated into the cellular membrane and we propose that these form part of transport systems for the uptake of oligosaccharides. On the basis of Psort and SignalP results, two HUS-encoded proteins are predicted to be secreted. The protein sequences were compared against the Carbohydrate Active enzymes (CAZY) database using the dbCAN server’s BLAST tool [41, 42]. This showed that these two proteins represent glycoside hydrolases belonging to the Glycoside Hydrolase (GH) families GH10 and GH43, respectively.
Orthologs of a thermostable extracellular GH10 family xylanase XynA1 that degrades xylan to its component xylooligosaccharides have been identified in a number of Geobacillus spp. [15, 16, 43–45]. An orthologous xynA1 gene is found in the hemicellulose utilization locus of 15/18 HUS+ strains. This gene is ~1,2 kb in size and is transcribed on the opposite strand to all other genes in the HUS loci. Truncated genes (223 nt) showing sequence homology to the 3′ end of the xynA1 gene are also present in the HUS loci of Geobacillus sp. A8 and WSUCF1, while no orthologs is present in G. caldoxylolyticus CIC9 HDI, suggesting that the latter three strains lack the ability to degrade xylan extracellularly into its component xylooligosaccharides. The translated protein products of the complete xynA1 genes share an average amino acid identity of 90.9% among the HDI+ and 90.5% to XynA1 of G. stearothermophilus T-6 (P40943; Bitscore: 765; e-value: 0.0). An extracellular GH43 endo α-1,5-arabinanase (abnA) has been shown to hydrolyze pectin-associated L-arabinan polymers into shorter arabino-saccharides and arabinose in G. stearothermophilus T-6 [21] and forms part of the AUS locus in this strain. Orthologs of abnA are also found in the other three AUS+ strains and their encoded proteins share 97.8% average amino acid identity to the T-6 arabinanase.
Additional predicted orthologous glycoside hydrolases are encoded in the HUS loci of Geobacillus sp. JF8, MAS1, and G. thermoglucosidasius C56 YS93, which share 93.9% average amino acid identity among them. The ~1.5 kb gene for this enzyme is localized in a non-conserved region of the HUS locus between the genes coding for a predicted acetyl-esterase (axe2) and a β-xylosidase (xynB3) (Figure 2). A truncated copy of this gene is also present within this region in Geobacillus sp. CAMR12739. Comparison of the translated protein products of the three complete gene copies against the CAZY database using the dbCAN Blast tool [41, 42] showed that they code for a GH5 glycoside hydrolase, showing greatest orthology to a glycoside hydrolase family protein in Verminephrobacter eiseniae EF01-2 (ABM56721.1; average amino acid identity: 57%; Bitscore: 593; e-value: 1e-168). The GH5 family includes enzymes active on a wide range of carbohydrate substrates, including chitosanases, β-mannosidases, xyloglucan-specific endo- β-1,4-glucanases and cellulases [46]. The target carbohydrate for a glycoside hydrolases can sometimes be further elucidated by the identification of carbohydrate binding modules (CBM), which promote the association of the enzyme with a particular substrate, within the protein sequence of the enzyme [47]. Comparison against the CAZY database using the dbCAN Blast tool identified a module showing weak orthology to a CBM family 6 module of a predicted endoglucanase in Ignavibacterium album JCM16511 (IALB_2578; Average amino acid identity: 31.4%; Bitscore: 157; e-value: 2e-35) at the N-terminal end (amino acid positions 32–285) of the GH5 protein orthologs of all four Geobacillus strains. The CBM6 modules have been demonstrated to have binding specificities towards a number of substrates, including both branched and debranched xylan, β-1,4-glucan (cellulose), and agarose [48]. It is thus difficult to distinguish the target carbohydrate substrate for the Geobacillus GH5 enzyme. Similarly, it is difficult to determine the subcellular localizations of these enzymes. SignalP analysis shows that no typical Gram-positive signal peptides are present. The PsortB SubCellular Localization SCL-BLAST + tool, however, showed that the Geobacillus proteins share orthology with the cellodextrinase CelA of Ruminococcus flavefaciens FD-1 (P16169), for which an extracellular localization has been determined experimentally [49].
The limited number of secreted hydrolytic enzymes has been proposed to provide a selective advantage to Geobacillus spp. and related hemicellulolytic organisms, as the relatively large oligosaccharides which result from extracellular hemicellulose hydrolysis will require further cleavage by intracellular enzymes and are thus not readily available to competing, non-hemicellulolytic, microorganisms [21]. The assimilation of xylo- and arabinoxylo-oligosaccharides may furthermore serve in the conservation of energy.
Variability in the membrane transport systems encoded on the Geobacillus HUS locus
Following the extracellular degradation of xylan into its component oligosaccharides, these intermediates are typically transported into the cell by means of specific ABC sugar transporters [21]. Carbohydrate Uptake Transporters (CUT) have been classified into two distinct families, with CUT1 signifying those transporters that facilitate the uptake of di- and oligosaccharides as well as glycerol phosphate and polyols, while members of the CUT2 family transport only monosaccharides [50, 51]. Eight gene clusters encoding distinct CUT1 transporters could be identified among the HUS+ strains, with between two and four of these present in each of the strains, while a single CUT2 family transporter is encoded in most of the HUS+ as well as three HUS- strains.
Three CUT1 transporters with a role in transport of hemicellulose/pectic polymer degradation products have been described and functionally characterized in G. stearothermophilus T-6. These are required for the uptake of arabinosaccharides (abnEFJ), xylooligosaccharides (xynEFG) and aldotetraouronic acid (2-O-α-(4-O-methyl-D-glucuronosyl)-xylotriose – MeGlcAXyl3) (aguEFG) [19–21]. Orthologs of the xynEFG xylooligosaccharide transporter, which preferentially transports xylotriose, are found in all eighteen HUS+ strains, while the aguEFG aldotetraouronic acid transporter is present in sixteen HUS+ strains. The reading frame of the aguE gene coding for an aldotetraouronic acid extracellular sugar-binding protein in T-6 is disrupted by a transposon, suggesting that this transporter is non-functional in this strain. In contrast to the prevalent aguEFG and xynEFG transport systems, the abnEFJ arabinosaccharide transporter, which forms part of the L-arabinan utilization system, is restricted in distribution to three strains, namely T-6, Y412MC52 and Y412MC52. One CUT2 transporter has also been identified in the HUS locus of G. stearothermophilus T-6, which is predicted to transport arabinose monosaccharide [21]. The polycistronic operon (araEGH) coding for this transporter is also disrupted in T-6 by a transposon insertion. Orthologs are present in ten out of the 18 HDI+ strains, and also in three HDI- strains, suggesting that this transporter is not a characteristic of hemicelluloses degrading strains, but may form a part of an arabinose metabolic pathway present in most Geobacillus spp.
