Abstract
The genomes of three strains of Listeria monocytogenes that have been associated with food-borne illness in the USA were subjected to whole genome comparative analysis. A total of 51, 97 and 69 strain-specific genes were identified in L.monocytogenes strains F2365 (serotype 4b, cheese isolate), F6854 (serotype 1/2a, frankfurter isolate) and H7858 (serotype 4b, meat isolate), respectively. Eighty-three genes were restricted to serotype 1/2a and 51 to serotype 4b strains. These strain- and serotype-specific genes probably contribute to observed differences in pathogenicity, and the ability of the organisms to survive and grow in their respective environmental niches. The serotype 1/2a-specific genes include an operon that encodes the rhamnose biosynthetic pathway that is associated with teichoic acid biosynthesis, as well as operons for five glycosyl transferases and an adenine-specific DNA methyltransferase. A total of 8603 and 105 050 high quality single nucleotide polymorphisms (SNPs) were found on the draft genome sequences of strain H7858 and strain F6854, respectively, when compared with strain F2365. Whole genome comparative analyses revealed that the L.monocytogenes genomes are essentially syntenic, with the majority of genomic differences consisting of phage insertions, transposable elements and SNPs.
INTRODUCTION
Listeria monocytogenes is a Gram-positive bacterium that can cause life-threatening infections for humans and more than 40 animal species. Immunocompromised individuals, pregnant women, the elderly and neonates are at high risk for listeriosis. Outbreaks of listeriosis have been associated with the consumption of ready-to-eat foods, especially meat and dairy products (1). The disease can result in abortion, stillbirths, septicemia, meningitis, encephalitis and death. The ubiquity of L.monocytogenes in food processing, distribution and retail environments, coupled with its inherent resistances and ability to grow in many foods, including those stored refrigerated, makes this pathogen particularly difficult to both manage and regulate (1). In the USA, L.monocytogenes is responsible for about 2500 cases of listeriosis each year, with a hospitalization rate of 91% and a case fatality rate of 20% (2). Despite appreciable efforts worldwide by research organizations, regulatory-action agencies and the food industry to reduce the incidence of listeriosis, this pathogen, quite arguably, remains the most critical threat to the safety of our food supply.
There are 13 described serotypes of L.monocytogenes, with serotypes 1/2a, 1/2b and 4b accounting for 95% of human infections (3). Among strains recovered from foods or food processing plants, serotype 1/2a strains are over-represented. Serotype 4b strains are, however, over-represented when compared with other serotypes among strains responsible for outbreaks and sporadic cases of listeriosis (4). The species also exists in two major genomic divisions, with substantial linkage disequilibrium and apparently limited gene flow between the two. Numerous molecular subtyping data indicate that the divisions fall along serotypic cluster lines, division I consisting of serotypes 1/2a, 1/2c, 3a and 3c, and division II of serotypes 1/2b, 4b and 3b (5–7). The clonality of the pathogen remains poorly described, and descriptions of diversity at the global genomic level have been lacking.
To date, only L.monocytogenes strain EGD-e (serotype 1/2a) and Listeria innocua CLIP 11262 (serotype 6a) have been fully sequenced (8). Although the initial comparison between these two strains provided considerable insight on the virulence attributes of this pathogen, the sequencing and comparative genomic analysis of additional strains was necessary if a core set of L.monocytogenes-specific genes was to be defined.
To better understand the molecular mechanisms of L.monocytogenes virulence in humans and survival of this bacterium in food and in the environment, a genomic survey of three strains of L.monocytogenes was conducted. These strains were chosen as they are food isolates associated with human listeriosis, and they represent the two main genomic divisions. More specifically, L.monocytogenes strain F2365 is a serotype 4b (genomic division II) cheese isolate from the Jalisco cheese outbreak of 1985 in California (9), L.monocytogenes strain F6854 is a serotype 1/2a (genomic division I) turkey frankfurter isolate from a sporadic case in 1988 in Oklahoma (10), and L.monocytogenes strain H7858 is a serotype 4b frankfurter isolate from the multistate outbreak of 1998–1999 in the USA (11). The strains were used in a comparative genomics study that includes a comparison with the two previously published strains: L.monocytogenes strain EDG-e (serotype 1/2a) and L.innocua strain CLIP 11262 (8). The analyses of the newly sequenced L.monocytogenes genomes have provided novel information that improves on current understanding of this species.
