Abstract
Sen1p in Saccharomyces cerevisiae is a Type I DNA/RNA helicase. Mutations in the helicase domain perturb accumulation of diverse RNA classes, and Sen1p has been implicated in 3′ end formation of non-coding RNAs. Using a combination of global and candidate-specific two hybrid screens, eight proteins were identified that interact with Sen1p. Interactions with three of the proteins were analyzed further: Rpo21p(Rpb1p), a subunit of RNA polymerase II, Rad2p, a deoxyribonuclease required in DNA repair, and Rnt1p (RNase III), an endoribonuclease required for RNA maturation. For all three interactions, the two-hybrid results were confirmed by co-immunoprecipitation experiments. Genetic tests designed to assess the biological significance of the interactions indicate that Sen1p plays functionally significant roles in transcription and transcription-coupled DNA repair. To investigate the potential role of Sen1p in RNA processing and to assess the functional significance of the Sen1p/Rnt1p interaction, we examined U5 snRNA biogenesis. We provide evidence that Sen1p functions in concert with Rnt1p and the exosome at a late step in 3′ end formation of one of the two mature forms of U5 snRNA but not the other. The protein–protein and protein–RNA interactions reported here suggest that the DNA/RNA helicase activity of Sen1p is utilized for several different purposes in multiple gene expression pathways.
INTRODUCTION
Eukaryotic gene expression involves a complex interplay between proteins devoted to the synthesis of primary transcripts and those involved in transcript maturation including 5′ capping, splicing, transcription termination, 3′ end processing and ribonucleoprotein (RNP) export. Transcription is accomplished by a holoenzyme whose complexity is much greater than previously anticipated. The RNA polymerase II holoenzyme forms a cytologically visible ‘transcriptosome’ several megadaltons in size (1) that interacts with multiple sub-complexes allowing for regulated coupling of transcription with the maturation of transcripts (2).
Many of the proteins that are part of the transcriptosome associate with the C-terminal domain (CTD) of the largest subunit of eukaryotic RNA polymerase II, which corresponds to Rpb220p/Sua8p/Rpo21p/Rpb1p (referred to throughout as Rpo21p) in Saccharomyces cerevisiae (2,3). CTD-binding proteins include those of the Ser-Arg-rich (SR) protein family called CASPs (CTD-associated SR-like proteins) (3). These proteins form a bridge between the polymerase and RNA. We have been studying a yeast DNA/RNA helicase called Sen1p (4–8) that has been shown by others to co-purify with evolutionarily conserved transcriptosomal components involved in polymerase-associated formation of the 5′ cap (Sto1p/Cbc1p/Cbp80p/Grc3p) (9), cleavage and polyadenylation of mRNAs (Dis2p/Cid1p/Glc7p) (10,11), capping and maturation of snRNAs and formation of snRNPs (Sto1p and Smd3p) (9,12), and nucleotide excision repair associated with transcription factor TFIIH (Rad1p/Lpb9p) (10). In this paper we show that one of the underlying bases for co-purification of Sen1p with transcriptosomal components is a physical interaction between Sen1p and the CTD of the Rpo21p subunit of RNA polymerase II. Furthermore, we show that Sen1p physically interacts with Rad2p, a single-strand endonuclease that acts during transcription-coupled nucleotide excision repair (13,14).
Saccharomyces cerevisiae Sen1p and its ortholog from Schizosaccharomyces pombe are 5′→3′ ATP-dependent helicases of the helicase I family, which can unwind both DNA and RNA substrates (5,15). Sen1p helicase activity is required for the expression of diverse classes of non-protein-coding RNAs (4–8,16). In S.cerevisiae, the SEN1 gene codes for a 252 kDa protein that localizes to the nucleus (6) and is essential for growth (4). Strains carrying the sen1-2 allele, which lacks the first 975 N-terminal amino acids, but retains the ATP-helicase and nuclear localization domains, are viable but the growth rate is reduced. Also, the diffuse nuclear distribution of truncated sen1-2p differs from the granular nuclear distribution typical of wild-type Sen1p (6).
In S.cerevisiae, genetic studies of temperature-sensitive mutations causing amino acid substitutions in the helicase domain of Sen1p indicate that loss of function causes altered accumulation of intron-containing tRNA precursors (4), ribosomal RNA precursors (7) and 3′-extended forms of some snoRNAs and snRNAs (7,8,16), as well as mislocalization of core snoRNPs (6). A study of the snoRNA snR13 showed that impaired function of Sen1p causes aberrant forms of snR13 RNA to accumulate that have truncated 5′ ends and long 3′ ends extending beyond the site of normal transcription termination into the next adjacent gene on the chromosome (7,8). Mutations in SEN1 or NRD1, which codes for a CTD-binding SR protein, cause transcription termination read-through for some snoRNA transcripts, including snR13 (7,8,16,17).
In addition to physical interactions with Rpo21p and Rad2p, we show in this report that Sen1p interacts with Rnt1p (RNase III), an endonuclease involved in 3′ end processing of snRNAs (18,19), snoRNAs (20,21) and pre-rRNAs (22,23). The relationship between Sen1p and Rnt1p was investigated by examining the maturation pathway for U5 snRNA as a paradigm for how Sen1p might function in snRNA 3′ end formation. Our results suggest a close functional relationship between Sen1p, Rnt1p and exosomal nucleases.
MATERIALS AND METHODS
Strains and growth conditions
Strains FWY1 (ura3-52 leu2-3, –112 pep4-3 trp1 sen1-1 MATa) and 1971 (isogenic with FWY1 but carries SEN1) (7) were used in genetic crosses to construct double mutants carrying sen1-1 and other mutations of interest. To examine genetic interactions between mutant sen1 and rad2, strains differing by the presence or absence of functional SEN1 and RAD2 genes were constructed by transforming strain KCY2 (his3Δ1 leu2Δ met15Δ ura3Δ sen1-1 rad2::kanB:kanC MATα) with a centromeric plasmid carrying the wild-type SEN1 gene. A sen1-1 rad2 sibling from a cross involving strain KCY2 was transformed with a plasmid carrying the wild-type SEN1 gene. Similar strategies were used to construct sets of strains to analyze molecular and genetic interactions between mutant sen1 and rpo21, rnt1 and rrp6 starting with double mutant strains DUY1339 (leu2 ura3-52 sen1-1 rpb1-1 MATα), DUY749 (leu2-3, -112 his3 trp1 prc1 HIS3:pet56:rnt1 sen1-1 MATα), and DUY982 (leu2-3,-112 ura3-52 pep4-3 sen1-1 rrp6::URA3 MATα), respectively. Genetic interactions were also examined by combining sen1-1 with nup1::LEU2; nup100–1::URA3, nup116–6::URA3, nup145–1::URA3, sen2-1, lig1-2, rna1-1, and prp20–1.
Strains and plasmids used for immunoprecipitation of Sen1p with Rpo21p, Rad2p and Rnt1p were as follows: Sen1p and Rpo21p-HA: strain DDY1222 (leu2-Δ1 ura3-52 trp1-Δ1 lys2-801 his3-Δ200 ade2-101 sen1-Δ3::TRP1 MATa) carrying centromeric plasmids FW21 (SEN1) (7) and pY1At (RPO21-HA) (24); sen1-2p and Rpo21p-HA: strain DDY59 (leu2-Δ1 ura3-52 trp1-Δ1 lys2-801 his3-Δ200 ade2-101 sen1-Δ3::TRP1 MATa) carrying centromeric plasmid pY1At (RPO21-HA) and HR5 (sen1-2) (5); sen1-2 without Rpo21-HA: strain DDY59 carrying centromeric plasmids HR5 and the empty vector pRS315; cMyc–Sen1p and GST-Rad2p: strain PJ69–4A (trp1-901 leu2-3, -112 ura3-52 his3-200, gal4Δ, gal80Δ, lys2-GAL1-HIS3 GAL2-ADE2 met2:GAL7-lacZ MATa) (25) carrying centromeric plasmids pJF9 (cMyc–SEN1) and pKC10 (GST-RAD2); cMyc–Sen1p and GST-Rnt1p: strain M10 (leu2-3, -112 ura3-52 trp1-7 MATa) carrying centromeric plasmids pJF9 (cMyc–SEN1) and pYEX4T-1 (GST–RNT1). pNE118 (URA3) and pNE103 (TRP1) (E.Neeno-Eckwall, unpublished), which were used in cloning SEN1 and RAD2, and pYEX4T-1 (Invitrogen) were designed for expression of inserted ORFs from a CUP1 promoter. Co-immunoprecipitation of RNA with Sen1p was performed using strain 1971 (7).
