Abstract
In the absence of the viral vif gene, human immunodeficiency virus (HIV) may be restricted by the APOBEC3G gene on chromosome 22. The role of the HIV Vif protein is to exclude host cell APOBEC3G from the budding virion. As APOBEC3G shows sequence homology to cytidine deaminases, it is presumed that in the absence of Vif, cytidine residues in the cDNA are deaminated yielding uracil. It is not known if additional proteins mediate APOBEC3G function or if deamination occurs in concert with reverse transcription. This report describes an in vitro assay showing that Baculovirus derived APOBEC3G alone extensively deaminates cDNA independently of reverse transcriptase. It reproduces the dinucleotide context typical of G → A hypermutants derived from a Δvif virus. By using an RNaseH– form of reverse transcriptase, it was shown that the cDNA has to be free of its RNA template to allow deamination. APOBEC3G deamination of dC or dCTP was not detected. In short, APOBEC3G is a single-stranded DNA cytidine deaminase capable of restricting retroviral replication.
INTRODUCTION
Despite nearly 20 years of HIV molecular biology, only recently has the role of the viral vif gene surfaced (1–6). The gene can be found in all but one of the lentiviruses — the subset of retroviruses harbouring the AIDS viruses, HIV-1 and HIV-2. Albeit referred to as an accessory gene, the Δvif phenotype is unequivocal: the virus cannot replicate in vivo (7). When Δvif viruses are grown in culture, the results are a little different. Some cell lines are permissive for growth of Δvif derivatives. However, others are non-permissive, suggesting restriction by a host gene product. Through a subtractive mRNA screen, it was shown that a single cellular gene restricted HIV replication in the absence of Vif (1). The gene sequence revealed itself to be APOBEC3G which shows strong homology to cytidine deaminases (1,8).
APOBEC3G is a member of a cluster of proteins of which APOBEC1 is the prototype. The protein is the catalytic component of the apolipoprotein B editing complex which specifically deaminates C6666 to U in the mRNA of apolipoprotein B (9). The most widely known member is activation induced cytidine deaminase (AID) which is intimately linked with rearrangement and somatic hypermutation of the immunoglobulin genes (10–12). The APOBEC3 cluster comprises at least seven genes on human chromosome 22 some of which are probably pseudogenes (8). In conjunction with accessory proteins, APOBEC1 specifically deaminates a single residue in an RNA molecule, apparently its physiological function. However, when expressed in E.coli, it can extensively edit DNA in a non-specific manner (13). Likewise, AID can extensively deaminate single-stranded DNA (14–17).
A recent flurry of papers have unambiguously linked the HIV Δvif phenotype to the genesis of G → A hypermutated genomes (2–6). G → A hypermutants are sequences in which G residues on the viral plus strand are monotonously substituted by A (18–27). As many as 680 Gs (31%) within a complete genome may be substituted (28). In a normal setting, the HIV Vif protein interacts with APOBEC3G and prevents it from becoming incorporated into the virion, the complex being routed to the proteasome (29–33). In the absence of Vif, APOBEC3G is packaged into the virion by an unknown mechanism. Given the kinship to cytidine deaminases, it is presumed, but not formally shown, that cytidine residues on the neo-synthesized minus DNA strand are massively deaminated yielding uracil. Copying the edited minus strand into plus strand DNA would copy U into A giving rise to the so-called G → A hypermutants (34–40).
When expressed in E.coli, human APOBEC3G acts as a DNA mutator, as do the phylogenetically related proteins APOBEC1, APOBEC3C and AID (13), although the relevance of this assay has been questioned recently (41). Recombinant E.coli derived APOBEC3G could partially deaminate cytidine residues in deoxyoligonucleotides (5,42). In contrast, Zhang et al. failed to find any deamination upon incubation of any polynucleic acid, whether it be dsDNA, ssDNA, RNA–DNA hybrids or RNA, again using human APOBEC3G purified from E.coli (3). While these findings may partly reflect experimental variables, it is possible that cofactors are necessary, as is the case for APOBEC1 (43), or that efficient deamination occurs in concert with reverse transcription. Certainly, everything is in place within the virion for an endogenous cDNA reaction made using a Δvif virus from a non-permissive cell line, yielding G → A hypermutants (2,4).
MATERIALS AND METHODS
RNA was in vitro transcribed from a 342 bp target sequence under the T7 promoter (44). The sequence corresponds to the hypervariable V1 and V2 regions of the gp120 coding sequence of HIV-1 Lai. Following DNase treatment and extraction, 2.4 µg RNA was reverse transcribed in a buffer reflecting the intracellular environment, notably 50 mM HEPES pH 7.0, 15 mM NaCl, 15 mM Mg(Aspartate)2, 130 mM KAcetate, 1 mM dithiothreitol and 5% PEG6000 (45). Reaction volumes, times and temperatures were 50 µl, 3 h and 37°C, respectively. Sufficient material for subsequent cloning was recovered by PCR in 12 cycles. The internal PCR fragment yielded a 202 bp target sequence. PCR products were cloned into the TOPO TA cloning vector and DNA sequenced using Big Dye terminators. HIV-1 reverse transcriptase (RT) was purchased from Amersham, MoMLV RNaseH+ RT from Invitrogen and Moloney murine leukemia virus (MoMLV) RNaseH– RT from Promega. Human APOBEC3G was expressed as a GST fusion protein using a recombinant Baculovirus and purified as described previously (8).