MATERIALS AND METHODS
Three strains of L.monocytogenes were sequenced by the random shotgun method, with cloning, sequencing and assembly conducted as described previously for genomes sequenced at The Institute for Genomic Research (TIGR) (12). The genome of strain F2365 was sequenced to closure, whereas the genomes of strains F6854 and H7858 were sequenced to 8-fold coverage of an estimated 3.5 Mbp genome without gap closure. Basically, one small insert plasmid library (1.5–2.5 kb) and one medium insert plasmid library (10–12 kb) were constructed for each strain by random mechanical shearing and cloning of genomic DNA. In the initial random sequencing phase, 8-fold sequence coverage was achieved from the two libraries (sequenced to 5- and 3-fold coverage, respectively). The sequences from the respective strains were assembled separately using TIGR Assembler or Celera Assembler (www.tigr.org). All sequence and physical gaps in strain F2365 were closed by editing the ends of sequence traces, primer walking on plasmid clones, and combinatorial PCR followed by sequencing of the PCR product. Pseudomolecules for strains F6854 and H7858 were constructed by first determining the order of the contigs relative to strain F2365 (for H7858) or to strain EGD-e (for F6854) using NUCmer (13). This information was then fed into BAMBUS (www.tigr.org) for scaffolding based on mate-pair information, repeat information and alignment to the reference genome.
An initial set of open reading frames (ORFs) that probably encode proteins was identified using GLIMMER (14), and those shorter than 90 bp as well as some of those with overlaps eliminated. For the closed F2365 genome, a region containing the likely origin of replication was identified and bp 1 was designated adjacent to the dnaA gene located in this region. For all three genomes, ORFs were searched against a non-redundant protein database as previously described (12). Frameshifts and point mutations were detected and corrected where appropriate. Remaining frameshifts and point mutations are considered authentic, and corresponding regions were annotated as ‘authentic frameshift’ or ‘authentic point mutation’, respectively. The ORF prediction and gene family identifications were completed using methodology described previously (12). Two sets of hidden Markov models (HMMs) were used to determine ORF membership in families and superfamilies. These included 721 HMMs from Pfam v2.0 and 631 HMMs from the TIGR ortholog resource. TMHMM (15) was used to identify membrane-spanning domains (MSDs) in proteins.
Comparative genomics
All genes and predicted proteins from the three sequenced L.monocytogenes genomes, as well as from all other published microbial genomes, were compared using BLAST. For the identification of strain-specific sequences, the genes from all five Listeria genomes were compared against each other. [A second filtering step was performed to determine the true uniqueness of these genes. The nucleotide sequence of each ‘unique’ gene (from the closed F2365 strains) was used as the query for BLASTN analysis against a WUBLAST-formatted database of the complete nucleotide sequence from each Listeria strain. Those genes that matched a non-self genomic sequence >90% of its length and with >90% identity were considered non-unique.] Newly identified genes that were not identified in the original comparisons of L.monocytogenes strain EGD-e and L.innocua strain CLIP 11262 (8) are available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession numbers TPA: BK005164–BK005176. Single nucleotide polymorphisms (SNPs) were identified by comparing the closed genome of strain F2365 with those of strains H7858 (178 contigs) and F6854 (133 contigs) using MUMer (13). A polymorphic site was considered of high quality when its underlying sequence comprised at least three sequencing reads with an average Phred score greater than 30 (16,17). (Strain EGD-e was not included in this analysis due to the fact that we did not have access to the underlying sequence files that are necessary for this analysis.) By mapping the position of the SNP to the annotation in the strain F2365 genome, it was possible to determine the location of the SNP (intergenic versus intragenic) and its effect on the deduced polypeptide (synonymous versus non-synonymous). For each deduced polypeptide, the degree of relatedness across strains was calculated using a BLAST score ratio. The BLASTP raw score was obtained for the alignment against itself (REF_SCORE) and the most similar protein in strains H7858, F6854 and EGD-e as well as for L.innocua CLIP 11262 (QUE_SCORE). Scores were normalized by dividing the QUE_SCORE for each query genome by the REF_SCORE. Normalized scores were plotted as xy coordinates.