Strains used in sen1-2p depletion experiments were constructed by transforming plasmids into strain DUY1222 (leu2-Δ1 ura3-52 trp1-Δ1 lys2-801 his3-Δ200 ade2-101 sen1-Δ3::TRP1 MATa) or by matings to form diploids as follows: DUY1222 {SEN1 low copy: [SEN1 URA3 CEN4 (FW21)]}; DUY1223 {SEN1 multi-copy: [SEN1 URA3 2 µM (FW20)]}; DD44-N3 [heterozygous diploid: SEN/sen1-Δ3::TRP1; DDY86 (chromosomally integrated sen1-2: leu2-Δ1::YIP351:sen1-2); DDY59 {multi-copy sen1-2: [sen1-2 URA3 2 µM (HR5)]}; and DUY291 {GAL1–Sen1-2 [GAL1::sen1-2 URA3 CEN4 (pDU112)]}.
General growth media and conditions were described previously (26). Growth rates were assayed by drop tests using serial dilutions of mid-log-phase cultures. Five microliter drops of undiluted and 5-, 25- and 125-fold diluted cultures were placed on solid medium and incubated at various temperatures and times as indicated in the figures. To test for the effects of UV light on growth, serial dilutions of cells as described above were placed on plates containing SD medium lacking uracil and were exposed to UV radiation using a Stratalinker UV Crosslinker 2400 (Stratagene). After exposure, the cells were allowed to grow at 30°C. Percent survival was calculated by averaging the number of colonies obtained by plating from different dilutions of the same culture. The average error per data point was ±9%.
To examine depletion of sen1-2p, cells from strain DUY291 were grown to mid-log-phase in synthetic medium lacking uracil and glucose and containing 2% galactose. At time zero, cells were collected, washed in synthetic dextrose (SD) medium (–uracil) containing 2% glucose and allowed to grow at 30°C, taking samples at time points as indicated in Figure 5. Cultures of cells were assayed for growth by diluting a fraction of the culture into fresh SD medium and following growth over time. The number of viable cells was determined by colony forming ability on SD (–uracil) medium containing 2% galactose at 30°C and by staining for viable cells with methylene blue. Rates of growth in presence or absence of galactose were comparable for about 16.5 h, after which the cells in glucose-containing medium continued to grow but at reduced rates due to depletion of sen1-2p.
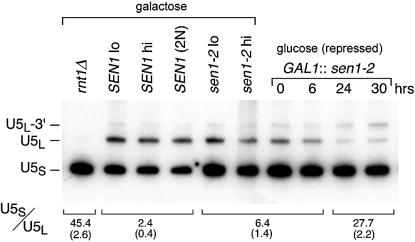
Figure 5.
Differential effects of N- and C-terminal segments of Sen1p on the U5S/U5L snRNA ratio. The panels on the left side show a comparison of U5 RNA accumulation in strains expressing N-terminally truncated sen1-2p (Fig. 1A) and wild type Sen1p. Isogenic strains include SEN1 expressed from a low copy (SEN1 lo) and a high copy (SEN1 hi) plasmid, a heterozygous SEN1/sen1Δ diploid [SEN1 (2N)], and sen1-2 expressed from a low (sen1-2 lo) and high copy (sen1-2 hi) plasmid (Materials and Methods). The effects of depleting sen1-2p are shown in the panels on the right. GAL1::sen1-2 (sen1-2 ORF fused to the GAL1 promoter) (6) was induced from a centromeric plasmid in the presence of galactose. Depletion was achieved when GAL1::sen1-2 expression was repressed by shifting cells from galactose to glucose-containing medium. U5 RNA accumulation was monitored at intervals and detected by northern blotting using a U5-specific probe (Materials and Methods). The U5S/U5L accumulation ratio and standard deviation (parentheses) indicated below each lane was calculated from three independent experiments.
Two-hybrid analyses
Global two-hybrid screens were performed using library C1 as described by James et al. (25). The N-terminal segment of Sen1p amino acids 1–975 was used as bait. SEN1-GAL4 binding domain (GBD) fusions containing DNA coding for the Sen1p segments shown in Figure 1 were constructed using the plasmid pOBD-2 (S.Fields, University of Washington) and pGBD-C(x) (25). Fusions containing RNT1 and the GAL4 activating domain (GAD) were obtained from S. A. Elela (29).
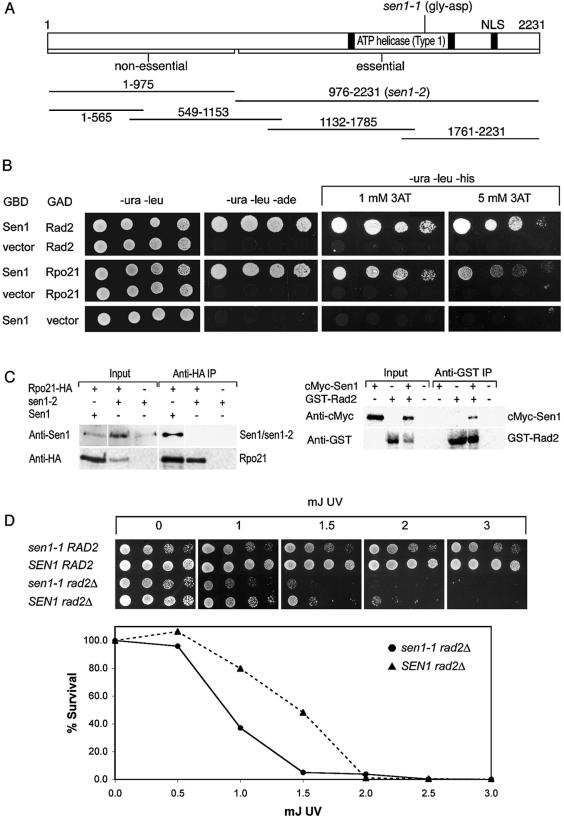
Figure 1.
Sen1p physically interacts with Rpo21p and Rad2p. (A) Schematic representation of Sen1p including the position of the temperature-sensitive sen1-1 mutation, a map of the truncated sen1-2 allele (5), and the segments used for two-hybrid analyses. Strains remain viable when the N-terminal 975 amino acids are deleted in sen1-2, but a complete deletion of SEN1 is lethal. (B) Two-hybrid interactions between the first 565 amino acids of Sen1p with the CTD of Rpo21p (amino acids 1497–1733) and Rad2p (amino acids 327–672). GBD indicates a fusion with the GAL4 DNA binding domain and GAD indicates a fusion with the GAL4 transcriptional activation domain. The figure shows growth after 3 days at 30°C resulting from activation of the reporters GAL2-ADE2 on SD medium lacking uracil, leucine and adenine and GAL1-HIS3 on SD medium lacking uracil, leucine and histidine. Growth is expected for all strains on medium lacking uracil and leucine, which selects for the presence of the two-hybrid vectors. The extent of growth was monitored by drop tests using increasingly dilute cell cultures from left to right in each row (Materials and Methods). 3AT, which inhibits His3p enzyme activity, was added to growth media as indicated. The strength of the interaction correlates with the extent of resistance to 3AT. (C) Co-immunoprecipitation of Sen1p with Rpo21p (left) and Rad2p (right). Proteins were immunoprecipitated from cell lysates using anti-HA or anti-GST immune serum. Bound proteins were gel-fractionated and analyzed on western blots probed with Anti-1 antibodies that recognize Sen1p and sen1-2p, anti-HA antibodies that recognize Rpo21p-HA, anti-GST antibodies that recognize GST-Rad2p or anti-cMyc antibodies that recognize cMyc–Sen1p. Input lanes represent 1/10 (left panel) and 1/5 (right panel) of the total cell lysates used for immunoprecipitation. In the left panel, a composite of Sen1p and sen1-2p are shown. The two proteins do not co-migrate (Sen1p is 252 kDa and sen1-2p is 140 kDa). (D) Synthetic reduction of growth rate in sen1-1 rad2Δ double mutants compared to single mutants with and without exposure to UV irradiation (Materials and Methods). Growth was monitored as described above on SD medium lacking uracil for 2 days. 30°C is semi-permissive for the growth of strains carrying sen1-1. In the graph, the average error for each data point was ±9%. One hundred percent survival corresponds to 1.5 × 106 cells.