HPLC-based cytidine deamination assay
dC (500 µM) was incubated at 37°C for 2, 5 or 18 h in a total volume of 20 µl of Tris–HCl buffer (45 mM, pH 7.5) or buffer R (50 mM Tris, pH 7.5, 1 µM EDTA in the presence of 5 mM MgCl2 or 5 mM ZnCl2) or buffer S (50 mM HEPES, pH 7.5, 1 mM EDTA in the presence of 5 mM MgCl2 or 5 mM ZnCl2) or physiological buffer used above containing various amounts of APOBEC3G (0.5, 1 or 2 µg). Incubation of dC in the absence of enzyme was made to control for possible spontaneous deamination. Five to 10 µl samples were withdrawn and analysed by reverse phase HPLC on a C18 column (Kromasil, 5 µm–10 nm, 150 × 4.5 mm) using a Perkin-Elmer instrument and a diode array detector. The signal-to-noise ratio was ∼200:1. Samples (5–10 µl) were analysed using a 0–8% linear gradient of acetonitrile in 10 mM triethylammonium acetate buffer (pH 7.8) at 1 ml/min over 30 min. Under these conditions, retention times for dC and dU were 6.9 and 10.2 min, respectively. In the same manner, dCTP (1 mM) was incubated at 37°C in 20 µl of Tris–HCl or physiologic buffer containing various amounts of APOBEC3G (0.7 or 1.4 µg). Samples (5–10 µl) after 2, 5 and 18 h incubation time were analysed by HPLC on a C18 column (Kromasil, 5 µm–10 nm, 150 × 4.5 mm) using a linear gradient from 60 to 40% of buffer A in B at 1 ml/min over 30 min, then 40% buffer A for 30 min. Buffer A contained 10 mM NaH2PO4 pH 6.8, 100 mM tetrabutylammonium hydroxide while buffer B contained 100 mM NaH2PO4 pH 5.5, 3 mM tetrabutylammonium hydroxide and 25% methanol. Retention times for dCTP and dUTP were 28 and 32 min, respectively. dC, dU and dUTP were purchased from Sigma–Aldrich while ultrapure dCTP was from Amersham.
RESULTS
As a starting point, we sought to explore the hypothesis that APOBEC3G deamination occurred in concert with reverse transcription. RNA was in vitro transcribed from a DNA fragment corresponding to the V1V2 hypervariable regions of the HIV-1 LAI envelope protein, while cDNA was synthesized using commercially available cloned HIV-1 RT as previously described (44). Baculovirus derived human APOBEC3G (8) was added at the same time as RT. cDNA was then recovered by PCR.
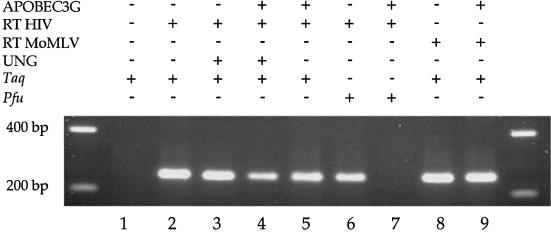
If cytidine was deaminated in the context of DNA yielding uracil then there are two ways to test this. Firstly, if PCR is performed with Taq polymerase then product should be readily recovered. However, if Pfu polymerase is used, which does not readily copy templates bearing uracil (46), then amplification should be inefficient. Secondly, the product should be sensitive to uracil DNA N-glycosylase (UNG). Accordingly prior treatment with UNG should reduce the efficiency of PCR due to gaps in the template (47).
Figure 1 shows that although PCR product was readily obtained when Taq polymerase was used to amplify APOBEC3G-treated cDNA (lane 5), Pfu polymerase had great difficulty in amplifying cDNA following APOBEC3G treatment (cf. lanes 6 and 7). Furthermore, prior treatment of the cDNA by UNG resulted in reduced recovery of PCR product by Taq polymerase (cf. lanes 3 and 4). That some product was obtained using Taq polymerase after UNG treatment (lane 4) may be expected given that PCR allows re-assembly of fragmented DNA (48). These observations indicate that APOBEC3G-treated cDNA harboured uracil residues. If RT from the MoMLV was used, cDNA synthesis in the presence of APOBEC3G was just as efficient as the HIV-1 RT (cf. lanes 5 and 9).
Figure 1.
Agarose gel of PCR products following reverse transcription in the presence or absence of APOBEC3G.
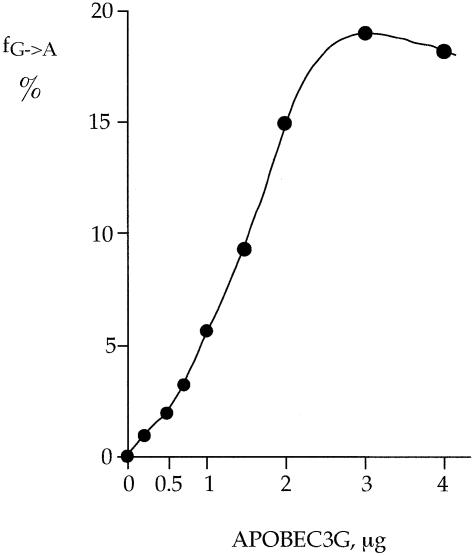
Taq PCR products were cloned and ∼17–70 clones sequenced from each sample, a representative selection of sequences being shown in Figure 2. As can be seen, the sequences were peppered by G → A substitutions with respect to the retroviral plus strand taken as reference. Mechanistically, they correspond to multiple deamination of cytidines in the minus DNA strand. The frequency of deamination increased in a dose-dependent manner reaching a plateau of around 20% deamination of all target cytidines at 3 µg of APOBEC3G per reaction (Fig. 3 and Table 1, reactions 3–11). Sequencing of products having undergone UNG treatment (Fig. 1, lane 4) showed a much reduced C → U substitution frequency reflecting contra-selection of templates lacking uracil (data not shown).