A comparative database of Listeria genes was generated for position effect determination by identifying all matches between the five sequenced genomes using a BLAST-Extend-Repraze (BER) search (P-value <0.1; bit score >50). These BER matches were then run through position effect software (TIGR) to determine conservation of gene order. The query and hit gene from each match were defined as anchor points in gene sets composed of adjacent genes, with up to 10 genes upstream and downstream from each anchor gene used in creating the gene sets. An optimal alignment was calculated between the ordered gene sets using percentage similarity from BER and applying a linear gap penalty of 100. Positive scoring optimal alignments containing gene sets of four or more matching genes were stored in the database. The genome sequences and the annotation of the three TIGR-sequenced strains are available in the Listeria-specific comparative database at www.tigr.org/tdb/listeria. The nucleotide sequence for the closed genome of strain F2365 has been deposited at DDBJ/EMBL/GenBank under accession number AE017262. The genomes of strains F6854 and H7858 that were sequenced to 8-fold coverage were deposited at DDBJ/EMBL/GenBank under accession numbers AADQ00000000 and AADR00000000, respectively. The versions described in this paper are the first versions, AADQ01000000 and AADR01000000, for strains F6854 and H7858, respectively. The contigs separator that was used to create the pseudomolecules for the 8X strains is NNNNNTTAATTAATTAANNNNN.
RESULTS
Genome features and mobile elements
The completely sequenced genome of strain F2365 is a single, circular chromosome, 2 905 310 bp in length with an average G + C content of 38%. There are a total of 2847 predicted coding regions in the genome, and putative role assignments could be made for 1710 (60%) of the ORFs. Genome summary information on the sequenced strains is presented in Table 1 and Figure 1.
Table 1. Summarized features of three sequenced L.monocytogenes genomes.
| Strain | F2365 | F6854 | H7858 |
|---|---|---|---|
| Serotype |
4b |
1/2a |
4b |
| Isolated from |
Jalisco cheese |
Turkey frankfurter |
Hot dogs/meat products |
| Chromosome |
|
|
|
| Length (bp) |
2 905 310 |
∼2 953 211 |
∼2 893 921 |
| G + C content |
38% |
37.8% |
38% |
| No. of ORFs |
2847 |
2973 |
3024 |
| Assigned function |
1710 |
1792 |
1780 |
| Conserved hypothetical |
616 |
676 |
725 |
| Unknown function |
375 |
370 |
372 |
| Hypothetical |
146 |
82 |
112 |
| Unassigned |
0 |
53 |
35 |
| Phage regions |
2 |
3 |
2 |
| Monocins |
1 |
1 |
1 |
| Plasmid |
None |
None |
1 |
| Length (bp) |
– |
– |
82 270 |
| G + C content |
– |
– |
37.5% |
| No. of ORFs |
– |
– |
94 |
| Assigned function |
– |
– |
37 |
| Conserved hypothetical |
– |
– |
25 |
| Unknown function |
– |
|
1 |
| Hypothetical |
|
|
24 |
| Unassigned | - | 7 |
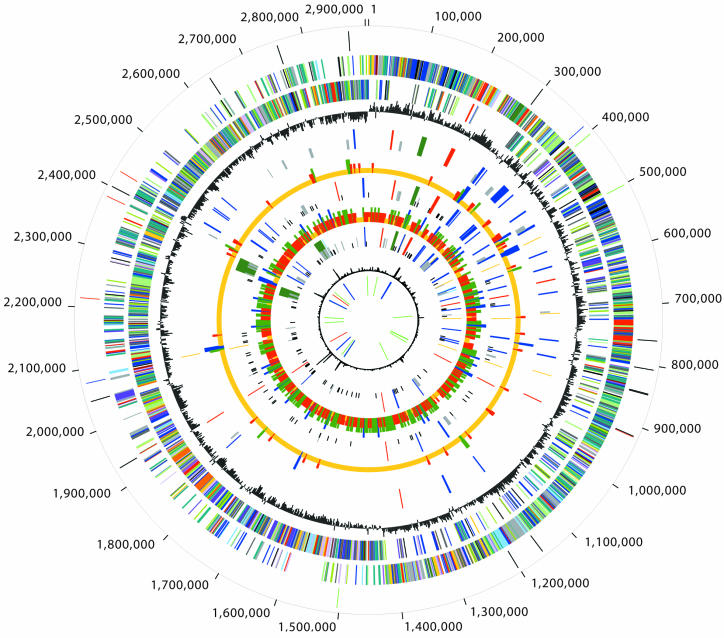
Figure 1.