The five remaining distinct CUT1 transporters encoded in the HUS loci of the different sequenced Geobacillus strains share only limited orthology to characterized transporters. For further reference, the gene clusters encoding these transporters were named cutABC, cutDEF, cutGHI, cutJKL, cutMNO in order to distinguish between them and to avoid duplicity of gene names (Figure 2; Additional file 1: Table S1). A locus in Bacillus subtilis encodes a CUT1 transporter for the uptake of α-1,5-arabinooligosaccharides [52, 53]. Orthologous loci (cutABC) are present in 10/18 HUS+ strains, all of which are missing abnEFJ orthologs, with the translated protein products sharing 58.4% average amino acid identity with the AraNPQ proteins of B. subtilis 168. In Geobacillus sp. CAMR5420, only a partial cutB and complete cutC gene are present, while an ortholog of cutA is missing. The B. subtilis AraNPQ transport system is required for the uptake of linear α-1,5-arabinosaccharide homopolymers consisting of three or four arabinosyl units residues, but not the branched arabinooligosaccharides decorated with α-1,2- and α-1,3-linked arabinosyl side chains. These branched arabinooligosaccharides are transported into the cell by an as yet unknown mechanism [53]. The nature of the substrate(s) transported by the Geobacillus CutABC system is yet to be experimentally determined. A distinct CUT1 transporter (cutJKL) is encoded within the AUS locus of Geobacillus sp. WSUCF1 in the location occupied by abnEFG in the other AUS+ strains. The translated products for these genes share only 43.7% average amino acid identity to G. stearothermophilus T-6 AbnEFJ, but share greater orthology (71.7% average amino acid identity) with an ABC transporter of an unknown sugar substrate in Bacillus halodurans C-125 (BH1864-1866). The location of the CUT-1 encoding genes between the arabinanase-encoding abnA and abnB genes in WSUCF1, however, suggests that as in the case of T-6, they encode an arabinosaccharide transporter, although whether this CUT1 system facilitates the uptake of linear or branched saccharides is unknown.
The cutDEF genes encoding a CUT1 transporter are located between the axe2 and xynB3 genes in the loci of 5/18 HUS+ strains. Their translated products share 63.7% average amino acid identity with a characterized aldotetrouronate transporter in Paenibacillus JDR-2 [54], but share only 33.4% average amino acid identity with the aguEFG aldotetrouronate transporter of G. stearothermophilus T-6. Four of the strains with a cutABC cluster also encode aguEFG orthologs, suggesting they possess two alternative pathways for the uptake of aldotetraouronic acid. On the other hand, C56-T2 lacks aguEFG orthologs, but does appear to carry a functional cutDEF system. The translated protein products of the predicted cutMNO CUT1 transporter (Figure 2) in the HUS loci of in Geobacillus sp. MAS1 and JF8 share 96.2% amino acid identity between the two strains but show limited orthology (36.4% average amino acid identity) to the xynEFG transporter in G. stearothermophilus T-6. The localization of the cutMNO genes adjacent to the gene encoding the predicted GH5 hydrolytic enzyme in these two strains, suggests they likely code for a transporter for the uptake of the degraded saccharide products of the latter enzyme. The other two strains which encode a GH5 ortholog, namely Geobacillus sp. CAMR12739 and G. thermoglucosidasius C56YS93 do not contain an orthologous transporter. Three genes encoding an additional orthologous CUT1 transporter (cutGHI) are localized between araRDBA and abfA-araJKLMN in Geobacillus sp. HH01 and between araM and araN in G. caldoxylolyticus CIC9, respectively (Figure 2). The encoded proteins share 85.8% average amino acid identity between these two strains and 72.5% average amino acid identity with a predicted transporter of an unknown sugar substrate in Paenibacillus mucilaginosis 3013 (PMI3016_2562-2564).
Variability in the intracellular enzyme complement of HUS+ Geobacillus spp
Typically, following the intracellular transport of the component oligosaccharides they are further degraded into monomeric sugars through the concerted action of a battery of intracellular glycosyl hydrolases [19, 20]. The intracellular enzyme complement for the cleavage of xylooligosacharides, methylglucuronate side chains and L-arabinan arabinosaccharides have been characterized in G. stearothermophilus T-6 [19–21]. The AguEFG-transported substrate MeGlcAXyl3 is cleaved into D-glucuronic acid and xylotriose by the GH67 family α-glucuronidase AguA [19, 55]. Orthologs of the T-6 aguA gene are located in the loci of all HUS+ strains, with the exception of Geobacillus sp. JF8. As this strain is also missing orthologs of aguEFG it is likely that this strain cannot utilize this substrate. The Geobacillus sp. C56-T2 HUS locus does encode an AguA ortholog, but is also missing orthologs of the aguEFG genes, suggesting this strain can also not utilize aldotetraouronic acid as a metabolic intermediate, although it cannot be excluded that an alternative transporter for this substrate may exist in this strain.
The xylotriose derived from cleavage of MeGlcUAX3, as well as the xylooligosaccharides transported into the cell via the xynEFG transporter, are degraded to D-xylose through the concerted action of an intracellular xylanase and β-xylosidases [19, 56]. The xynA2 gene in G. stearothermophilus T-6 encodes an intracellular GH10 family xylanase that degrades xylooligosaccharides, including xylotriose, into xylose and xylobiose subunits [18]. Orthologs are encoded in all HUS+ strains with the exception of Geobacillus sp. JF8 and G. caldoxylolyticus CIC9. The structure, biochemistry and mechanism of action of three distinct β-xylosidases encoded in the HUS locus of G. stearothermophilus T-6, XynB1 (GH39), XynB2 (GH52) and XynB3 (GH43), have been described [17, 56–59]. The natural substrate of XynB2 and XynB3 is xylobiose, while that of XynB1 is xylotriose. The xynB2 gene forms part of the xylooligosaccharide degradation cluster (Figure 1 - cluster G) of the loci of all eighteen HUS+Geobacillus strains, and as such probably degrades xylobiose released through the intracellular hydrolysis of xynEFG-transported xylooligosaccharides by xynA2. The xynB1 gene forms part of the aldotetraouronic acid utilization cluster (Figure 1 – cluster I) and the ability of XynB1 to degrade xylotriose, the cleavage product of MeGlcUAX3, may preclude the requirement of an additional intracellular xylanase for the cleavage of this substrate. A copy of xynB1 is present in 17/18 HUS+ strains. The presence of a xynB1 ortholog in G. caldoxylolyticus CIC9 suggests its protein product may complement the xylotriose cleaving action of the missing xynA2 enzyme. In Geobacillus sp. JF8, both xynB1 and xynA2 orthologs are missing, implying that this strain is incapable of utilizing the xylotriose substrate. Complete copies of xynB3 are found in 14/18 HUS+Geobacillus strains. In MAS1, Y412MC52, Y412MC61and C56T3 the xynB3 reading frame is disrupted by a transposon insertion, but given the similar function of the two β-xylosidases XynB2 and XynB3 in the hydrolysis of xylobiose, the complete xynB2 copy in these strains may complement the disrupted XynB3 product.