Figure 2.
A selection of APOBEC3G hypermutated sequences. Only a 90 bp segment of the 202 bp target sequence is shown for clarity. However, it included 25 of the 42 cytidine targets (60%). The sequence is given with respect to the retroviral plus strand even though the complementary, or minus strand, is modified. Potential targets are indicated by dots above the sequence. Only differences among hypermutated sequences are shown. Three representative sequences from eight samples with increasing amounts of APOBEC3G per reaction are given. The number of substitutions per sequence is indicated to the right. Some sites are exquisitely sensitive to deamination while others are highly refractory.
Figure 3.
Deoxycytidine deamination is proportional to APOBEC3G concentration. Deamination reaches a plateau by 3 µg APOBEC3G per reaction. For each sample, the substitution frequency (fG→A) is the average over 42 targets for between 17 and 70 sequences.
Table 1. Deoxycytidine deamination frequencies as a function of reaction conditions.
| Reaction no. | Reaction conditions | No. of clones analysed | No. of G → A subs | fG → A/base | Fold increase |
|---|---|---|---|---|---|
| 1 |
RNA + 1 µg APOBEC3G, heated/ice then + HIV RT |
19 |
0 |
<1.2 × 10–3 |
∼1 |
| 2 |
RNA+HIV RT + 1 µg APOBEC3G, heated/ice then + HIV RT |
19 |
0 |
<1.2 × 10–3 |
∼1 |
| 3 |
HIV RT cDNA |
27 |
0 |
<9 × 10–4 |
∼1 |
| 4 |
HIV RT cDNA + 0.25 µg APOBEC3G |
56 |
27 |
1.1 × 10–2 |
12 |
| 5 |
HIV RT cDNA + 0.50 µg APOBEC3G |
58 |
50 |
2.1 × 10–2 |
23 |
| 6 |
HIV RT cDNA + 0.75 µg APOBEC3G |
52 |
78 |
3.6 × 10–2 |
40 |
| 7 |
HIV RT cDNA + 1 µg APOBEC3G |
57 |
134 |
5.6 × 10–2 |
62 |
| 8 |
HIV RT cDNA + 1.5 µg APOBEC3G |
70 |
256 |
8.7 × 10–2 |
97 |
| 9 |
HIV RT cDNA + 2 µg APOBEC3G |
53 |
307 |
1.4 × 10–1 |
156 |
| 10 |
HIV RT cDNA + 3 µg APOBEC3G |
41 |
320 |
1.8 × 10–1 |
200 |
| 11 |
HIV RT cDNA + 4 µg APOBEC3G |
44 |
313 |
1.7 × 10–1 |
189 |
| 12 |
HIV RT cDNA, heated/ice then + 1 µg APOBEC3G |
25 |
65 |
6.1 × 10–2 |
68 |
| 13 |
HIV RT cDNA, then + 1 µg APOBEC3G |
26 |
80 |
7.3 × 10–2 |
81 |
| |
|
|
|
|
|
| 14 |
MoMLV RT cDNA |
19 |
0 |
<1.2 × 10–3 |
∼1 |
| 15 |
MoMLV RT cDNA + 1 µg APOBEC3G |
48 |
132 |
6.5 × 10–2 |
54 |
| 16 |
MoMLV RT RNaseH– cDNA |
19 |
1 |
1.2 × 10–3 |
1 |
| 17 |
MoMLV RT RNaseH– cDNA + 1 µg APOBEC3G |
19 |
0 |
<1.2 × 10–3 |
∼1 |
| 18 |
MoMLV RT cDNA, heated/ice then + 1 µg APOBEC3G |
19 |
37 |
4.6 × 10–2 |
38 |
| 19 |
MoMLV RT cDNA, then + 1 µg APOBEC3G |
23 |
72 |
7.4 × 10–2 |
61 |
| 20 |
MoMLV RT RNaseH– cDNA, heated/ice then + 1 µg APOBEC3G |
17 |
3 |
4.2 × 10–3 |
3.5 |
| 21 | MoMLV RT RNaseH– cDNA, then + 1 µg APOBEC3G | 19 | 0 | <1.2 × 10–3 | ∼1 |
Cytosine deamination occurs preferentially in CpC
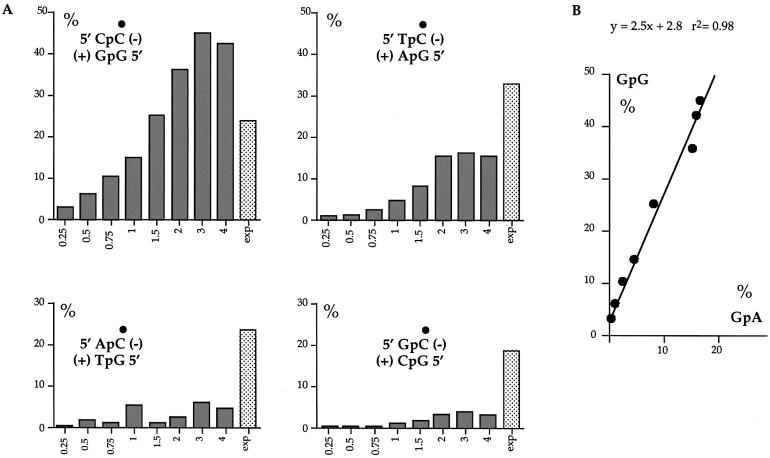
Not all target Cs were deaminated at the same efficiency as is apparent from Figure 2. Indeed some were highly refractory. Analysis of the local sequence context showed that substitutions occurred preferentially in the context 5′GpG/CpC5′ (plus/minus DNA strand, where the underlined base indicated the modified C or its complementary G, Fig. 4A). The hierarchy 5′GpG > GpA > GpT ∼ GpC was apparent at all concentrations of APOBEC3G. Furthermore, there was an overall linear relationship between the substitution frequencies in 5′GpG/CpC5′ and 5′GpA/CpT5′ with the former being ∼2.5× more frequent (Fig. 4B). Informative analysis of the 3′ nucleotide context was precluded given the single CpG target. These findings recapitulate the dinucleotide preference seen among G → A hypermutated genomes on a HIV-1 Δvif background (2–6).