Sets of GAD and GBD fusions as well as plasmids representing empty vectors were introduced into strain PJ69–4A carrying three reporter genes ADE2, HIS3 and lacZ that are under the control of Gal4p inducible GAL2, GAL1 and GAL7 promoters, respectively. Cells were assayed for growth using drop tests as described above. Growth in the presence of 1 mM 3-aminotriazole (3AT) was used to estimate the strength of the activation of the HIS3 gene, where growth in the presence of 3AT indicates a strong interaction. The interactions were verified by plasmid loss after plating on 5-FOA containing medium and by plasmid rescue followed by sequence analysis.
Immunoprecipitation
To examine co-immunoprecipitation of Sen1p with Rpo21p, Rad2p and Rnt1p, the relevant proteins were precipitated and/or detected using commercially available antibodies that recognize HA influenza virus (hemaglutinin) or c-Myc epitope tags, or GST (glutathione-S-transferase). We also used affinity-purified rabbit Anti-1 antibodies directed against an internal peptide in Sen1p (peptide 1, amino acids 1539–1551) (6) or rabbit anti-SEN1 antibodies directed against a β-galactosidase–Sen1p fusion (C-terminal 108 amino acids of Sen1p) (5). For each experiment the appropriate antibodies were immobilized by binding to protein A Sepharose 4 Fast Flow (Amersham Biosciences, Inc.).
Cell lysates were prepared by glass bead extraction of exponentially growing cells (OD600 ≈ 0.5) and disrupted in lysis buffer (50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.1% NP-40 containing 150 or 250 mM NaCl for protein or RNA analysis, respectively) and protease/RNase inhibitors aprotinin, pepstatin, chymostatin, leupeptin, antipain (final concentration of 1 µg/ml), 100 mM phenylmethyl-sulfonyl fluoride and 10 mM vanadyl ribonucleoside complex (Gibco BRL). For each set of immunoprecipitation experiments equal amounts of cells based on OD600 (for Sen1-RNA co-IP) or equal amounts of protein in lysates (for Sen1-protein co-IP) were analyzed. Lysates were allowed to bind to immobilized antibodies followed by a 3× wash with 500 µl lysis buffer (0°C). For immuno-competition, competitor Sen1 Peptide 1 or Sen1-lacZ polypeptide (50 µg) was pre-bound for 30 min to antibodies (10 µg) as described by Ursic et al. (7). Proteins were extracted by boiling for 5 min in 4× Laemmli buffer and fractionated in a 5% (Sen1p, Rpo21p, Rad2p) or in a 10% (Rnt1p) polyacrylamide SDS gel.
Following transfer to immobilon-NC transfer membrane (Millipore), the blots were incubated in 5% milk TBST pH 7.5 for 1 h followed by overnight incubation with either anti-HA (12CA5 from mouse at 1 µg/ml; Berkeley Antibody Co., Inc.), anti-GST (from rabbit at 1/4000 dilution; Sigma) or anti-cMyc antibody (9E10 from mouse at 1/100 dilution; Santa Cruz Biotechnology) in TBS, pH 7.5. The blots were washed twice for 5 min in TBS pH 7.5, once for 15 min in TBS pH 9.5, and twice for 5 min in TBST pH 9.5. Western blots were probed with anti-mouse or anti-rabbit peroxidase conjugated antibodies (Pierce Biotechnology, following manufacturer’s recommendation) followed by two 5 min washes in TBST pH 9.5, one 15 min wash in TBS pH 9.5, and four 5 min washes in TBS pH 9.5. Proteins were detected by chemiluminescence (Super Signal Substrate; Pierce Biotechnology) using a Typhoon 9200 Variable Mode Imager (Amersham Biosciences, Inc.).
To visualize RNAs that co-immunoprecipiate with Sen1p, the final pellet from immuno-purification was extracted following deproteination using 0.2 mg/ml proteinase K (Boehringer Mannheim) in PK buffer (0.1 M Tris–HCl pH 7.4, 12.5 mM EDTA, 150 mM NaCl, 1% SDS). The RNAs were isolated by phenol/chloroform extraction and ethanol precipitation in presence of 1 mg/ml glycogen carrier (Boehringer Mannheim). RNAs were 3′ end-labeled for 18 h at 4°C with [5′-32P]pCp in the presence of T4 RNA ligase (Promega) following manufacturer’s instructions. Labeled RNAs were purified using Sephadex G-25 (Sigma) spin columns followed by phenol/chloroform extraction and ethanol precipitation in presence of 1 mg/ml glycogen carrier. An equal amount of [5′-32P]pCp-labeled radioactive RNA was loaded in each lane of a 6% polyacrylamide gel. The fractionated RNAs were visualized by autoradiography and PhosphorImager analysis.
RNA preparation and northern hybridization
To prepare RNA for hybridization, RNA was isolated by hot acid phenol extraction. Cell pellets harvested at OD600 = 0.5 were suspended in 0.6 ml of buffer A (50 mM sodium acetate, 10 mM EDTA) containing 1% SDS and 1% diethylpyrocarbonate and were extracted twice with phenol saturated buffer A. Extractions were done at 65°C for 5 min. The aqueous phase was then extracted once with phenol/chloroform and precipitated twice with ethanol. The RNA pellets were suspended in water and the concentration measured by absorbance at 260 nm. RNAs (10 µg) were fractionated on 6% polyacrylamide (29:1), 8 M urea gel and transferred to GeneScreen Plus (DuPont, Inc.) using a BioRad Trans Blot Cell (Hercules, Inc.). The RNA was crosslinked using UV Stratalinker 2400. A U5-specific [α-32P]ATP-labeled DNA probe was used for northern blotting and was complementary to U5L plus an additional 85 nucleotides past the U5L 3′ end. Hybridization and washing conditions were as described by Ursic et al. (7).
RESULTS
Interactions identified in a global two-hybrid screen
Using the two-hybrid system (25,27), protein–protein interactions were detected using GAL2-ADE2 and GAL1-HIS3 reporters that are activated when GBD and GAD are brought together through a protein–protein interaction. The C-terminal half of 2231 amino acid Sen1p contains the conserved ATP-helicase domain. This region was excluded in the two-hybrid screen, limiting the proteins recovered to those that bind to the first 975 amino acids, which was used as the bait (Fig. 1A).
Approximately 500 000 colonies from Library C1 (25) were screened for the ability of fusion proteins to activate ADE2 and HIS3 reporters, which confer growth in the absence of exogenous adenine or histidine, respectively. Eighteen clones that gave rise to colonies in the initial screen were verified by DNA sequence analysis and retesting to express fusion proteins that interact with Sen1p(1–975). These clones corresponded to segments from seven different genes (Table 1). In this report, we focused on two of the interactions in greater detail: RPO21, which codes for the largest subunit of RNA polymerase II; and RAD2, an endonuclease involved in DNA repair.
Table 1. Proteins recovered from a global two-hybrid screen that interact with residues 1–975 of Sen1p.
| Protein | Length (amino acids) | No. times recovered | Residues in clone | Function | Cellular location |
|---|---|---|---|---|---|
| Gcr2p |
534 |
2 |
163–534 |
Transcription activator |
Nucleus |
| Nup49p |
472 |
1 |
21–472 |
Nuclear export |
Nuclear pore |
| Pyc2p |
1180 |
1 |
696–979 |
Pyruvate carboxylase |
Cytoplasm |
| Rad2p |
1031 |
3 |
327–672 |
DNA repair |
Nucleus |
| Rad2p |
|
1 |
327–544 |
|
|
| Rad2p |
|
1 |
491–606 |
|
|
| Rpo21p(Rpb1p) |
1733 |
3 |
1497–1733 |
RNA polymerase |
Nucleus |
| YPR148C |
435 |
2 |
95–435 |
Function unknown |
Cytoplasm |
| Prs3p |
320 |
1 |
142–204 |
|
Cytoplasm |
| Prs3p | 3 | 142–320 | Ribose-phosphate pyrophosphokinase 3 |
Information on the genes and proteins can be found in the Saccharomyces Genome Database (http://www.yeastgenome.org/).