Circular representation of the three sequenced Listeria genomes. Each concentric circle represents genomic data and is numbered from the outermost to the innermost circle. The outermost circle indicates the AscI (black), NotI (red), SfiI (blue) and SrfI (green) restriction map of the closed L.monocytogenes serotype 4b F2365 strain. The second and third circles represent the predicted strain F2365 ORFs on the + and – strands, respectively, colored by role categories: salmon, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; green, energy metabolism; purple, fatty acid and phospholipid metabolism; pink, protein fate/synthesis; orange, purines, pyrimidines, nucleosides and nucleotides; blue, regulatory functions; gray, transcription; teal, transport and binding proteins; black, hypothetical and conserved hypothetical proteins. The fourth circle shows the GC-skew. The fifth (strain F2365), seventh (strain H7858) and ninth (F6854) circles indicate ORFs involved in virulence: ORFs other than internalins (red), internalins (blue), putative prophage or monocin regions (dark green), transposable elements (gold), CRISPR elements (light blue), strain-specific genes (half-sized light gray ticks) and contig breakpoints (quarter-sized black ticks) relative to the closed F2365 strain. The sixth circle represents the number of SNPs per 5 kb for the H7858 strain, relative to the F2365 strains: blank, no SNPs (or unsequenced region); quarter-sized gold ticks, 1–30 SNPs; half-sized red ticks, 31–50 SNPs; three quarter-sized dark green ticks, 51–80 SNPs; full-sized blue ticks, more than 81 SNPs. The eighth circle represents the number of SNPs per 5 kb for the F6854 strain, relative to the F2365 strain: blank, no SNPs (or unsequenced region); quarter-sized gold ticks, 1–75 SNPs; half-sized red ticks, 76–200 SNPs; three quarter-sized dark green ticks, 201–300 SNPs; full-sized blue ticks, more than 301 SNPs. The tenth circle denotes atypical regions (χ2 value). The eleventh circle depicts tRNA (green), rRNA (blue) and sRNA (red) for the F2365 strain.
The chromosomes of the serotype 4b strains (F2365 and H7858) lack intact insertion sequence (IS) elements, but do contain four copies of transposase ORFA of the IS3 family that are present in homologous locations in both strains. The serotype 1/2a strains (F6854 and EGD-e) contain three copies of the same transposase ORFA in the same location as three of the ORFA insertions in the serotype 4b strains. The additional copy of the transposase ORFA in the serotype 4b strains appears to have resulted from a complete and a partial duplication (along with the associated regions) in strains F2365 and H7858, respectively. In addition, an intact IS element (ISLmo1) is present in the serotype 1/2a strains F6854 (two copies) and EGD-e (three copies), respectively. Two of the copies are in the same chromosomal location in both strains, but in opposite orientations. None of these insertions physically disrupt any host genes, and there is no evidence that they interfere with the expression of flanking host genes. Although internalin-like genes flank these two ISLmo1 elements, the transposase gene is always positioned downstream. It is possible that the inversion of these IS elements with respect to the internalin-like genes is biologically significant, whereby an outward-facing promoter of ISLmo1 could produce antisense RNA and silence the translation of the nearby internalin, thereby altering the invasive properties of the strain (18,19). Since all currently sequenced L.monocytogenes genomes contain copies of transposase ORFA, this gene probably originated from the genome of an ancestral Listeria before the strains diverged, in contrast to ISLmo1, which appears to be a recent acquisition and may still be mobile in the chromosomes of the serotype 1/2a strains.
The five Listeria genomes were compared in order to identify polymorphisms in a class of short, extragenic DNA repeats called the clustered regularly interspaced palindromic repeats (CRISPRs). Each CRISPR locus is composed of a repeated DNA sequence that is spaced by unique intervening sequence, but the role of these elements in microbial genomes is still not known (20,21). CRISPR repeats could be identified at variable loci in the genomes of the L.monocytogenes serotype 1/2a strains and L.innocua CLIP 11262 (Supplementary table 1 available at NAR Online), but not in the genomes of the serotype 4b strains. The repeat sequence differs by only one nucleotide between L.innocua CLIP 11262 and L.monocytogenes strain F6854 at locus 1, but is more variable at the two additional loci within strain F6854. The variable presence and absence of CRISPR elements in the Listeria lineage suggests that the presence of these elements is the result of gene transfer events. One possible application of the observed heterogeneity is the use of CRISPR repeats as markers to differentiate Listeria strains.