Figure 4.
Sequence context among sites of APOBEC3G deamination. (A) The ordinates represent the substitution frequency as a function of the 5′ nucleotide. The dot indicates the deaminated C residue. Deamination in the minus strand occurs preferentially in local sequence context in the order 5′CpC > TpC > ApC ∼ GpC, or on the plus strand 5′GpG > GpA > GpT ∼ GpC. This sequence context is typical for hypermutated genomes derived from Δvif viruses grown on APOBEC3G producing cells (2–6,13). The lightly stippled bars indicate the frequency expected assuming no dinucleotide bias. A χ2 analysis showed that for reactions with 2, 3 and 4 µg of APOBEC3G, the observed 5′CpC frequencies proved highly significant (P < 0.001) compared to the expected values. Informative analysis of the 3′ nucleotide context is precluded given that the single CpG target is also a hot spot for deamination. (B) Overall GpG substitution frequencies increase linearly with those in GpA sites.
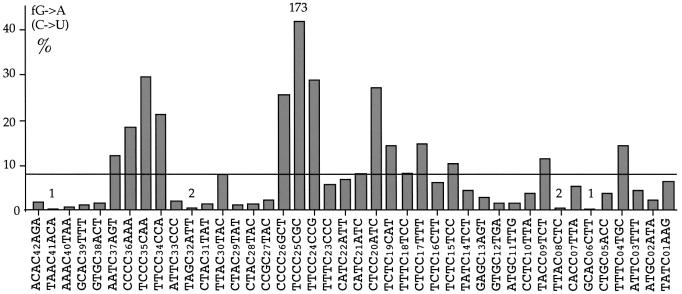
When a large number of clones was sequenced, none of the 42 potential cytidine targets in template cDNA was totally refractory, although two targets (C06 and C41) were deaminated but once among 431 sequences (Fig. 5). As can be seen, the deamination frequencies varied by up to a factor of 173 (cf. C25 versus C06 or C41). Although deamination was particularly evident within oligocytidine tracts, a more detailed analysis of the most frequent hot and cold spots revealed more subtle context effects (Table 2). The region 5′ to the modified base tended to be rich in pyrimidine residues (HHYYYC where H = T, C or A, modified base underlined) which is an extension of the 5′GpG/CpC5′ and 5′GpA/CpT5′ bias seen in Figure 4A. In contrast, cold spots were characterized by the motif HRCH which recapitulates the aversion for deamination within the 5′GpT/CpA5′ and 5′GpC/CpG5′ dinucleotides modulated by surrounding pyrimidine residues (Table 2). Interestingly, these hot and cold spot motifs are generally the inverse of those seen for the phylogenically related cytidine deaminase AID [Table 2, (15)].
Figure 5.
Deamination frequencies across the target sequence. The base specific deamination frequencies among a collection of 431 sequences from all the reactions are given as a function of local sequence context. No site was totally refractory. Only two sequences harboured a single substituted cytidine at positions C06 and C41, indicated by 1 over the site, while C08 and C32 were substituted only twice in different clones. In contrast, the maximum site deamination frequency was ∼40% for C25 (173 substitutions). Hence, the maximum ratio of site deamination frequencies is 173:1. The horizontal bar indicates the average site deamination frequency assuming no effect of sequence context. Oligocytidine tracts are particularly vulnerable.
Table 2. Sequence contexts for cytidine deamination.
| Hot spots |
|
Cold spots |
|
|---|---|---|---|
| Site | No. C → U | Site | No. C → U |
| TTTCCCCGC |
173 |
TAACACA |
1 |
| ATTCCCCAA |
120 |
GCACCTT |
1 |
| ATTTCCCCG |
117 |
TAGCATT |
2 |
| TTCTCCATC |
111 |
TTACCTC |
2 |
| TTCCCCGCT |
106 |
GCACTTT |
4 |
| CATTCCCCA |
86 |
CTACTAT |
4 |
| TTCCCCAAA |
75 |
AAACTAA |
5 |
| CTCTCCTTT |
63 |
CTACTAC |
5 |
| TTTCTCCAT |
60 |
CTACTAT |
6 |
| TCTTTCTGC |
59 |
GTGCTGA |
7 |
| |
|
ATGCTTG |
7 |
| |
|
|
|
| HHYYYCNNN |
|
NHRCHNN |
|
| WRC | SYC |
The 10 hottest and 11 coldest spots are given along with their substitution frequencies among 431 sequences. The consensus sequence motifs are given where R = G or A; Y = T or C; H = T, C or A; W = T or A; S = C or G; N = any base. Given separately at the bottom are the corresponding sequence motifs for the phylogenetically related AID cytidine deaminase derived from an in vitro assay. From Pham et al. (15).