RPO21 codes for a protein 1733 amino acids in length. Among three independent RPO21 clones, the plasmids recovered expressed identical fusion proteins containing amino acid residues 1497–1733, which correspond to the CTD of Rpo21p (Fig. 1B). Segment 1–565 (Fig. 1B) and 1–975 of Sen1p (not shown) activated the ADE2 and HIS3 reporters in the presence of an Rpo21p(1497–1733) fusion protein. Another segment of Sen1p(549–1153) that overlaps with segment 1–565 by 17 amino acids failed to activate reporters in the presence of Rpo21p(1497–1733) (not shown). These results indicate that the Sen1p–Rpo21p interaction requires the N-terminal 565 amino acids of Sen1p. To test the strength of the interaction between Sen1p(1–565) and Rpo21p(1497–1733), growth was assessed in the presence of 3AT, which inhibits the activity of the His3p reporter enzyme. Growth persisted in the presence of 5 mM 3AT, indicating a relatively strong interaction (Fig. 1B).
Protein extracts from strains expressing Rpo21p-HA, which contains a heamaglutinin (HA) epitope tag at the C-terminus, and either full-length Sen1p or truncated sen1-2p (Fig. 1A) were used to test whether Sen1p and Rpo21p co-immunoprecipitate. The proteins were bound to immobilized anti-HA antibodies (Materials and Methods), and the eluates were analyzed on western blots probed with anti-Sen1 antibodies (Materials and Methods) that recognize full-length Sen1p and truncated sen1-2p. The truncated protein lacks the region that interacts with Rpo21p in the two-hybrid system (Fig. 1C). Sen1p was detected when Rpo21p-HA and full-length Sen1p were expressed, but was not detected in the absence of Rpo21p-HA or when sen1-2p was expressed instead of full-length Sen1p. The RNA binding domain for type I helicases, including Sen1p, is located in C-terminal motif VI in the helicase domain (28). This region is not required for the two-hybrid interaction between Sen1p and Rpo21p. Furthermore, the region required for the interaction does not resemble any known RNA binding motif. Similar arguments can be made for the Sen1p/Rad2p interaction described below. The results are consistent with the conclusion that Sen1p interacts with the CTD of Rpo21p through a protein–protein interaction.
We examined the growth phenotype that resulted when the function of both proteins was compromised. A single haploid strain containing two temperature-sensitive mutations, one in SEN1 (sen1-1) and one in RPO21 (rpb1-1), exhibited extremely slow growth at temperatures that are permissive for each single mutation (not shown). The synthetic slow growth phenotype of the double mutant suggests that Sen1p may function in the context of an association with the CTD of Rpo21p. The close physical proximity to the CTD-binding protein Nrd1p in the transcriptosome complex may explain the similarity of sen1 and nrd1 mutant phenotypes with respect to read-through of snoRNA transcription termination (7,8,16). However, NRD1 was not recovered in the two-hybrid screen using Sen1p segment 1–975 as bait, and full-length Nrd1p failed to interact with any of the Sen1p segments shown in Figure 1A.
RAD2, which codes for a DNA endonuclease 1031 amino acids in length, is required for nucleotide excision repair of damage from several agents including UV irradiation (13,14). Among five independent RAD2 clones, the plasmids recovered expressed fusions containing different but overlapping segments spanning residues 327–672 of Rad2p (Table 1). The minimal Rad2p segment responsible for the interaction includes residues 491–544. Clones carrying this segment activated the ADE2 and HIS3 reporters in the presence of the Sen1p(1–565)GBD (Fig. 1B) and Sen1p(1–975)GBD (not shown). The growth rates on media lacking adenine or histidine (+1 and 5 mM 3AT) were similar for all fusions where positive interactions were detected. By comparison with the Rpo21p/Sen1p interaction, the Rad2p–Sen1p interaction was stronger.
Protein extracts from strains expressing GST-Rad2p (Rad2p fused to glutathione-S-transferase) and N-terminal epitope-tagged cMyc–Sen1p were used to test whether Rad2p and Sen1p co-immunoprecipitate. The proteins were bound to immobilized anti-GST antibodies, and the eluates were analyzed on western blots probed with anti-GST and anti-cMyc antibodies (Fig. 1C). cMyc–Sen1p was detected only when GST-Rad2p was co-expressed with the eptiope-tagged cMyc–Sen1p fusion.
We examined the extent to which isogenic strains carrying sen1-1 alone and sen1-1 combined with a rad2 null allele (rad2::kanB:kanC, referred to as rad2Δ) are sensitive to UV-induced DNA damage. The growth rates for single and double mutant strains were monitored at 30°C using serial dilution drop tests after irradiation with 0–3 mJ of UV light (Materials and Methods). The sen1-1 mutation prevents growth at 33°C and above, while at 30°C, function is partially impaired but growth is permitted. At 30°C the growth of the single mutant carrying sen1-1 was about the same with or without exposure to UV light (Fig. 1D). By contrast, deletion of the N-terminal 975 amino acids (sen1-2p) had a slight effect on UV sensitivity (not shown). The growth of the single mutant carrying rad2Δ was diminished after exposure to UV light. Growth was completely inhibited at an exposure of 5 mJ of UV. When cells were plated after UV exposure, we found that the sen1-1 rad2Δ double mutant lost viability more rapidly than the rad2Δ single mutant (Fig. 1D). At doses of 1–1.5 mJ, cell viability of the double mutant was ∼60% less than the rad2Δ single mutant.
Candidate-specific screens: Sen1p interaction with Rnt1p (RNaseIII)
We screened individual proteins for interaction with Sen1p based on previous information about potential co-participation in the synthesis of non-coding RNAs (7,8). An interaction was detected with Rnt1p, the S.cerevisiae ortholog of bacterial RNase III, which is required for cleavage of numerous precursor RNAs (20,21). The 471 amino-acid-long Rnt1 protein consists of several regions including an N-terminal domain that affects the efficiency of RNA binding and cleavage, a central nuclease domain required for site-specific cleavage, and a C-terminal domain for dsRNA binding (29).
Two-hybrid assays were conducted using three RNT1 segments coding for amino acid residues 1–191, 1–321 and the full-length protein (1–471). The Rnt1p hybrid fusions were examined in combination with four SEN1 segments coding for amino acid residues 1–565, 549–1153, 1132–1785 and 1761–2231 (Fig. 1A). The N-terminal region of Rnt1p (residues 1–191) interacted the strongest with Sen1p(1–565), but also interacted weakly with Sen1p(549–1153) (Fig. 2). The weaker interaction between Rnt1 fusions and Sen1p(549–1153) might be due to the overlapping residues shared in common between Sen1p(1–565) and Sen1p(549–1153). No interaction was detected between any of the Rnt1 fusions and Sen1p(1132–1785) or Sen1p(1761–2231) (not shown).
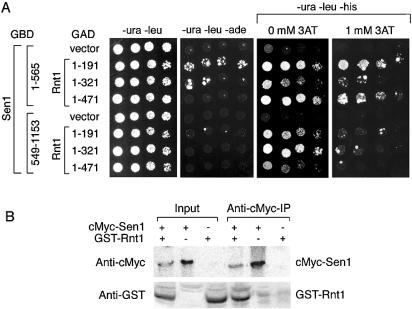
Figure 2.
In vivo and in vitro interactions between Sen1p and endoribonuclease Rnt1p. (A) Two-hybrid interactions between Sen1p amino acid segments 1–565 and 549–1153 with three segments of Rnt1p, including amino acid segments 1–191, 1–321 and 1–471 (full-length Rnt1p). Growth was monitored as described in Figure 1B except that plates are shown after 5 days of incubation at 30°C. (B) Co-immunoprecipitation of Sen1p and Rnt1p. Proteins were immunoprecipitated from cell lysates using anti-cMyc (cMyc–Sen1p) immune serum. Bound proteins were gel-fractionated and analyzed on a western blot probed with anti-GST antibodies that recognize GST–Rnt1p and anti-cMyc antibodies that recognize cMyc–Sen1p. Input lanes represent 1/5 of the total cell lysate used for immunoprecipitation. Upper and lower panels show different gels due to the large difference in size and distance of gel migration for Sen1p and Rnt1p. Control experiments indicate that anti-cMyc antibodies do not co-IP GST unless it is fused to Sen1p (not shown).
The interactions were best detected using the GAL1-HIS3 reporter. Rnt1p(1–191) and full-length Rnt1p conferred growth in the presence of 1 mM 3AT when expressed in combination with Sen1p(1–565), but no growth was observed at higher 3AT concentrations. The interactions between the Rnt1p(1–191) and full-length Rnt1p fusions with Sen1p(549–1153) were not detected in the presence of 1 mM 3AT, indicating that the most likely domain of interaction resides in the Sen1 segment spanning residues 1–565. Interactions between Sen1p(1–565), Rnt1p(1–191) and a weaker interaction with Rnt1p(1–321) were also detected using the GAL2-ADE2 reporter. Analysis of a third reporter, GAL7-lacZ, revealed that the Rnt1p(1–191)–Sen1p(1–565) and the Rnt1p(1–471)–Sen1p(1–565) interactions caused a 6–8-fold induction of lacZ and a corresponding increase in β-galactosidase activity. A weaker 2-fold increase was observed for the Rnt1p(321)–Sen1p(1–565) interaction. The remaining Sen1p–Rnt1p combinations had β-galactosidase activities that were comparable to vector-only controls.