Three intracellular enzymes in the AUS locus of G. stearothermophilus T-6 play a role in the concerted release of component arabinose monosaccharides from the arabinan-derived oligosaccharides [21]. The GH51 arabinofuranosidase Abf2 of B. subtilis 168 (P94552) has been shown to remove O-2 and O-3-linked arabinofuranosyl side chains of branched arabinan polymers [60]. The translated protein products of the abfB gene found in all four AUS+Geobacillus strains share 74.9% average amino acid identity with Abf2, suggesting a similar function for AbfB proteins. While the majority of arabinose in plant cell wall polymers exists in the furanose (five-member ring) conformation, a small fraction of arabinose in the side-chain terminal ends of arabinan is in the six-membered ring pyranose form [1, 61]. Orthologs of a GH27 family arabinopyranosidase (Abp) which can cleave the terminal arabinopyranose residues [62] are encoded in the AUS loci of all four strains, but the presence of a transposon integrated in the T-6 abp gene suggests this capacity is disrupted in this strain. Finally, the resultant debranched linear α-1,5-arabinofuranose chain is cleaved into its component arabinose monomers by the intracellular GH43 arabinanase AbnB [21, 63], orthologs of which are encoded in all four AUS+ strains. Orthologs of a GH51 family arabinofuranosidase (AbfA) are encoded in seventeen HUS+ strains. The absence of additional encoded enzymes for the utilization of L-arabinan polymers in most strains suggests that this enzyme plays a role in the liberation of O-2 and O-3-linked arabinofuranosyl residues from arabinoxylan [21]. The presence of both abfA and abfB orthologs in the AUS+ strains thus suggests that they may have a capacity for debranching both arabinan and arabinoxylan polymers. The HUS locus of C56-T2 lacks orthologs of both abfA and abfB, but instead carries a gene encoding a distinct GH43 family glycoside hydrolase. The encoded protein shares only 13.5% and 10.7% amino acid identity with the AbfA and AbfB proteins of G. stearothermophilus T-6, respectively. However, it shares orthology with the Abf43B protein of Paenibacillus sp. E18 (AFC38437; Amino acid identity: 62.8%; Bitscore: 824; E-value: 0.0), which was characterized as a GH43 family α-arabinofuranosidase with a role in degradation of arabinoxylans [64].
In nature, hemicellulose polymers are frequently substituted with acetyl side chains. For example, the main hemicellulose of hardwoods, 4-O-methyl-D-glucuronoxylan, contains acetyl side chains at positions two or three of 70% of the xylose backbone subunits [65]. These acetyl side chains contribute towards the recalcitrance of hemicelluloses by restricting access of hydrolytic enzymes to the backbone sugars. As a ‘counteractive’ measure, many microorganisms produce acetylesterase enzymes, which can cleave the acetyl side chains [66, 67]. Two genes encoding predicted acetylesterases have been identified within the G. stearothermophilus T-6 hemicellulose utilization locus, axe1 and axe2 [20], with orthologous genes present in 16/18 and 17/18 HUS+ strains, respectively. The axe1 gene forms part of xylooligosaccharide utilization cluster (Figure 1 - cluster G) and the encoded Axe1 protein products share 86.7% average amino acid identity among the HUS+ strains. Comparison against the CAZY database with the dbCAN Blast tool shows that they belong to Carbohydrate Esterase family CE4 [41, 68], which incorporates acetylxylan esterases and chitin deacetylases, and show greatest orthology with the predicted polysaccharide deacetylase of Paenibacillus sp. JDR-2 (ACT03992.1; Average amino acid identity: 53.4%; Bitscore: 230; e-value: 4.5e-58). The incorporation of the axe1 gene in the xylooligosaccharide utilization cluster (Figure 1 – cluster G) suggests that this esterase plays a role in the deacetylation of xylan or its internalized derivative oligosaccharides. The crystal structure of the intracellular Axe2 protein of G. stearothermophilus T-6 has been determined, and this functionally characterized acetylxylan esterase has been shown to belong to a novel CE family [23, 67]. Orthologs of the Axe2 proteins of sixteen sequenced HUS+ strains share 91.2% average amino acid identity with the T-6 Axe2 protein, and negligible sequence similarity to the Axe1 protein in this strain. The axe2 gene forms part of the second intracellular xylooligosaccharide degradation cluster (Figure 1 – cluster L). While the Axe2 protein has been shown to be active on short acetylated xylo-oligmers [23], the natural target xylooligosaccharide is unknown. The native substrate of Axe1 is also unknown, but it can be hypothesized that these two esterases act in concert to completely deacetylate the xylooligomers resulting from xylan degradation.
A further putative intracellular hydrolytic enzyme is encoded in the HUS locus of JF8, while a transposon-disrupted copy is also found in MAS1. The translated JF8 protein product lacks a recognizable Gram-positive signal peptide, suggesting an intracellular localization, and shares 73% amino acid identity with a GH3 family glycosyl hydrolase of Thermoanaerobacterium thermosaccharolyticum DSM571 (Tthe_2487). Furthermore, the amino acid sequence contains a β-glucosidase conserved domain (PRK15098; Bitscore: 284; e-value: 3.7e-84). The predicted JF8 and MAS1 β-glucosidase-encoding (bgl) gene is located downstream of gh5 and cutDEF genes, suggesting the encoded enzyme is likely involved in the intracellular hydrolysis of the substrate derived through the extracellular hydrolysis by the GH5 enzyme and intracellular transport by the potentially associated cutABC transporter. Orthologs of the JF8 bgl gene and the cutDEF transporter are, however, absent in Geobacillus sp. CAMR12739 and G. thermoglucosidasius C56YS93, which contain a partial and complete copy of the GH5-encoding gene, respectively. Members of the GH3 family include glucosidases and xylosidases, and as such the target substrate of this gene cluster and its encoded proteins, whether it is a glucose- or xylose-based polymer, remains undetermined.