APOBEC3G functions independently of RT
To resolve the question as to whether cytidine deamination occurs in concert with reverse transcription, or whether RT was a cofactor, cDNA was first made, the RT inactivated by heating at 95°C for 15 min and then incubated with APOBEC3G at 37°C for 3 h. The substitution frequency was indistinguishable to that determined without inactivation (Table 1, cf. reactions 12 and 13) indicating that APOBEC3G acted independently of RT. When the RT from MoMLV was used, the substitution frequency was comparable to that obtained with the HIV-1 RT (Table 1, cf. reactions 7 and 15). This result would be expected if APOBEC3G acted independently of cDNA synthesis. That APOBEC3G function is not coupled to reverse transcription agrees with some reports showing that DNA synthesis in a HIV-1 Δvif background is unimpeded (49).
Single-stranded DNA is the substrate
During reverse transcription, the RNaseH activity of RT degrades template RNA in the context of RNA/DNA hybrids. To test whether RNA template degradation was a prerequisite for APOBEC3G deamination, cDNA was synthesized using a commercially available RNaseH– form of MoMLV RT. The sequence data indicated that, while the normal MoMLV RT gave rise to G → A hypermutants, the RNaseH– form did not (Table 1, reactions 15 and 17). Using this in vitro assay, it was possible to reproduce the previous observation that APOBEC3G does not deaminate RNA (Table 1, reactions 1 and 2). Together, these observations show that APOBEC3G is active only on ssDNA.
PCR product from reaction 1 (Table 1) was heated to 95°C for 15 min, quenched on ice after which 1 µg of APOBEC3G was added for 3 h at 37°C. As can be seen from Figure 6, both G → A and C → T hypermutants were produced as expected were APOBEC3G to deaminate complementary single DNA strands. In the literature, there is a single case of C → T hypermutation on the HIV-1 DNA plus strand (27). In view of the present findings, it would seem that this occurred via APOBEC3G deamination of a single-stranded plus strand.
Figure 6.
Deamination of complementary ssDNA strands. dsDNA was denatured at 95°C for 15 min then quenched on ice. APOBEC3G was then added. Potential C targets are denoted by dots and asterisks corresponding to the minus and plus strands, respectively. The combined dinucleotide preferences for the two strands were GpG 51%, GpA 28%, GpT 8%, GpC 13%, which is typical of data in Figure 4.
dC or dCTP are poor substrates for APOBEC3G
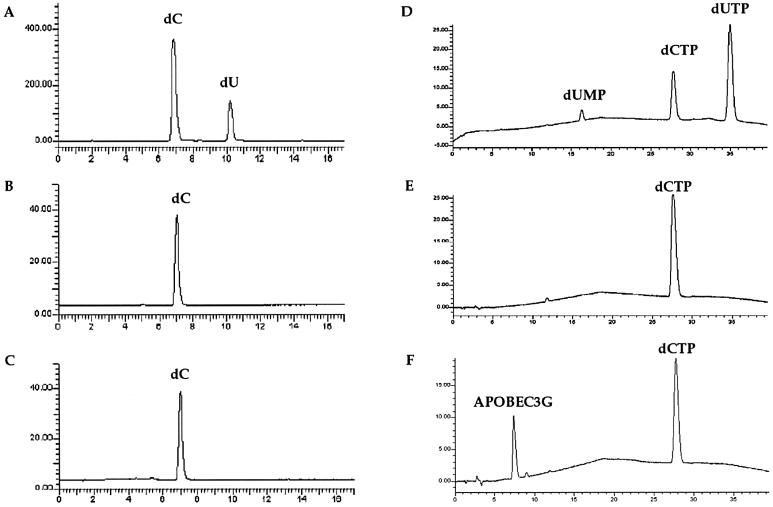
Some of the APOBEC proteins are able to deaminate deoxycytidine in addition to RNA or DNA deamination. One report indicated that E.coli-purified APOBEC3G could deaminate dC (3), although this could be due to contaminating E.coli deoxycytidine deaminase (42). Given the evident activity of the Baculovirus-purified APOBEC3G used here, a potential dC deamination activity was sought in a variety of different buffer conditions, including those used above and in other reports (3). As can be seen in Figure 7, no cytidine deamination activity was detectable when the reaction products were analysed by HPLC with a signal-to-noise ratio of ∼200:1. Similarly, dCTP proved not to be a substrate either. When cDNA was treated by APOBEC3G in the presence of 1 mM dCTP, there was no change in the deamination frequency (data not shown), in keeping with the above finding. While these are negative findings, it would appear that the major activity of APOBEC3G is ssDNA cytidine deamination.
Figure 7.
APOBEC3G does not efficiently deaminate dC or dCTP. HPLC elution profiles for reaction products, the ordinate describes product detection (mV), the abscissa the elution time (min). (A and D) Control samples; (B and E) 500 µM dC and 1 mM dCTP incubation controls for spontaneous deamination in 45 mM Tris–HCl pH 7.5 for 18 and 5 h, respectively; (C) 500 µM dC incubation with 1 µg APOBEC3G for 18 h in 45 mM Tris–HCl pH 7.5; (F) 1 mM dCTP incubation with 1.4 µg APOBEC3G for 5 h in 45 mM Tris–HCl pH 7.5. Identical results were obtained using buffer R, S with or without 5 mM MgCl2 or 5 mM ZnCl2.