Protein extracts from strains expressing GST-Rnt1p and N-terminal epitope-tagged cMyc–Sen1 were used to test whether Rnt1p and Sen1p co-immunoprecipitate. The proteins were bound to immobilized anti-cMyc antibodies, and the eluates were analyzed on western blots probed with anti-GST and anti-cMyc antibodies (Fig. 2B). GST-Rnt1p was detected only when cMyc–Sen1p was co-expressed with the GST-Rnt1p fusion. These results indicate that Sen1p interacts in vivo and in vitro with Rnt1p.
Interactions and epistatic effects of Sen1p and Rnt1p in U5 snRNA 3′ end formation
We performed an experiment to assess the overall number and extent to which different RNAs co-purify with Sen1p (Fig. 3A). RNAs were purified in the presence (+) and absence (–) of the Sen1–β-galactosidase fusion to which anti-SEN1 antibodies were directed. The fusion protein competitively inhibits the formation of the Sen1p/anti-SEN1 antigen–antibody complex. The bound RNAs were 3′ end-labeled with [5′-32P]pCp and fractionated on a 6% polyacrylamide gel. Nine RNAs detected in this experiment (identified with an asterisk in Fig. 3A) were shown previously to co-IP with Sen1p or are considered likely candidates for binding to Sen1p based on genetic evidence (7,8,16). The nine RNAs include C/D box snoRNAs snR13, snR45 and U3 as well as H/ACA snoRNAs snR5, snR10, snR11, snR30, snR31 and snR42.
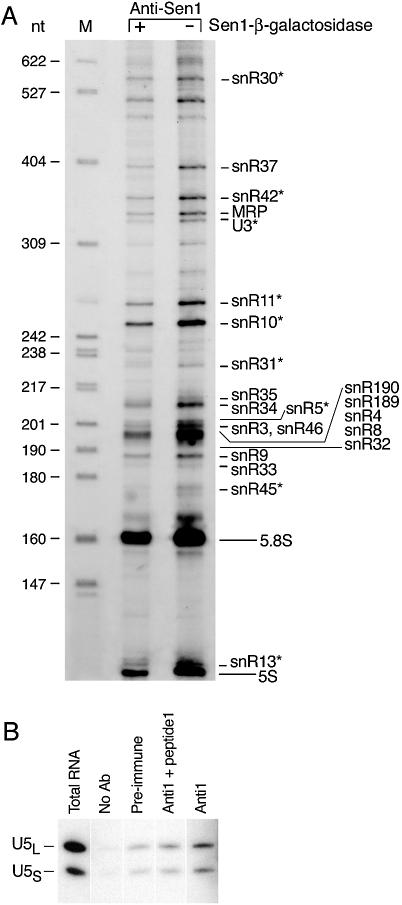
Figure 3.
Co-immunoprecipitation of non-coding RNAs with Sen1p. (A) RNAs were precipitated from total cell extracts using anti-SEN1 antibodies that recognize a Sen1–β-galactosidase fusion. Bound RNAs were end- labeled with [5′-32P]pCp prior to electrophoresis and equal cpm were loaded in each lane of a 6% gel. The specificity of protein–RNA binding was assessed using a Sen1–β-galactosidase competitive inhibitor, where diminished abundance of an RNA in the presence of the inhibitor was taken as evidence of specific binding. The identities of the RNAs were determined previously (6,7,39). RNAs marked with an asterisk (*) are known or suspected to associate with Sen1p based on previously published evidence (see text) (6,7,16). HpaII-digested pBR322 DNA (lane M) serve as size markers. (B) U5S and U5L snRNA co-immunoprecipitate with Sen1p. Sen1p was immunoprecipitated from a cell lysate of wild-type SEN1 strain 1971 using antibodies directed against an internal peptide of Sen1p (Materials and Methods). As controls, immunoprecipitation was carried out in the absence of antibodies, with pre-immune serum, or in the presence of a competitor that binds to the antibody. RNAs were fractionated on a 6% polyacrylamide gel and analyzed by northern blotting using a probe specific for U5 snRNAs. The lane labeled ‘Total RNA’ represents ∼1% of the lysate used for immunoprecipitation.
The complexity of the pattern and large differences in the efficiency of in vitro 3′ end-labeling of different RNAs may have obscured detection of snRNAs involved in splicing. Since Sen1p and Rnt1p were shown previously to affect the accumulation of RNAs related to snRNA U5 (7,18), we performed an experiment similar to the one described above, but instead of using 3′ end-labeling for detection, we used a probe specific for U5 RNA. Two forms of mature U5 snRNA exist called U5S (180 nt) and U5L (214 nt), which differ in length at their 3′ ends (30). Sen1p was immunoprecipitated using anti-1 antibodies directed against an internal peptide (residues 1539–1551) (4–8). The precipitates were analyzed on northern blots using a U5-specific probe (Fig. 3B). A small amount of U5L and U5S RNA were bound non-specifically to the agarose beads using pre-immune serum or in the absence of antibodies. Significantly more RNA was bound in the presence of antibodies, and the binding was specific as indicated by the diminished amount of the U5 RNAs detected in the presence of a competing peptide. Less than 1% of the amount of U5S and U5L present in total RNA extracts was bound specifically in Sen1p precipitates. The relatively inefficient immuno-precipitation of the U5 snRNAs suggests a transient interaction. Consistent with this, Sen1p is not a structural subunit of the U5 snRNP particle (31), but may nonetheless participate in the formation of mature U5 RNAs through transient association with the U5 snRNP and through its interaction with Rnt1p.
The largest detectable U5-related RNA is ∼690 nucleotides in length (21). Rnt1p cleaves this RNA or possibly a shorter derivative of this RNA at two alternative sites, λ and σ, leading to the two alternative mature forms of U5 snRNA, U5L and U5S (Fig. 4A). The cleavages at the λ and σ sites produce two RNA intermediates, designated U5L-3′ and U5-3′a, that undergo further processing by the exosomal and Rex nucleases to produce U5L and U5S (18,21,32).
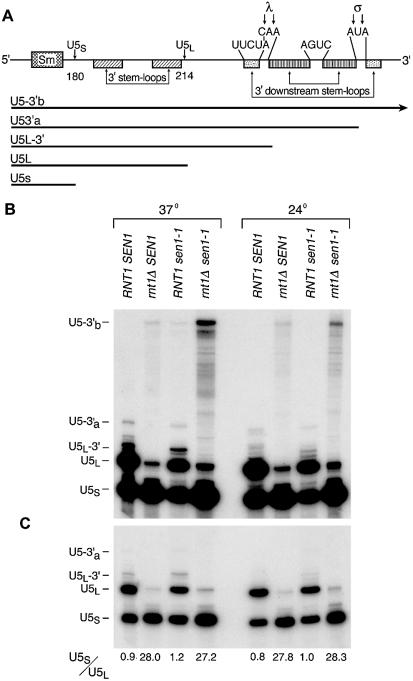
Figure 4.
Accumulation of U5 snRNAs in sen1 rnt1Δ double mutants. (A) Schematic diagram of the 3′ region of U5 snRNA (18,21). ‘Sm’ indicates the region where U5 snRNA interacts with the Sm protein complex. λ and σ indicate two alternative sites for cleavage in stem–loop regions by Rnt1p that lead to formation of the two mature forms of U5 RNA called U5L (214 nt) and U5S (180 nt). The bars beneath the diagram indicate the lengths of major RNAs that accumulate during maturation of the 3′ ends. When Rnt1p cleaves at the λ site, the product, U5L-3′, is processed by exosomal nucleases to mature U5L. When Rnt1p cleaves at the σ site, the product, U5L-3′a, is processed by exosomal nucleases to mature U5S. (B) RNAs from four congenic strains differing only at SEN1 and RNT1 were analyzed by northern blotting using a U5-specific probe (Materials and Methods). The strains carry wild-type SEN1 and RNT1 genes, a single mutant carrying sen1-1, a single mutant carrying null allele rnt1Δ, and the double mutant. RNA was extracted either from cells grown at 24°C (semi-permissive for sen1-1) or 5 h following a shift of cells to 37°C (restrictive for sen1-1). (C) The same blot is shown at reduced exposure. The U5S/U5L accumulation ratio for the experiment shown is indicated below each lane.