Variability in the metabolic genes in the HUS loci of Geobacillus spp
The monosaccharides derived from intracellular oligosaccharide cleavage are typically catabolised into intermediate substrates that enter the pentose phosphate and Embden Meyerhof Parnas (EMP) glycolysis pathways [19, 20]. The catabolic systems for D-xylose (xylAB), uronic acids (kdgKA-uxaC-uxuAB) and L-arabinose (araRDBA) monosaccharides have been well-characterized in Geobacillus stearothermophilus and its phylogenetic relative B. subtilis [19, 52, 69]. Orthologs of the xylAB and kdgKA-uxaC-uxuAB loci are present in all eighteen HUS+ strains, while araRDBA orthologs are present in seventeen of the strains. The latter locus is absent from G. thermoglucosidasius C56Y93, which is also missing orthologs of all additional genes encoding enzymes required for the transport and catabolism of arabinosaccharides, suggesting that the hemicellulose degradation system functions on arabinofuranose-free hemicellulose substrates. In Y412MC52 and Y412MC61, a transposase is inserted into the reading frame of uxaC, which encodes a uronate isomerase that catalyzes the first step in the conversion of uronic acids to glycerol-3-phosphate and pyruvate, which can enter the EMP pathway [19]. This suggests that these two strains are unable to catabolise this substrate.
An additional gene cluster, araJKLMN, has been identified in the G. stearothermophilus T-6 HUS locus. Complete araJKLMN loci are present in eleven of the eighteen HUS+ strains, while partial or transposon-disrupted loci are present in 6/18 HUS+ strains. Both araK and araN orthologs are absent from strains NG-80, G11MC16 and DSM465, while an araN ortholog is missing from the C56-T2 HUS locus. In C56-T3 and CAMR5420, the araN reading frame is disrupted by a transposon. A second copy of the araN gene, araNO is located downstream of the cutABC locus of GHH01, with its translated protein product sharing 91.3% amino acid identity with the AraN protein in this strain. Similarly, a second copy (araNO) is located upstream of araPST in the G. caldoxylolyticus CIC9 HUS locus, but its translated product shares lower sequence similarity (57.7% amino acid identity) with the CIC9 AraN protein. The genes in this cluster have been shown to encode orthologs of a NADP sugar dehydrogenase (araJ), aldose-1-epimerase (araK), sugar phosphatase (araL), NADP-dependent glycerol-1-dehydrogenase (araM) and hypothetical protein (araN) and have been postulated to constitute an alternative pathway for the catabolism of pentose sugars [21].
Additional genes in the Geobacillus echD-npdislands may play a role in hemicellulose utilization and alcohol fermentation
Additional non-conserved genes can be observed at the 5′ end of the echD-npd islands of several strains, flanking the hemicellulose utilization locus (Figure 2). Their translated protein products were annotated by BlastP comparison against the NCBI non-redundant protein database to identify orthologs for which functions have been predicted or experimentally determined (Additional file 3: Table S3). A gene cluster (dppABCDFE) at the 5′ end of the echD-npd island of five HUS+Geobacillus strains, namely Y412MC52, Y412MC61, MAS1, GHH01 and C56YS93, encodes a predicted oligopeptide transporter. Orthologs could furthermore be identified in the three HUS- strains, Geobacillus sp. Y4.1MC1, G. thermoglucosidasius TNO-09.20 and M10EXG. The echD-npd island 5′ end regions of G. caldoxylolyticus CIC9, Geobacillus sp. G11MC16, and G. thermodenitrificans NG80-2 and DSM465T harbor large inserts, 26.6 to 28 kb in size. The genes in these inserts encode a system for the catabolism of the polyol myo-inositol (iolG/IDEBCA), as well as two predicted ABC sugar transporters. Strains G11MC16, NG80-2 and DSM465T also encode a 1,044 amino acid protein which, by comparison against the CAZY database, was identified as a putative GH38 family glycoside hydrolase. The GH38 protein products show extensive orthology to the functionally characterized α-mannosidase (ManA) of Bacillus sp. GL1 (BAB40444.2; Average amino acid identity: 55.6%; Bitscore: 1299; e-value: 0.0; [70]. Mannose monosaccharides frequently form part of the backbone of hemicellulose polymers, such as the galactomannans found in the seeds of members of the Fabaceae family, and galactoglucomannans in conifer wood [1]. As such, the presence of a manA ortholog and a possible mannose-specific ABC transport system adjacent to the hemicellulose utilization locus, suggests that they form part of the hemicellulolytic arsenal in these strains. However, an ortholog showing 87.9% average amino acid identity to the ManA protein of the three HUS+ strains is also incorporated into the echD-npd island of the HUS- strain G. kaustophilus HTA426. The ManA protein may therefore rather form part of the general sugar utilization structure in Geobacillus spp. We suggest that the echD-npd island represents an insertion point for the centralization, and potentially co-regulation, of multiple sugar metabolic pathways in Geobacillus spp.
An additional cluster of orthologous genes is also integrated into the HUS loci of Geobacillus sp. JF8, MAS1 and CAMR12739 in the non-conserved region harboring the GH5 family glycoside hydrolase-encoding gene (Figure 2; Additional file 1: Table S1). One of these genes codes for an ortholog of the aldehyde dehydrogenase AldH-T of G. stearothermophilus SIC1 (BAA02975; Average amino acid identity: 92.2%; Bitscore: 935; e-value: 0.0). This enzyme has been shown to play a role in the conversion of acetyl CoA to aldehydes which are further converted into alcohols by alcohol dehydrogenases [11, 71]. Furthermore, another gene in this cluster encodes an orthologs of the aldo/keto-reductase YtbE of Bacillus sp. ECU0013 (ADP24600; Average amino acid identity: 68%; Bitscore: 400; e-value: 4e-114), which has been shown to catalyze the NADH-dependent reduction of aliphatic and aromatic aldehydes into their corresponding alcohols [72, 73]. The co-localization of genes with a putative role in alcohol fermentation and hemicellulose utilization further suggests a role for the echD-npd island in the centralization, and potentially maintenance and regulation, of the metabolic and fermentative pathways of Geobacillus spp.