DISCUSSION
RNA viruses and retroviruses replicate with little or no proof reading. Consequently, their genome sizes are limited by an error threshold beyond which there is a collapse of information (50). By using drugs, it has been possible to push some viruses ‘over the edge’ resulting in inactivation (51,52). The observation of hypermutated RNA virus and retroviral genomes suggests that nature has exploited the vulnerability of viral genomes unable to undertake proof reading (27,53). As such, the ssDNA cytidine deamination activity of APOBEC3G can be seen as an innate immune response to retroviruses. The present findings recapitulate the mutagenic effect of human APOBEC3G when over-expressed in ung-1 E.coli (13).
Recently, the related enzyme AID was shown to specifically deaminate cytidine residues in the context of ssDNA but not dsDNA or RNA (14–17). Thus, despite 54 and 66% amino acid variation between AID and the amino and carboxy domains of APOBEC3G respectively (8), these two human proteins have the same general polynucleotide substrate specificity. A difference between the two is the local sequence contexts for hot and cold spots which are almost the exact opposites [Table 2, (15)]. APOBEC3G deaminates efficiently in the context of 5′CpC and to a lesser extent 5′TpC, while AID favours deamination in 5′GpC and 5′ApC dinucleotides. Hence, the efficiency of deamination depends on both the sequence and the enzyme.
With this in mind, there is small discordance between the sequence context surrounding APOBEC3G deamination in vitro and hypermutants generated in vivo. Among naturally arising G → A retroviral hypermutants, there appears to be two types that can be distinguished by local sequence context. Most frequently substitutions occur in GpA > GpG (TpC > CpC) (18,20,22,23,26–28,54). Somewhat less frequently are hypermutants whereby GpG > GpA (CpC > TpC) (21,24,25). Both observations hold for the same virus, i.e. HIV-1, and can be found in the same DNA sample. For the non-immunodeficiency retroviruses, the data bases are much smaller. However, for the moment G → A hypermutants among CAEV (23), EIAV (19), HTLV (55) and HBV (20), all show a GpA > GpG substitution preference. For HIV-1 Δvif viruses grown on non-permissive cells, the preferred context is GpG > GpA (2–6), which finds expression in the in vitro data described above. As deamination efficiency depends upon both sequence context and the enzyme, naturally arising G → A hypermutants in which substitutions occur preferentially within GpA rather than GpG might reflect APOBEC3G allelic variation or, more interestingly, the activity of other APOBEC3 family members.
The dose–response relationship between substitution frequency and APOBEC3G concentration, as well as the finding that deamination occurs post cDNA synthesis, indicate that the lower G → A hypermutation frequencies observed for three other retroviruses, MoMLV, spleen necrosis virus and human T cell leukemia virus (HTLV) compared to HIV-1 (26,55,56) are likely due to a smaller number of APOBEC3G-like molecules packaged within the virions. In contrast, the heavily G → A hypermutated hepatitis B virus (HBV) sequences argue that large numbers of such molecules can, under certain circumstances, be packaged into the HBV virion (20). That such G → A hypermutated genomes are so rare for these four viruses remains to be elucidated. The case of HTLV (55) is particularly intriguing for it infects the same target cell as HIV, the CD4+ T lymphocyte. Presumably, it too must have evolved a strategy to overcome the effects of APOBEC3G.
Given that APOBEC3G acts on neo-synthesized minus strand DNA on a lentiviral Δvif background, any G → A hypermutants must be excluded when establishing RT point mutation rates for they have nothing to do with RT fidelity. Inspection of the retroviral mutants used to derive mutation rates indicates that there are a few hypermutants while G → A substitutions in GpG and GpA are the dominant mutations in the mutant spectrum (57,58). In view of this, it is likely that some retroviral mutation rates may be somewhat less than initially thought. Whether APOBEC3G deamination contributes to HIV sequence evolution as has been suggested (3), even in a small way, is a more complicated issue in as much as it requires maintenance of a defective phenotype (i.e. Δvif). Nevertheless, some genes are surprisingly robust to G → A hypermutation (59).
In conclusion, APOBEC3G is indeed a highly potent single-stranded DNA cytidine deaminase that functions independently of RT. That its function may, by analogy with APOBEC1, also be modulated by other proteins does not preclude it from constituting a phenomenal barrier itself to reverse transcription in some cell types.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Pasteur Institute. R.S. is a recipient of a Boehringer Ingelheim Fonds Fellowship.