We examined the accumulation of U5-related RNAs in strains carrying single mutations in RNT1 (HIS3:pet56:rnt1, referred to as rnt1Δ) or SEN1 (temperature-sensitive sen1-1) and in a double mutant strain carrying rnt1Δ and sen1-1. The U5L-3′ and U5-3′a intermediates were undetectable in the strain lacking a functional RNT1 gene (Fig. 4B and C). The accumulation of U5L was severely reduced, whereas U5S was present at a level similar to wild type (the U5S/U5L ratio was 28 compared to 0.9 for wild type). Except for a small increase in U5-3′b RNA, no significant accumulation of other large U5-related RNAs was observed. The lack of accumulation of the precursors of Rnt1p cleavage in the rntΔ strain suggests that the uncleaved precursors are alternatively processed or degraded. The pattern of RNA accumulation is consistent with the pattern previously reported for rnt1 strains (7,8,21).
The sen1-1 mutant accumulated 2–3-fold more of the U5L-3′ precursor than wild type at the restrictive temperature of 37°C (7). The levels of accumulation of U5S and U5L were similar, but the ratio was slightly altered in favor of U5S (0.9 for wild type versus 1.2 for sen1-1). Similar to the rnt1Δ strain, a small amount of longest detectable RNA, U5-3′b, was evident in the sen1-1 strain at 37°C, whereas this RNA species was below detectable levels in wild type (Fig. 4B).
The accumulation of RNAs in the sen1-1 rnt1Δ double mutant exhibited some features typical of the rnt1Δ single mutant and some features unique to the double mutant. Similar to the rnt1Δ single mutant, the precursor to U5L (U5-3′a) was not detected and very low levels of U5L were present. However, unlike the rnt1Δ or sen1-1 single mutants, numerous U5 RNAs were detected that extend in size up to that of U5-3′b, which accumulated to a relatively high level compared to either single mutant. These longer RNAs were detected at much lower levels in both of the single mutants but were not detected in wild type. U5-3′b RNA, which is a precursor to Rnt1p cleavage, is presumably acted upon by alternate nucleases in the rnt1Δ single mutant strain, but U5-3′b is partially stabilized in the double mutant. These results suggest an epistatic relationship between Sen1p, Rnt1p and the alternative nucleases that degrade the Rnt1p substrate when Rnt1p is absent.
Effects of Sen1p and truncated sen1-2p on the accumulation of U5-related RNAs
We compared the accumulation of U5-related RNAs in a wild-type RNT1 genetic background in strains carrying SEN1 and sen1-2 which were either chromosomally integrated in a single copy or carried on multi-copy 2 µm plasmids (Fig. 5). The truncated sen1-2 allele is missing the domains that mediate binding of Sen1p to Rpo21p, Rad2p and Rnt1p as well as the native SEN1 promoter, and is expressed from a cryptic promoter instead (Fig. 1A). Strains carrying a lethal null allele of SEN1 are viable when they express truncated sen1-2p. The average U5S/U5L ratio was 2.4 for SEN1 strains and 6.4 for sen1-2 strains (Fig. 5). Gene copy number had little effect on the ratio. The 2.5–3-fold difference in the ratio indicates that the N-terminal segment of Sen1p has a demonstrable effect on the accumulation of U5L.
To compare the effects of deleting the N-terminal region of Sen1p with the effects that result from loss of ATP-helicase function, a depletion experiment was performed using GAL1::sen1-2 which expresses the N-terminally truncated protein from a galactose inducible, glucose-repressible promoter (Fig. 5). After a shift from galactose to glucose, growth slowed starting at 16 h, but 80–90% of the cells remained viable for 30 h as measured by colony formation and resistance to methylene blue staining (not shown). By 24 h, the U5S/U5L ratio was 27.7 compared to an initial ratio of 6.4, indicating a 4–4.5-fold reduction in the accumulation of U5L. In addition, we observed a time-dependent increase in the accumulation of U5L-3′, the precursor to U5L (Fig. 4). After 30 h, a 2.3-fold increase in U5L-3′ was observed. Compared to isogenic wild-type strains grown in galactose, the overall change in the U5S/U5L ratio was 11–12-fold. No other larger RNAs were observed to accumulate in the upper portion of the northern blot (not shown). The results indicate that the overall reduction in the accumulation of U5L is partly due to the absence of the N-terminal segment in sen1-2p and partly due to depletion of the truncated protein containing the ATP-helicase domain. These results suggest that, like Rnt1p, Sen1p is required for the accumulation of U5L RNA but not U5S RNA and that at least two regions of Sen1p contribute to the formation of U5L.
Synthetic growth rate reduction in double mutants
Since Sen1p, Rnt1p and the exosome all participate in 3′ end formation of numerous RNAs (7,8,21), we examined the overall effects on growth when the sen1-1 mutation was combined with a rnt1Δ null allele and an rrp6Δ null allele. Rrp6p is a nuclease associated with the exosome (7,8,21).
The synthetic slow growth of a sen1-1 rnt1Δ double mutant strain was most evident when cells were incubated at 30° and 24°C, which are permissive growth temperatures for sen1-1 (Fig. 6). The corresponding single mutant strains carrying either sen1-1 or rnt1Δ grew much better than the double mutant, although these strains still did not grow as well as the wild-type strain. Although U5 snRNA is essential for growth (30), the synthetic slow growth of the double mutant is probably not the result of reduced U5L snRNA synthesis since U5S accumulates to levels comparable to wild type and because strains carrying rnt1Δ by itself exhibit levels of U5L and U5S accumulation similar to the double mutant and are viable.
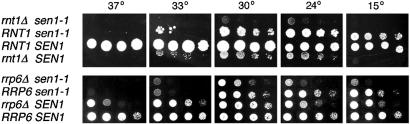
Figure 6.
Growth of sen1-1 rnt1Δ and sen1-1 rrp6Δ double mutants. The growth of isogenic sets of strains (Materials and Methods) was monitored by serial drop tests (Materials and Methods). Cells were incubated for 3 days at the temperatures indicated.
The nuclease Rrp6p, an ortholog of Escherichia coli RNase D (33) is a nuclear component of the exosome known to participate in the maturation of U5 snRNA. Exosomal nucleases trim nucleotides from the 3′ ends of U5 RNAs following cleavage by Rnt1p (21,32). We examined double mutants carrying sen1-1 and the null allele rrp6Δ to look for synthetic phenotypes with regard to both U5 snRNA synthesis and growth. The pattern of accumulation of U5 RNAs most closely resembled the pattern observed for an rrp6Δ single mutant (not shown). This is most likely because there are alternate routes for the production of substrates for Rrp6p and for the accumulation of U5L and U5S in the absence of functional Sen1p. When growth rates were examined, it was found that growth of the double mutant was reduced at 30°, 24° and 15°C compared to either single mutant (Fig. 6).
The slow growth phenotypes of sen1-1 rnt1Δ and sen1-1 rrp6Δ double mutants suggest an underlying functional relationship among the proteins because Sen1p, Rnt1p and the exosome are needed in a common subset of RNA maturation pathways. It is unlikely that synthetic slow growth is due to non-specific, additive effects because no synthetic growth phenotypes were observed when sen1-1 was combined with mutations that severely affect nuclear cytoplasmic transport (nup1::LEU2; nup100–1::URA3, nup116–6::URA3, nup145–1::URA), tRNA maturation (sen2-1, lig1-2) or nuclear-cytoplasmic RNA shuttling (rna1-1, prp20–1) (not shown). Synthetic slow growth could be due to reduced accumulation of a single essential RNA other than U5 snRNA or to the cumulative effects of multiple RNAs that require Sen1p, Rnt1p and exosomal nucleases for maturation.
DISCUSSION
Seven proteins were identified in a global two-hybrid screen that interact with Sen1p. Since Sen1p is exclusively nuclear (6), we focused on further characterization of two of the nuclear-localized interacting proteins that were recovered: the RNA polymerase II subunit Rpo21p and the single-strand DNA endonuclease Rad2p. Both interactions are mediated by an N-terminal segment of Sen1p spanning amino acids 1–565. Sen1p interacts with the CTD of Rpo21p and an internal segment of Rad2p.