Conclusions
Our comparative genomic analyses have revealed the presence of a centralized hemicellulose utilization locus in most sequenced Geobacillus strains. However, this locus, which is localized on a common genomic island, displays extensive genetic variability. Genes or pathways central to the utilization of the hemicellulose backbone and substituents are absent from the HUS loci of many strains, while other genes, or operons, are frequently disrupted by transposon insertion. From this we may deduce that different Geobacillus strains have a highly variable capacity to degrade hemicellulose polymers. It is possible that, in their natural environments, Geobacillus strains form part of consortia, whereby strains deficient in genes or pathways required for hemicellulose degradation and utilization may be complemented by other members of the consortia. However, given the limited number of extracellular hydrolytic enzymes produced by Geobacillus, which are generally conserved across their HUS loci, and the specific trans-membrane transport systems they utilize for the internalization of relatively large oligosaccharides, we argue that such complementation is unlikely. Comparisons of the HUS loci of the sequenced strains to that of the well-characterized G. stearothermophilus T-6 revealed the presence of a number of additional genes, some of which have orthologs in related genera and code for both extra- and intracellular hydrolytic enzymes and transmembrane transporters. These findings further support the concept of variable hemicellulolytic capacities among Geobacillus strains, and imply that they have evolved to degrade distinct hemicellulose substrates, such as are found in different plant species and tissues. For example, the lack of genes encoding an α-glucuronidase (aguA), its associated transport system (aguEFG) and uronate metabolic genes in Geobacillus sp. JF8 implies that this strain utilizes a xylan which is not substituted with glucuronic acid, while a xylan polymer devoid of arabinofuranosyl substituents is the likely target of G. thermoglucosidasius C56YS93. This novel substrate-guided ‘host-parasite’ relationship has not been demonstrated experimentally, but is potentially testable. A recent review highlighted the extensive geographical and environmental diversity of members of the genus Geobacillus and their important ecological roles in composting of plant biomass under thermophilic conditions [74]. Their ability to degrade distinct hemicellulose polymers may thus reflect their environmental exposure to particular hemicelluloses and potentially, their role in carbon cycling in places where hot conditions persist.
The extensive genetic variability among the Geobacillus hemicellulose utilization systems highlighted in this study can potentially serve as a backbone for the development of recombinant Geobacillus strain(s) with improved hemicellulolytic capacities or with the capacity to degrade structurally diverse hemicellulose substrates. Such engineered strains, with the added advantage of the thermostability of many of the Geobacillus extra- and intracellular hydrolytic enzymes, could then be exploited in various biotechnological applications, such as the production of bioethanol and other value-added products.
Methods
Elucidation and characterization of the Geobacillushemicellulose utilization loci
The G. stearothermophilus T-6 hemicellulose utilization locus (NCBI Acc. # DQ868502) was subdivided into thirteen gene clusters on the basis of their predicted function. The nucleotide sequences of these clusters, as well as the translated protein products for the genes were compared by localized BlastN and tBlastN using the Bioedit v. 7.1.11 software package [75] to identify orthologs in the partial or complete genome sequences of twenty-four Geobacillus strains (Table 1). The Genbank sequences of thirteen complete Geobacillus genomes were submitted to the Islandviewer server, which integrates several tools for the prediction of genomic islands [36]. Localized BlastN analysis was subsequently used to elucidate the complete echD-npd islands in the eleven Geobacillus strains for which only partial genome sequences are available and G. stearothermophilus T-6. Mean G + C contents for the genome sequences, the echD-npd islands and for individual genes were determined using Bioedit [75]. Open reading frames were predicted using the Softberry FgenesB server [38] and the resultant CDS sets were standardized by local BlastN and BlastP comparison.
In silicoanalyses of the HUS locus and echD-npdisland protein coding sequence sets
The translated protein products encoded in the echD-npd islands and HUS loci were compared to the NCBI non-redundant protein database to identify orthologs which have been characterized functionally and/or in silico (Additional file 1: Table S1). Comparisons between the proteins sets of the different Geobacillus strains were performed using Localized BlastP analysis in Bioedit [75]. Conserved protein domains were identified by comparison against the NCBI conserved domain database using CD-Search [76, 77]. Signal peptides were predicted using the SignalP 4.1 server [40] and the subcellular localizations of the proteins were determined with the PsortB 3.0 server [39]. Proteins showing orthology to transport-related proteins were compared to the Transport Classification Database (TCDB) [78]. Glycoside hydrolases and carbohydrate esterases encoded in the HUS loci were characterized and classified into their respective families by comparison against the CAZY database using the dbCAN Blast tool [41, 42, 68].
Phylogenetic analyses
The nucleotide sequences of the recN gene, which codes for DNA repair protein RecN, of the twenty-four sequenced Geobacillus strains as well as twelve Geobacillus spp. type strains were used to demonstrate the phylogenetic relationships among the HUS+ and HUS-Geobacillus strains. Similarly, the RecN amino acid sequences of the HUS+ strains and twelve strains belonging to related genera, as well as the concatenated XynDCEFG-XylAB amino acid sequenced were used to construct phylogenies. Sequence alignment was performed using Clustal Omega v. 1.2.0 [79], with default parameters. Phylogenetic trees were constructed with the Molecular Evolutionary Genetics Analysis (MEGA) v.5.0.3 software package [80], using the neighbour-joining method with the following parameters: Poisson correction, complete gap deletion and bootstrapping (n = 1,000).
Availability of supporting data
The complete genome sequences of all the Geobacillus strains incorporated in this study are available on the NCBI database under the NCBI accession numbers listed in Table 1. The phylogenies included in the manuscript (Figures 3 and 4) have been deposited and available in TreeBASE [81]. The amino acid sequence sets for each hemicellulose utilization locus and the Genbank files for each of the representative hemicellulose loci are available in the LabArchives repository [82].
Electronic supplementary material
Additional file 1: Table S1: Mean G + C contents for the conserved and non-conserved genes in the echD-npd islands of Geobacillus sp. A8 and G. caldoxylolyticus CIC9. The mean G + C contents for forty-one genes conserved in both strains and seven and twenty-eight non-conserved genes in strains A8 and CIC9, respectively, were calculated. (DOCX 13 KB)
Additional file 2: Table S2: Annotations of the genes/proteins encoded in the HUS loci of the Geobacillus strains. The gene names, as well as the size and predicted functions of the encoded proteins are shown. The presence of an ortholog in a given Geobacillus strain is indicated by a black block, while absence of orthologs is denoted by a '-'. The Geobacillus strain numbers are those indicated in Table 2. Orthology to proteins in non-Geobacillus organisms was determined. (XLSX 130 KB)
Additional file 3: Table S3: Annotations of the additional genes/proteins encoded at the 5′ end of the echD-npd islands of Geobacillus strains. The gene names, as well as the size and predicted functions of the encoded proteins are shown. The presence of an ortholog in a given Geobacillus strain is indicated by a black block, while absence of orthologs is denoted by a '-'. The Geobacillus strain numbers are those indicated in Table 2. Orthology to proteins in non-Geobacillus organisms was determined. (XLSX 31 KB)
Acknowledgements
This project was partially funded through the University of Pretoria Postdoctoral Fellowship Programme and the National Research Foundation (NRF) Research Career Advancement (RCA) Fellowship Programme. The authors wish to acknowledge Prof Peter Biely for his crictical review of the manuscript.