REFERENCES
- 1.Sheehy A.M., Gaddis,N.C., Choi,J.D. and Malim,M.H. (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature, 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 2.Lecossier D., Bouchonnet,F., Clavel,F. and Hance,A.J. (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science, 300, 1112. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H., Yang,B., Pomerantz,R.J., Zhang,C., Arunachalam,S.C. and Gao,L. (2003) The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature, 424, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangeat B., Turelli,P., Caron,G., Friedli,M., Perrin,L. and Trono,D. (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature, 424, 99–103. [DOI] [PubMed] [Google Scholar]
- 5.Harris R.S., Bishop,K.N., Sheehy,A.M., Craig,H.M., Petersen-Mahrt,S.K., Watt,I.N., Neuberger,M.S. and Malim,M.H. (2003) DNA deamination mediates innate immunity to retroviral infection. Cell, 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 6.Mariani R., Chen,D., Schrofelbauer,B., Navarro,F., Konig,R., Bollman,B., Munk,C., Nymark-McMahon,H. and Landau,N.R. (2003) Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell, 114, 21–31. [DOI] [PubMed] [Google Scholar]
- 7.Desrosiers R.C., Lifson,J.D., Gibbs,J.S., Czajak,S.C., Howe,A.Y., Arthur,L.O. and Johnson,R.P. (1998) Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol., 72, 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarmuz A., Chester,A., Bayliss,J., Gisbourne,J., Dunham,I., Scott,J. and Navaratnam,N. (2002) An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics, 79, 285–296. [DOI] [PubMed] [Google Scholar]
- 9.Teng B., Burant,C.F. and Davidson,N.O. (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science, 260, 1816–1819. [DOI] [PubMed] [Google Scholar]
- 10.Muramatsu M., Kinoshita,K., Fagarasan,S., Yamada,S., Shinkai,Y. and Honjo,T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- 11.Revy P., Muto,T., Levy,Y., Geissmann,F., Plebani,A., Sanal,O., Catalan,N., Forveille,M., Dufourcq-Labelouse,R., Gennery,A., Tezcan,I., Ersoy,F., Kayserili,H., Ugazio,A.G., Brousse,N., Muramatsu,M., Notarangelo,L.D., Kinoshita,K., Honjo,T., Fischer,A. and Durandy,A. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell, 102, 565–575. [DOI] [PubMed] [Google Scholar]
- 12.Petersen-Mahrt S.K., Harris,R.S. and Neuberger,M.S. (2002) AID mutates E.coli suggesting a DNA deamination mechanism for antibody diversification. Nature, 418, 99–103. [DOI] [PubMed] [Google Scholar]
- 13.Harris R.S., Petersen-Mahrt,S.K. and Neuberger,M.S. (2002) RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell, 10, 1247–1253. [DOI] [PubMed] [Google Scholar]
- 14.Bransteitter R., Pham,P., Scharff,M.D. and Goodman,M.F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA, 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham P., Bransteitter,R., Petruska,J. and Goodman,M.F. (2003) Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature, 424, 103–107. [DOI] [PubMed] [Google Scholar]
- 16.Ramiro A.R., Stavropoulos,P., Jankovic,M. and Nussenzweig,M.C. (2003) Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immun., 4, 452–456. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri J., Tian,M., Khuong,C., Chua,K., Pinaud,E. and Alt,F.W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature, 422, 726–730. [DOI] [PubMed] [Google Scholar]
- 18.Janini M., Rogers,M., Birx,D.R. and McCutchan,F.E. (2001) Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol., 75, 7973–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry S.T., Flaherty,M.T., Kelley,M.J., Clabough,D.L., Tronick,S.R., Coggins,L., Whetter,L., Lengel,C.R. and Fuller,F. (1992) The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J. Virol., 66, 4085–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther S., Sommer,G., Plikat,U., Iwanska,A., Wain-Hobson,S., Will,H. and Meyerhans,A. (1997) Naturally occurring hepatitis B virus genomes bearing the hallmarks of retroviral G → A hypermutation. Virology, 235, 104–108. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgibbon J.E., Mazar,S. and Dubin,D.T. (1993) A new type of G → A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retroviruses, 9, 833–838. [DOI] [PubMed] [Google Scholar]
- 22.Gao F., Yue,L., White,A.T., Pappas,P.G., Barchue,J., Hanson,A.P., Greene,B.M., Sharp,P.M., Shaw,G.M. and Hahn,B.H. (1992) Human infection by genetically diverse SIVsm-related HIV-2 in West Africa. Nature, 358, 495–499. [DOI] [PubMed] [Google Scholar]
- 23.Wain-Hobson S., Sonigo,P., Guyader,M., Gazit,A. and Henry,M. (1995) Erratic G → A hypermutation within a complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology, 209, 297–303. [DOI] [PubMed] [Google Scholar]
- 24.Vartanian J.P., Meyerhans,A., Sala,M. and Wain-Hobson,S. (1994) G → A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc. Natl Acad. Sci. USA, 91, 3092–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch V., Riedel,N. and Mullins,J.I. (1987) The genome organization of STLV-3 is similar to that of the AIDS virus except for a truncated transmembrane protein. Cell, 49, 307–319. [DOI] [PubMed] [Google Scholar]
- 26.Pathak V.K. and Temin,H.M. (1990) Broad spectrum of in vivo forward mutations, hypermutations and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts and hypermutations. Proc. Natl Acad. Sci. USA, 87, 6019–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartanian J.P., Meyerhans,A., Asjo,B. and Wain-Hobson,S. (1991) Selection, recombination and G → A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol., 65, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vartanian J.P., Henry,M. and Wain-Hobson,S. (2002) Sustained G → A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol., 83, 801–805. [DOI] [PubMed] [Google Scholar]