Comparative genome analysis of all five Listeria genomes revealed nine putative prophage and five putative monocins/defective or satellite prophage (Table 2). In addition to harboring the published prophage regions (8), the genomes of strain EGD-e and L.innocua CLIP 11262 possess a phage-related region that was not previously recognized (Figures 1 and 2, and Table 2). With φEGDe.1 as a reference, the nucleotide identity in the other Listeria phage-related regions was calculated as: φF2365.1 (96.29%), φF6854.1 (97.42%), φH7858.1 (96.22%) and φ111262.2 (85.7%). Six of the nine putative prophages have at least 11 ORFs that are homologous to ORFs of φA118 (>35% amino acid identity; less than 1 × 10–5 P-value over >75% of the length of the hit). In L.innocua CLIP 11262 and all the L.monocytogenes strains except strain F2365, a prophage has inserted into comK, the known target for integration of the serotype 1/2a-specific typing phage φA118 (22). Using NUCmer (13), the nucleotide percentage identity of these prophages to φA118 was determined as follows: φEGDe.2 (55.6%), φF6854.2 (59.2%), φH7858.2 (16.6%) and φ11262.5 (15.9%). Surprisingly, in L.innocua, a putative prophage (φ11262.1) with 43.5% nucleotide identity to φA118 (greater similarity than the comK-specific prophage) has inserted into tRNA-Lys-4. This prophage may be a relatively recent acquisition in L.innocua CLIP 11262. This prophage also appears to have swapped integrase regions with a phage that inserted into tRNA-Lys-4, possibly by recombination with an existing prophage. It should be noted that only strain F2365 did not contain an intact prophage in the genome.
Table 2. Genome properties for predicted prophage and monocin regions in the genomes of five Listeria.
| Name | Type | 5′ end | 3′ end | Size (bp) | % GC | ORFs | Span | Target | att site |
|---|---|---|---|---|---|---|---|---|---|
|
L.monocytogenes F2365 φF2365.1 |
Monocin |
132 412 |
143 134 |
10 723 |
37.31 |
17 |
LMOf2365_0131–LMOf2365_0147 |
None |
None |
|
L.monocytogenes H7858 φH7858.1 |
Monocin |
159 262 |
169 984 |
10 723 |
37.30 |
17 |
LMOh7858_0138–LMOh7858_0154 |
None |
None |
|
L.monocytogenes H7858 φH7858.2 |
Prophage |
2 387 007 |
2 346 385 |
40 623 |
35.46 |
66 |
LMOh7858_2410–LMOh7858_2475 |
comK |
ggacg |
|
L.monocytogenes F6854 φF6854.1 |
Monocin |
142 468 |
153 188 |
107 21 |
37.20 |
17 |
LMOf6854_0126–LMOf6854_0142 |
None |
None |
|
L.monocytogenes F6854 φF6854.2 |
Prophage |
2 384 184 |
2 342 944 |
41 241 |
36.10 |
48 |
LMOf6854_2344–LMOf6854_2391 |
comK |
gga |
|
L.monocytogenes F6854 φF6854.3 |
Prophage |
2 697 390 |
2 658 558 |
38 833 |
35.68 |
52 |
LMOf6854_2652–LMOf6854_2703 |
tRNA-Thr-4 |
ttaagccacttgtcggatttgaaccgacgacc ccttccttaccatggaag |
|
L.monocytogenes EGD-e φEGDe.1 |
Monocin |
120 657 |
131 377 |
10 721 |
37.28 |
17 (18) |
lmo0113–lmo0129 |
None |
None |
|
L.monocytogenes EGD-e φEGDe.2 |
Prophage |
2 402 413 |
2 360 621 |
41 793 |
36.11 |
62 (68) |
lmo2271–lmo2332 |
comK |
gga |
|
L.innocua 11262 φ11262.1 |
Prophage |
76 060 |
115 548 |
39 489 |
37.28 |
58 (63) |
lin0071–lin0129 |
tRNA-Lys-4 |
actcttaatcagcgggtcgggggttcgaaaccctcacaacccatatat |
|
L.innocua 11262 φ11262.2 |
Monocin |
155 934 |
166 658 |
10 725 |
36.26 |
17 |
lin0160–lin0176 |
None |
None |
|
L.innocua 11262 φ11262.3 |
Prophage |
1 246 499 |
1 297 065 |
50 567 |
34.61 |
71 (81) |
lin1231–lin1302 |
Similar to lmo1263 |
aagtacacatca |
|
L.innocua 11262 φ11262.4 |
Prophage |
1 762 142 |
1 713 123 |
49 020 |
36.06 |
69 (81) |
lin1697–lin1765 |
Intergenic |
tatcccacaaaa[a/aa]tcccacaa |
|
L innocua 11262 φ11262.5 |
Prophage |
2 445 938 |
2 406 614 |
39 325 |
35.86 |
54 (64) |
lin2372–lin2426 |
comK |
gga |
| L.innocua 11262 φ11262.6 | Prophage | 2 625 922 | 2 587 434 | 38 489 | 35.13 | 50 (63) | lin2561–lin2610 | tRNA-Arg-4 | atgccctcggaggga |
Note that the coordinates and predicted sizes of each region include sequences for putative core att sites. The number of ORFs was derived from the GenBank accessions for the two published genomes (8), but the number in parentheses reflects the number of predicted ORFs derived from a TIGR automated ORF prediction and annotation.