Footnotes
Competing interests
DAM (Lucigen Corp.) and PJB (C5-6 Technologies Inc.) are employed at biotechnology companies with a vested interest in developing commercial products for cloning, expressing, and characterizing carbohydrate active enzymes. The authors declare that they have no competing interests.
Authors’ contributions
PDM, PJB, DAM and DAC conceived the study. PDM performed experiments and analyses, PDM, PJB, DAM and DAC wrote the original manuscript. All authors contributed to the final version.
Contributor Information
Pieter De Maayer, Email: Pieter.DeMaayer@up.ac.za.
Phillip J Brumm, Email: Pbrumm@C56technologies.com.
David A Mead, Email: Dmead@lucigen.com.
Don A Cowan, Email: Don.Cowan@up.ac.za.
References
- 1.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 2.Gibson LJ. The hierarchical structure and mechanics of plant materials. J R Soc Interface. 2012;9:2749–2766. doi: 10.1098/rsif.2012.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebringerova AZH, Heinze T. Hemicellulose. Adv Polymer Sci. 2005;185:1–67. doi: 10.1007/b136816. [DOI] [Google Scholar]
- 4.Ridley B, O'Neill M, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-relating signalling. Phytochemistry. 2001;57:929–967. doi: 10.1016/S0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 5.Shallom D, Shoham Y. Microbial hemicellulases. Curr Opin Microbiol. 2003;6:219–228. doi: 10.1016/S1369-5274(03)00056-0. [DOI] [PubMed] [Google Scholar]
- 6.Suurnäkki A, Tenkanen M, Buchert J, Viikari L. Hemicellulases in the bleaching of chemical pulps. Adv Biochem Eng Biotechnol. 1997;57:261–287. doi: 10.1007/BFb0102077. [DOI] [PubMed] [Google Scholar]
- 7.Mathlouthi N, Lallès JP, Lepercq P, Juste C, Larbier M. Xylanase and beta-glucanase supplementation improve conjugated bile acid fraction in intestinal contents and increase villus size of small intestine wall in broiler chickens fed a rye-based diet. J Anim Sci. 2002;80:2773–2779. doi: 10.2527/2002.80112773x. [DOI] [PubMed] [Google Scholar]
- 8.Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Mielenz JR. Ethanol production from biomass: technology and commercialization status. Curr Opin Microbiol. 2001;4:324–329. doi: 10.1016/S1369-5274(00)00211-3. [DOI] [PubMed] [Google Scholar]
- 10.Otieno DO, Ahring BK. The potential for oligosaccharide production from the hemicellulose fraction of biomasses through pretreatment processes: xylooligosaccharides (XOS), arabinooligosaccharides (AOS), and mannooligosaccharides (MOS) Carbohydr Res. 2012;360:84–92. doi: 10.1016/j.carres.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Cripps RE, Eley K, Leak DJ, Rudd B, Taylor M, Todd M, Boakes S, Martin S, Atkinson T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab Eng. 2009;11:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Balyaev, Ivanov MV. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int J Syst Evol Microbiol. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 13.Coorevits A, Dinsdale AE, Halket G, Lebbe L, De Vos P, Van Landschoot A, Logan NA. Taxonomic revision of the genus Geobacillus: emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. thermoglucosidans (nom. corrig., formerly 'thermoglucosidasius'); transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. Int J Syst Evol Microbiol. 2012;62:1470–1485. doi: 10.1099/ijs.0.030346-0. [DOI] [PubMed] [Google Scholar]
- 14.de Champdore M, Staiano M, Rossi M, D'Auria S. Proteins from extremophiles as stable tools for advanced biotechnological applications of high social interest. J R Soc Interface. 2007;4:183–191. doi: 10.1098/rsif.2006.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Zhang N, Zhao C, Lin B, Xie L, Huang Y. Characterization of a recombinant thermostable xylanase from hot spring thermophilic Geobacillus sp. TC-W7. J Microbiol Biotechnol. 2012;22:1388–1394. doi: 10.4014/jmb.1203.03045. [DOI] [PubMed] [Google Scholar]
- 16.Canakci S, Cevher Z, Inan K, Tokgoz M, Bahar F, Kacagan M, Sal FA, Belduz AO. Cloning, purification and characterization of an alkali-stable endoxylanase from thermophilic Geobacillus sp. 71. World J Microbiol Biotechnol. 2012;28:1981–1988. doi: 10.1007/s11274-011-1000-3. [DOI] [PubMed] [Google Scholar]
- 17.Barker IJ, Petersen L, Reilly PJ. Mechanism of xylobiose hydrolysis by GH43 beta-xylosidase. J Phys Chem B. 2010;114:15389–15393. doi: 10.1021/jp107886e. [DOI] [PubMed] [Google Scholar]
- 18.Teplitsky A, Shulami S, Moryles S, Shoham Y, Shoham G. Crystallization and preliminary X-ray analysis of an intracellular xylanase from Bacillus stearothermophilus T-6. Acta Crystallogr D Biol Crystallogr. 2000;56:181–184. doi: 10.1107/S0907444999013517. [DOI] [PubMed] [Google Scholar]
- 19.Shulami S, Gat O, Sonenshein AL, Shoham Y. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J Bacteriol. 1999;181:3695–3704. doi: 10.1128/jb.181.12.3695-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulami S, Zaide G, Zolotnitsky G, Langut Y, Feld G, Sonenshein AL, Shoham Y. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl Environ Microbiol. 2007;73:874–884. doi: 10.1128/AEM.02367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shulami S, Raz-Pasteur A, Tabachnikov O, Gilead-Gropper S, Shner I, Shoham Y. The L-arabinan utilization system of Geobacillus stearothermophilus. J Bacteriol. 2011;193:2838–2850. doi: 10.1128/JB.00222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hövel K, Shallom D, Niefind K, Belakhov V, Shoham G, Baasov T, Shoham Y, Schomburg D. Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase. EMBO J. 2003;22:4922–4932. doi: 10.1093/emboj/cdg494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alalouf O, Balazs Y, Volkinshtein M, Grimpel Y, Shoham G, Shoham Y. A new family of carbohydrate esterases is represented by a GDSL hydrolase/acetylxylan esterase from Geobacillus stearothermophilus. J Biol Chem. 2011;286:41993–42001. doi: 10.1074/jbc.M111.301051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boonmak C, Takahasi Y, Morikawa M. Draft genome sequence of Geobacillus thermoleovorans strain B23. Genome Announc. 2013;1:e0094413. doi: 10.1128/genomeA.00944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhd Sakaff MK, Abdul Rahman AY, Saito JA, Hou S, Alam M. Complete genome sequence of the thermophilic bacterium Geobacillus thermoleovorans CCB_US3_UF5. J Bacteriol. 2012;194:1239. doi: 10.1128/JB.06580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takami H, Takaki Y, Chee GJ, Nishi S, Shimamura S, Suzuki H, Matsui S, Uchiyama I. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 2004;32:6292–6303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Maayer P, Williamson CE, Vennard CT, Danson MJ, Cowan DA: The draft genomes of Geobacillus sp. CAMR5420 and CAMR12739.Genome Announc in press [DOI] [PMC free article] [PubMed]