Sen1p appears to be functionally related to the SR protein Nrd1p, which binds to the CTD of Rpo21p and also contains an RNA recognition motif (16,17). Loss-of-function mutations in NRD1 and SEN1 cause the accumulation of aberrant snoRNAs with 3′ ends extending past the site of transcription termination. The snoRNA snR13, which co-immunoprecipitates with Sen1p (Fig. 3A), is one of the best-studied examples of this. Mutant alleles of nrd1 and sen1 exhibit reduced accumulation of mature snR13 and increased accumulation of RNAs that extend through the snR13 transcription termination site and extend to the site of transcription termination of the next downstream gene on the chromosome (7,8,16). Despite the fact that two hybrid interactions between Sen1p and Nrd1p were not detected, the physical proximity of Sen1p and Nrd1p in the CTD-binding complex is consistent with a role for both proteins in transcription termination as proposed previously (16,17).
A strong interaction was detected between Sen1p and Rad2p, which is likely to serve a purpose in DNA repair, because sen1 rad2 double mutants are hypersensitive to UV irradiation. The nucleotide excision reactions needed to repair DNA damage are related to transcription in two respects. DNA coding strands are repaired at a faster rate than non-coding strands possibly because certain types of DNA damage inhibit the ability to transcribe through a damaged region (34). Secondly, repair of UV damage requires the transcription complex TFIIH (35), which consists of at least nine proteins (2,36). TFIIH acts in concert with two single-strand DNases, Rad1p/Rad10p and Rad2p, which cut on the 5′ and 3′ sides of a damaged region, respectively (13,14). Our results indicate that Sen1p interacts directly with Rad2p, but other studies show it also co-purifies with Rad1p (10). These results are suggestive of a role for Sen1p in transcription-coupled nucleotide excision repair. Redundancies of function may exist, however, because the sen1-1 mutation did not by itself affect UV sensitivity under semi-permissive growth conditions. UV hypersensitivity was only apparent in the sen1-1 rad2Δ double mutant. Further studies will be required to understand what role Sen1p plays in DNA repair.
It was shown previously that Sen1p interacts with Smd3p, a component of the Sm complex that assembles with snRNAs to form snRNPs (12). We did not recover Smd3p in our two-hybrid screens because the Smd3p binding domain in Sen1p was not included in the Sen1p segment used as bait. Since this interaction suggests a role for Sen1p in RNA processing, we examined the pathway for formation of two variant forms of mature U5 snRNA, U5L and U5S. It was known previously that the sen1-1 mutation causes excess accumulation of U5L-3′ RNA, a precursor to U5L RNA (Fig. 4) (7). We found that U5L and U5S RNAs co-immunoprecipitate with Sen1p and that residues 1–565 interact with N-terminal residues 1–191 of Rnt1p (RNase III), a nuclease involved in U5 snRNA maturation (22,29,37).
To examine the effects of Sen1p on U5 snRNA accumulation, we used two mutant alleles of SEN1 described previously (5,6). The first allele, sen1-1, contains a substitution of an evolutionarily conserved residue within a domain required for ATP-dependent DNA/RNA helicase activity. The sen1-1 mutation causes temperature-sensitive growth. The second allele, sen1-2, codes for a truncated version of Sen1p that is missing the first 975 N-terminal amino acids residues, including the binding domains for Rpo21p, Rad2p and Rnt1p.
The most noticeable U5-related phenotype of strains carrying sen1-1 is that the product of Rnt1p cleavage at the λ cleavage site (U5-3′, Fig. 4) accumulates to a higher level compared to wild type. Both U5L and U5S accumulate at near normal levels with slightly lower levels of U5L and slightly higher levels of U5S. When sen1-2p was expressed from a GAL1–sen1-2 fusion, the U5S/U5L ratio was increased 2.5–3-fold compared to Sen1p. This shows that the relative accumulation of U5L is decreased when the N-terminal 975 amino acids are deleted from Sen1p. We speculate that this may be due to absence of the Sen1-Rnt1 binding domain, which could help facilitate a Rnt1–RNA interaction.
When sen1-2p was depleted following transcriptional shut-off of the GAL1–sen1-2 fusion, the U5S/U5L ratio increased to about 28. The increased ratio reflects a much lower level of U5L RNA accumulation. By comparison, deletion of RNT1 resulted in a U5S/U5L ratio of about 45. Similar changes in U5L RNA accumulation were not evident in experiments using the sen1-1 allele because the time window between temperature-shift and cell death is small relative to the rate of decay of mature U5L. Stable U5L RNA made prior to the temperature shift probably masks a reduced rate of synthesis. Finally, the precursor to U5L, U5L-3′, shows a time-dependent increase in accumulation, indicating that the precursor U5L-3′ accumulates at the expense of the product U5L.
Since exosomal nucleases convert U5L-3′ to U5L, these nucleases appear to act less efficiently in the absence of Sen1p. We suggest that the Sen1p/Rnt1p interaction is required for normal formation of U5 snRNA, and that the absence of the Rnt1p binding domain in sen1-2p explains the phenotypes that occur upon depletion of sen1-2p. Since the product of Rnt1p cleavage at the λ site accumulates following depletion, we suggest that Sen1p may function at the interface between Rnt1p cleavage and subsequent processing by the exosome. The λ cleavage site occurs in a stem–loop, and Sen1p might help unwind the stem during or after Rnt1p cleavage to enhance access of the 3′ end to exosomal nucleases. Given that sen1-1 confers a synthetic growth defect when combined in double mutants with rnt1Δ or rrp6Δ, it is likely that Sen1p, Rnt1p and the exosome function together in other pathways in addition to the U5 snRNA maturation pathway.
The proposed function for Sen1p in U5 RNA processing described above is distinct from the role proposed for Sen1p in transcription termination, which may be mediated by the interaction between Sen1p and the CTD of Rpo21p. Sen1p may function twice in U5 snRNA expression. The accumulation of U5-related RNAs single or double mutants could be a composite consequence of separate roles for Sen1p in transcription and processing (Fig. 7). It is not yet clear whether the ladder of RNAs that accumulate in sen1-1 rnt1Δ double mutants are a consequence of defects in transcription termination or processing. Although the site of transcription termination for U5 RNA is not known, the largest detectable U5-related RNA, U5-3′b, is below detectable levels in wild type, accumulates to a small extent in rnt1Δ and sen1-1 strains, and accumulates to a much larger extent in a sen1-1 rnt1Δ double mutant. The double mutant also accumulates an extensive ladder of RNAs that may be partially degraded forms U5-3′b RNA. Interestingly, exosomal nucleases including Rrp41p, Rrp45p and Rrp6p, exhibit nearly identical ladders of RNAs compared to the sen1-1 rnt1Δ double mutant when the corresponding nuclease genes are disrupted or when the protein products are depleted in strains carrying rnt1Δ (21).
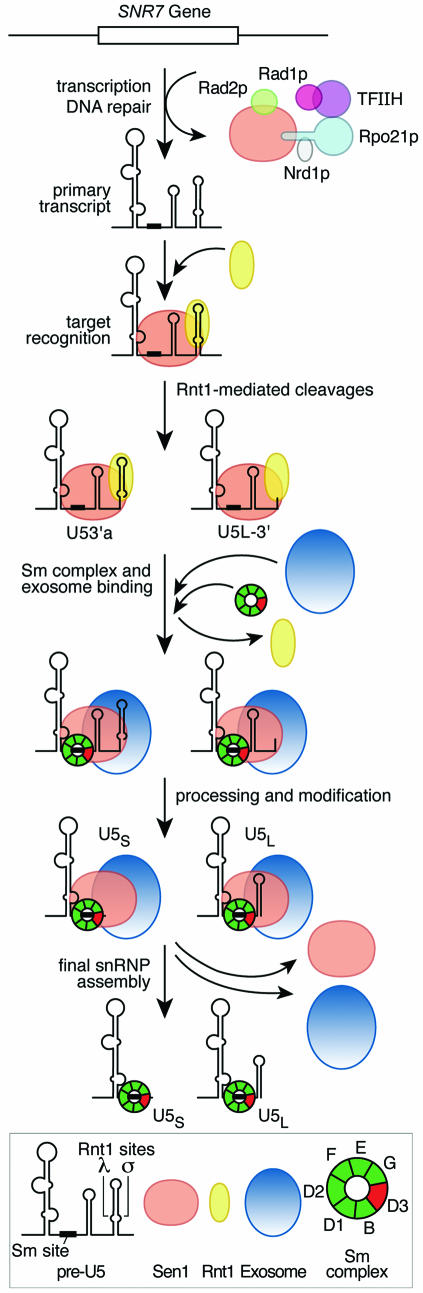
Figure 7.