- 29.Sheehy A.M., Gaddis,N.C. and Malim,M.H. (2003) The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med., 9, 1404–1407. [DOI] [PubMed] [Google Scholar]
- 30.Yu X., Yu,Y., Liu,B., Luo,K., Kong,W., Mao,P. and Yu,X.F. (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science, 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- 31.Marin M., Rose,K.M., Kozak,S.L. and Kabat,D. (2003) HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med., 9, 1398–1403. [DOI] [PubMed] [Google Scholar]
- 32.Stopak K., de Noronha,C., Yonemoto,W. and Greene,W.C. (2003) HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell, 12, 591–601. [DOI] [PubMed] [Google Scholar]
- 33.Conticello S.G., Harris,R.S. and Neuberger,M.S. (2003) The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol., 13, 2009–2013. [DOI] [PubMed] [Google Scholar]
- 34.Gu Y. and Sundquist,W.I. (2003) Good to CU. Nature, 424, 21–22. [DOI] [PubMed] [Google Scholar]
- 35.KewalRamani V.N. and Coffin,J.M. (2003) Weapons of mutational destruction. Science, 301, 923–925. [DOI] [PubMed] [Google Scholar]
- 36.Goff S.P. (2003) Death by deamination: a novel host restriction system for HIV-1. Cell, 114, 281–283. [DOI] [PubMed] [Google Scholar]
- 37.Harris R.S., Sheehy,A.M., Craig,H.M., Malim,M.H. and Neuberger,M.S. (2003) DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immun., 4, 641–643. [DOI] [PubMed] [Google Scholar]
- 38.Cullen B.R. (2003) HIV-1 Vif: countering innate antiretroviral defences. Mol. Ther., 8, 525–527. [DOI] [PubMed] [Google Scholar]
- 39.Neuberger M.S., Harris,R.S., Di Noia,J. and Petersen-Mahrt,S.K. (2003) Immunity through DNA deamination. Trends Biochem. Sci., 28, 305–312. [DOI] [PubMed] [Google Scholar]
- 40.Vartanian J.P., Sommer,P. and Wain-Hobson,S. (2003) Death and the retrovirus. Trends Mol. Med., 9, 409–413. [DOI] [PubMed] [Google Scholar]
- 41.Eto T., Kinoshita,K., Yoshikawa,K., Muramatsu,M. and Honjo,T. (2003) RNA-editing cytidine deaminase Apobec-1 is unable to induce somatic hypermutation in mammalian cells. Proc. Natl Acad. Sci. USA, 100, 12895–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beale R.C., Petersen-Mahrt,S.K., Watt,I.N., Harris,R.S., Rada,C. and Neuberger,M.S. (2004) Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol., 337, 585–596. [DOI] [PubMed] [Google Scholar]
- 43.Anant S., Henderson,J.O., Mukhopadhyay,D., Navaratnam,N., Kennedy,S., Min,J. and Davidson,N.O. (2001) Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J. Biol. Chem., 276, 47338–47351. [DOI] [PubMed] [Google Scholar]
- 44.Martinez M.A., Vartanian,J.P. and Wain-Hobson,S. (1994) Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc. Natl Acad. Sci. USA, 91, 11787–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sala M., Wain-Hobson,S. and Schaeffer,F. (1995) Human immunodeficiency virus type 1 reverse transcriptase tG:T mispair formation on RNA and DNA templates with mismatched primers: a kinetic and thermodynamic study. EMBO J., 14, 4622–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greagg M.A., Fogg,M.J., Panayotou,G., Evans,S.J., Connolly,B.A. and Pearl,L.H. (1999) A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA, 96, 9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pang J., Modlin,J. and Yolken,R. (1992) Use of modified nucleotides and uracil-DNA glycosylase (UNG) for the control of contamination in the PCR-based amplification of RNA. Mol. Cell. Probes, 6, 251–256. [DOI] [PubMed] [Google Scholar]
- 48.Stemmer W.P. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature, 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 49.Gaddis N.C., Chertova,E., Sheehy,A.M., Henderson,L.E. and Malim,M.H. (2003) Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol., 77, 5810–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eigen M. and Biebricher,C.K. (1988) Sequence space and quasispecies distribution. In Domingo,E., Holland,J.J. and Ahlquist,P. (eds), RNA Genetics, Volume 3: Variability of RNA Genomes. CRC Press Inc., Boca Raton, FL, pp. 211–245. [Google Scholar]
- 51.Crotty S., Maag,D., Arnold,J.J., Zhong,W., Lau,J.Y., Hong,Z., Andino,R. and Cameron,C.E. (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med., 6, 1375–1379. [DOI] [PubMed] [Google Scholar]
- 52.Loeb L.A., Essigmann,J.M., Kazazi,F., Zhang,J., Rose,K.D. and Mullins,J.I. (1999) Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl Acad. Sci. USA, 96, 1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cattaneo R., Schmid,A., Eschle,D., Baczko,K., ter Meulen,V. and Billeter,M.A. (1988) Biased hypermutation and other genetic changes in defective measles virus in human brain infections. Cell, 55, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borman A.M., Quillent,C., Charneau,P., Kean,C.M. and Clavel,F. (1995) A highly defective HIV group O provirus: evidence for the role of local sequence determinants in hypermutation during negative strand DNA synthesis. Virology, 208, 601–609. [DOI] [PubMed] [Google Scholar]
- 55.Vartanian J.P., Plikat,U., Henry,M., Mahieux,R., Guillemot,L., Meyerhans,A. and Wain-Hobson,S. (1997) HIV genetic variation is directed and restricted by DNA precursor availability. J. Mol. Biol., 270, 139–151. [DOI] [PubMed] [Google Scholar]
- 56.Mansky L.M. (2000) In vivo analysis of Human T-Cell Leukemia Virus Type 1 reverse transcription accuracy. J. Virol., 74, 9525–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansky L.M. (1996) The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology, 222, 391–400. [DOI] [PubMed] [Google Scholar]
- 58.Mansky L.M. and Temin,H.M. (1995) Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol., 69, 5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez M.A., Pezo,V., Marliere,P. and Wain-Hobson,S. (1996) Exploring the functional robustness of an enzyme by in vitro evolution. EMBO J., 15, 1203–1210. [PMC free article] [PubMed] [Google Scholar]