- 28.Doi K, Mori K, Martono H, Nagayoshi Y, Fujino Y, Tashiro K, Kuhara S, Ohshima T. Draft genome sequence of Geobacillus kaustophilus GBlys, a lysogenic strain with bacteriophage OH2. Genome Announc. 2013;1:e0063413. doi: 10.1128/genomeA.00634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhalla A, Kainth AS, Sani RK. Draft genome sequence of lignocellulose-degrading thermophilic bacterium Geobacillus sp. strain WSUCF1. Genome Announc. 2013;1:e0059513. doi: 10.1128/genomeA.00595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiegand S, Rabausch U, Chow J, Daniel R, Streit WR, Liesegang H. Complete genome sequence of Geobacillus sp. strain GHH01, a thermophilic lipase-secreting bacterium. Genome Announc. 2013;1:e0009213. doi: 10.1128/genomeA.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shintani M, Ohtsubo Y, Fukuda K, Hosoyama A, Ohji S, Yamazoe A, Fujita N, Nagata Y, Tsuda M, Hatta T, Kimbara K. Complete genome sequence of the thermophilic polychlorinated biphenyl degrader Geobacillus sp. strain JF8 (NBRC 109937) Genome Announc. 2014;2:e0121313. doi: 10.1128/genomeA.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A. 2007;104:5602–5607. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao N, Ren Y, Wang W. Genome sequence of a thermophilic Bacillus, Geobacillus thermodenitrificans DSM465. Genome Announc. 2013;1:e0104613. doi: 10.1128/genomeA.01046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Caspers MP, Abee T, Siezen RJ, Kort R. Complete genome sequence of Geobacillus thermoglucosidans TNO-09.020, a thermophilic sporeformer associated with a dairy-processing environment. J Bacteriol. 2012;194:4118. doi: 10.1128/JB.00318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeigler DR. Application of a recN sequence similarity analysis to the identification of species within the bacterial genus Geobacillus. Int J Syst Evol Microbiol. 2005;55:1171–1179. doi: 10.1099/ijs.0.63452-0. [DOI] [PubMed] [Google Scholar]
- 36.Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/PL00006158. [DOI] [PubMed] [Google Scholar]
- 38.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. New York: Nova Science Publishers; 2011; 2011. pp. 61–78. [Google Scholar]
- 39.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 41.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2011;40:W445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 43.Khasin A, Alchanati I, Shoham Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol. 1993;59:1725–1730. doi: 10.1128/aem.59.6.1725-1730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba T, Shinke R, Nanmori T. Identification and characterization of clustered genes for thermostable xylan-degrading enzymes, beta-xylosidase and xylanase, of Bacillus stearothermophilus 21. Appl Environ Microbiol. 1994;60:2252–2258. doi: 10.1128/aem.60.7.2252-2258.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gat O, Lapidot A, Alchanati I, Regueros C, Shoham Y. Cloning and DNA sequence of the gene coding for Bacillus stearothermophilus T-6 xylanase. Appl Environ Microbiol. 1994;60:1889–1896. doi: 10.1128/aem.60.6.1889-1896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evol Biol. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel G, Barbeyron T, Kloareg B, Czjzek M. The family 6 carbohydrate-binding modules have coevolved with their appended catalytic modules toward similar substrate specificity. Glycobiology. 2009;19:615–623. doi: 10.1093/glycob/cwp028. [DOI] [PubMed] [Google Scholar]
- 49.Wang WY, Thomson JA. Nucleotide sequence of the celA gene encoding a cellodextrinase of Ruminococcus flavefaciens FD-1. Mol Gen Genet. 1990;222:265–269. doi: 10.1007/BF00633827. [DOI] [PubMed] [Google Scholar]
- 50.Saier MH. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/MMBR.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider E. ABC transporters catalyzing carbohydrate uptake. Res Microbiol. 2001;152:303–310. doi: 10.1016/S0923-2508(01)01201-3. [DOI] [PubMed] [Google Scholar]
- 52.Sá-Nogueira I, Nogueira TV, Soares S, de Lencastre H. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. 1997;143:957–969. doi: 10.1099/00221287-143-3-957. [DOI] [PubMed] [Google Scholar]
- 53.Ferreira MJ, Sa-Nogueira I. A multitask ATPase serving different ABC-type sugar importers in Bacillus subtilis. J Bacteriol. 2010;192:5312–5318. doi: 10.1128/JB.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chow V, Nong G, Preston JF. Structure, function, and regulation of the aldouronate utilization gene cluster from Paenibacillus sp. strain JDR-2. J Bacteriol. 2007;189:8863–8870. doi: 10.1128/JB.01141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teplitsky A, Shulami S, Moryles S, Zaide G, Shoham Y, Shoham G. Crystallization and preliminary X-ray analysis of alpha-D-glucuronidase from Bacillus stearothermophilus T-6. Acta Crystallogr D Biol Crystallogr. 1999;55:869–872. doi: 10.1107/S0907444998012918. [DOI] [PubMed] [Google Scholar]
- 56.Czjzek M, Ben David A, Bravman T, Shoham G, Henrissat B, Shoham Y. Enzyme-substrate complex structures of a GH39 beta-xylosidase from Geobacillus stearothermophilus. J Mol Biol. 2005;353:838–846. doi: 10.1016/j.jmb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Bravman T, Belakhov V, Solomon D, Shoham G, Henrissat B, Baasov T, Shoham Y. Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus stearothermophilus T-6. J Biol Chem. 2003;278:26742–26749. doi: 10.1074/jbc.M304144200. [DOI] [PubMed] [Google Scholar]