Model for the role of Sen1p in the expression of the SNR7 gene coding for U5 RNA. The figure summarizes potential roles for Sen1p in transcription termination, transcription-coupled DNA repair, and RNA processing. Sen1p and Nrd1p have been implicated in transcription termination of non-coding RNAs (16,17). Rad1p and Rad2p are DNA endonucleases that cleave on the 5′ and 3′ sides of damaged DNA, and Rad1p is a subunit of transcription factor TFIIH (10,13,14). The primary events in the maturation of U5 RNA were described previously, including the involvement of Rnt1p (22,29,37), the exosomal and Rex nucleases (32,40), and the Sm complex (41,42). Data in this paper suggest a model in which a U5 post-transcriptional RNA complexes with Rnt1p and Sen1p; Rnt1p cleaves at the λ and σ cleavage sites and then dissociates; the seven subunit Sm complex associates with U5 RNA; Sen1p binds to one of the Sm subunits, Smd3p (12); and the exosome trims U5 RNAs yielding the two mature products, U5L and U5S. The Sm proteins and U5 RNA form the core U5 snRNP, but Sen1p dissociates and is not part of the final snRNP. Variations on the exact order of events are possible. It is not known whether the Sen1p that is bound to Rpo21p leaves with the polymerase after transcription is completed, in which case a second molecule of Sen1p must bind to the RNA, or whether the Sen1p molecule that functions in transcription remains bound to the RNA and functions with Rnt1p in subsequent processing events.
Either U5-3′b RNA is the primary transcript or it is a read-through transcript that accumulates when Sen1p, Rnt1p or the exosomal nucleases are inactivated. Since it is unlikely that Rnt1p and the exosomal nucleases promote read-through of transcription termination, we favor the view that U5-3′b is the primary transcript. However, this does not preclude the possibility that Sen1p influences termination of the primary U5 transcript through its interaction with the CTD of Rpo21p. Read-through transcripts may fail to accumulate because they may be rapidly degraded.
In conclusion, we note that mutations in Senataxin, the human ortholog of yeast SEN1, were recently identified in patients diagnosed with ataxia-ocular apraxia 2, a genetic disorder affecting the nervous system (38). Given that Sen1p appears to function in multiple pathways for gene expression in yeast, it is not yet clear in humans what role Sen1p might play in the neurological impairment associated with this disease. The link between Sen1p and human disease provides an additional incentive to continue efforts to understand what roles are played by helicases like Sen1p in gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by the University of Wisconsin Medical School and the College of Agricultural and Life Sciences, and Public Health Service Grant GM9019 (M.R.C.). We thank L. Olds for preparing the illustrations, K. Watts for technical assistance and M. Ares, C. Guthrie, S. A. Elela, S. Wente, L. Davis, A. Hopper, M. Wickens, E. Phizicky, P. James, P. Uetz, J. L. Corden, E. Neeno-Eckwall and J. Butler for providing strains and plasmids. This is Laboratory of Genetics paper no. 3610.
REFERENCES
- 1.Gall J.G., Bellini,M., Wu,Z. and Murphy,C. (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell, 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myer V.E. and Young,R.A. (1998) RNA polymerase II holoezyme and subcomplexes. J. Biol. Chem., 273, 27757–27760. [DOI] [PubMed] [Google Scholar]
- 3.Corden J.L. and Patturajan,M. (1997) A CTD function linking transcription to splicing. Trends Biochem. Sci., 22, 413–416. [DOI] [PubMed] [Google Scholar]
- 4.Winey M. and Culbertson,M.R. (1988) Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics, 118, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMarini D.J., Winey,M., Ursic,D., Webb,F. and Culbertson,M.R. (1992) SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2154–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ursic D., DeMarini,D.J. and Culbertson,M.R. (1995) Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol. Gen. Genet., 249, 571–584. [DOI] [PubMed] [Google Scholar]
- 7.Ursic D., Himmel,K.L., Gurley,K.A., Webb,F. and Culbertson,M.R. (1997) The yeast SEN1 gene is required for processing of diverse RNA classes. Nucleic Acids Res., 25, 4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen T.P. and Culbertson,M.R. (1998) The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 6885–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 10.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 123–124. [DOI] [PubMed] [Google Scholar]
- 11.Walsh E.P., Lamont,D.J., Beattie,K.A. and Stark,M.J.R. (2002) Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectroscopy. Biochemistry, 41, 2409–2420. [DOI] [PubMed] [Google Scholar]
- 12.Fromont-Racine M., Rain,J.-C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 216–217. [DOI] [PubMed] [Google Scholar]
- 13.Tomkinson A.E., Bardwell,A.J., Bardwell,L., Tappe,N.J. and Friedberg,E.C. (1993) Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature, 362, 860–862. [DOI] [PubMed] [Google Scholar]
- 14.Habraken Y., Sung,P., Prakash,L. and Prakash,S. (1993) Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature, 366, 365–368. [DOI] [PubMed] [Google Scholar]
- 15.Kim H.-D., Choe,J. and Seo,Y.-S. (1999) The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry, 38, 14697–14710. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz E.J., Conrad,N.K., Brow,D.A. and Cordon,J.,L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature, 413, 327–331. [DOI] [PubMed] [Google Scholar]
- 17.Steinmetz E.J. and Brow,D.A. (1998) Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl Acad. Sci. USA, 95, 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanfreau G., Elela,S.A., Ares,M.J. and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNAse III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elela S.A. and Ares,M. (1998) Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J., 17, 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chanfreau G., Legrain,P. and Jacquier,A. (1998) Yeast RNase III as a key processing enzyme in small nucleolar RNA metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- 21.Allmang A., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elela S.A., Igel,H. and Ares,M. (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- 23.Kufel J., Dichtl,B. and Tollervey,D. (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ETS but not the 5′ ETS. RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West M.L. and Corden,J.L. (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics, 140, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ursic D. and Culbertson,M.R. (1991) The yeast homolog to mouse Tcp1-1 affects microtubule-mediated processes. Mol. Cell. Biol., 11, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields S. and Song,O. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 28.Czaplinski K., Weng,Y., Hagan,K.W. and Peltz,S.W. (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA, 1, 610–623. [PMC free article] [PubMed] [Google Scholar]
- 29.Lamontagne B., Tremblay,A. and Elela,S.A. (2000) The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol., 20, 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson B. and Guthrie,C. (1987) An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell, 5, 613–624. [DOI] [PubMed] [Google Scholar]
- 31.Stevens S.W., Ryan,D.E., Ge,H.Y., Moore,R.E., Young,M.K., Lee,T.D. and Abelson,J. (2002) Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell, 1, 31–44. [DOI] [PubMed] [Google Scholar]
- 32.Van Hoof A., Lennertz,P. and Parker,R. (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNAse MRP and RNase P RNAs in yeast. EMBO J., 19, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briggs M.W., Burkard,K.T.D. and Butler,J.S. (1998) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNAse MRP and RNase P RNAs in yeast. J. Biol. Chem., 273, 13255–13263.9582370 [Google Scholar]
- 34.van Gool A.J., Verhage,R., Swagemakers,S.M., van de Putte,P., Brouwer,J., Troelstra,C., Bootsma,D. and Hoeijmakers,J.H. (1994) RAD26, the functional S.cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J., 13, 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash S. and Prakash,L. (2000) Nucleotide excision repair in yeast. Mutat. Res., 451, 13–24. [DOI] [PubMed] [Google Scholar]
- 36.Hoogstraten D., Nigg,A.L., Heath,H., Mullenders,L.H., van Driel,R., Hoeijmakers,J.H., Vermeulen,W. and Houtsmuller,A.B. (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell, 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 37.Nagel R. and Ares,M. (2000) Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA, 6, 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreira M.C., Klur,S., Watanabe,M., Nemeth,A.H., Le Ber,I., Moniz,J.C., Tranchant,C., Aubourg,P., Tazir,M., Schols,L. et al. (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nature Genet., 36, 225–227. [DOI] [PubMed] [Google Scholar]
- 39.Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 40.Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′ to 5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Will C.L. and Luhrmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- 42.Zhang D., Abovich,N. and Rosbash,M. (2001) A biochemical function for the Sm complex. Mol. Cell, 7, 319–329. [DOI] [PubMed] [Google Scholar]