- 58.Bravman T, Zolotnitsky G, Belakhov V, Shoham G, Henrissat B, Baasov T, Shoham Y. Detailed kinetic analysis of a family 52 glycoside hydrolase: a beta-xylosidase from Geobacillus stearothermophilus. Biochemistry. 2003;42:10528–10536. doi: 10.1021/bi034505o. [DOI] [PubMed] [Google Scholar]
- 59.Brüx C, Ben-David A, Shallom-Shezifi D, Leon M, Niefind K, Shoham G, Shoham Y, Schomburg D. The structure of an inverting GH43 beta-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J Mol Biol. 2006;359:97–109. doi: 10.1016/j.jmb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Inácio JM, Correia IL, de Sa-Nogueira I. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology. 2008;154:2719–2729. doi: 10.1099/mic.0.2008/018978-0. [DOI] [PubMed] [Google Scholar]
- 61.Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Lansky S, Salama R, Solomon VH, Belrhali H, Shoham Y, Shoham G. Crystallization and preliminary crystallographic analysis of Abp, a GH27 beta-L-arabinopyranosidase from Geobacillus stearothermophilus. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2013;69:695–699. doi: 10.1107/S1744309113013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alhassid A, Ben-David A, Tabachnikov O, Libster D, Naveh E, Zolotnitsky G, Shoham Y, Shoham G. Crystal structure of an inverting GH43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate. Biochem J. 2009;422:73–82. doi: 10.1042/BJ20090180. [DOI] [PubMed] [Google Scholar]
- 64.Shi P, Chen X, Meng K, Huang H, Bai Y, Luo H, Yang P, Yao B. Distinct actions by Paenibacillus sp. strain E18 alpha-L-arabinofuranosidases and xylanase in xylan degradation. Appl Environ Microbiol. 2013;79:1990–1995. doi: 10.1128/AEM.03276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sjöström E. Wood chemistry, fundamentals and applications. London: Academic Press; 1993. [Google Scholar]
- 66.Biely P, Mastihubová M, Tenkanen M, Eyzaguirre J, Li XL, Vršanská M. Action of xylan deacetylating enzymes on monoacetyl derivatives of 4-nitrophenyl glycosides of beta-D-xylopyranose and alpha-L-arabinofuranose. J Biotechnol. 2011;151:137–142. doi: 10.1016/j.jbiotec.2010.10.074. [DOI] [PubMed] [Google Scholar]
- 67.Lansky S, Alalouf O, Solomon V, Alhassid A, Govada L, Chayan NE, Belrhali H, Shoham Y, Shoham G. Crystallization and preliminary crystallographic analysis of Axe2, an acetylxylan esterase from Geobacillus stearothermophilus. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2013;69:430–434. doi: 10.1107/S1744309113004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 69.Wilhelm M, Hollenberg CP. Nucleotide sequence of the Bacillus subtilis xylose isomerase gene: extensive homology between the Bacillus and Escherichia coli enzyme. Nucleic Acids Res. 1985;13:5717–5722. doi: 10.1093/nar/13.15.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nankai H, Hashimoto W, Murata K. Molecular identification of family 38 alpha-mannosidase of Bacillus sp. strain GL1, responsible for complete depolymerization of xanthan. Appl Environ Microbiol. 2002;68:2731–2736. doi: 10.1128/AEM.68.6.2731-2736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imanaka T, Ohta T, Sakoda H, Widhyastitui N, Matsuoka M. Cloning, nucleotide sequence, and efficient expression of the gene coding for thermostable aldehyde dehydrogenase from Bacillus stearothermophilus, and characterization of the enzyme. J Ferment Bioeng. 1993;76:161–167. doi: 10.1016/0922-338X(93)90001-O. [DOI] [Google Scholar]
- 72.Ni Y, Li CX, Ma HM, Zhang J, Xu JH. Biocatalytic properties of a recombinant aldo-keto reductase with broad substrate spectrum and excellent stereoselectivity. Appl Microbiol Biotechnol. 2011;89:1111–1118. doi: 10.1007/s00253-010-2941-4. [DOI] [PubMed] [Google Scholar]
- 73.Lei J, Zhou YF, Li LF, Su XD. Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis. Protein Sci. 2009;18:1792–1800. doi: 10.1002/pro.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeigler DR. The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology. 2014;160:1–11. doi: 10.1099/mic.0.071696-0. [DOI] [PubMed] [Google Scholar]
- 75.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Ser. 1999;41:95–98. [Google Scholar]
- 76.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siever F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.TreeBASE repository. http://purl.org/phylo/treebase/phylows/study/TB2:S16328
- 82.LabArchive repository. https://mynotebook.labarchives.com/share/BMC_Genomics_Geobacillus_hemicellulose_degradation_locus/MjkuOXw1MjM0OC8yMy9UcmVlTm9kZS82MTgzNzU0MDJ8NzUuOQ== (http://dx.doi.org/10.6070/H4MK69V3)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Mean G + C contents for the conserved and non-conserved genes in the echD-npd islands of Geobacillus sp. A8 and G. caldoxylolyticus CIC9. The mean G + C contents for forty-one genes conserved in both strains and seven and twenty-eight non-conserved genes in strains A8 and CIC9, respectively, were calculated. (DOCX 13 KB)
Additional file 2: Table S2: Annotations of the genes/proteins encoded in the HUS loci of the Geobacillus strains. The gene names, as well as the size and predicted functions of the encoded proteins are shown. The presence of an ortholog in a given Geobacillus strain is indicated by a black block, while absence of orthologs is denoted by a '-'. The Geobacillus strain numbers are those indicated in Table 2. Orthology to proteins in non-Geobacillus organisms was determined. (XLSX 130 KB)
Additional file 3: Table S3: Annotations of the additional genes/proteins encoded at the 5′ end of the echD-npd islands of Geobacillus strains. The gene names, as well as the size and predicted functions of the encoded proteins are shown. The presence of an ortholog in a given Geobacillus strain is indicated by a black block, while absence of orthologs is denoted by a '-'. The Geobacillus strain numbers are those indicated in Table 2. Orthology to proteins in non-Geobacillus organisms was determined. (XLSX 31 KB)