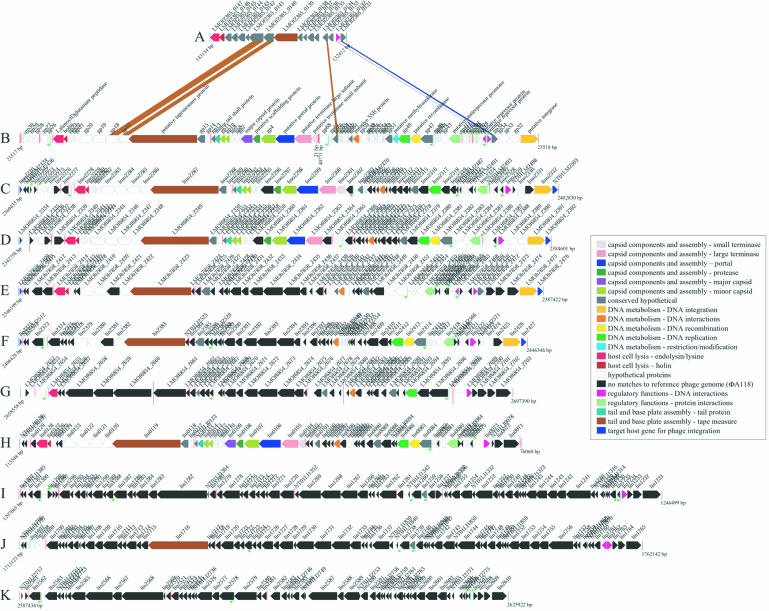
Figure 2.
Putative prophage regions within the genomes of five sequenced Listeria genomes. ORF colors are based on the annotation of the published sequence of L.monocytogenes 1/2a-specific typing phage φA118 (B). All putative prophage ORFs are colored to match φA118 if the protein sequence has a BLASTP P-value cut-off of ≤1 × 10–5, and a percentage identity ≥30% over 75% or more of the length of the query sequence. If there is no match to φA118 based on the above cut-offs, the ORF is colored black. The prophages are denoted as follows: (A) putative monocin region from L.monocytogenes F2365; (B) L.monocytogenes 1/2a-specific typing phage φA118 (Genbank accession NC_003216); (C) putative A118-like prophage φEGDe.2 from L.monocytogenes EGD-e inserted into comK; (D) putative A118-like prophage φF6854.2 from L.monocytogenes F6854 inserted into comK; (E) putative A118-like prophage φH7858.2 from L.monocytogenes H7854 inserted into comK; (F) putative A118-like prophage φ11262.5 from L.innocua CLIP 11262 inserted into comK; (G) putative prophage φF6854.3 from L.monocytogenes F6854 inserted into tRNA-Thr-4; (H) putative A118-like prophage φ11262.1 from L.innocua CLIP 11262 inserted into tRNA-Lys-4; (I) putative prophage φ11262.3 from L.innocua CLIP 11262 inserted into a previously undocumented gene similar to lmo1263; (J) putative prophage φ11262.4 from L.innocua CLIP 11262; and (K) putative PSA-like prophage φ11262.6 from L.innocua CLIP 11262 inserted into tRNA-Arg-4. Putative promoters (green bent arrows) were found in the Listeria putative prophages using the predicted promoter sequences from φA118 (22) and the EMBOSS program fuzznuc with a mismatch of 1. Putative transcriptional terminators (red lollypops) were found using the TIGR program TransTerm (www.tigr.org/software). Contig gaps (sequence or physical) are represented by vertical red lines.
In addition to the above listed mobile elements, we sequenced an L.monocytogenes plasmid identified in strain H7858. Named pLM80 to reflect its origin from L.monocytogenes and its approximate size, this 80 kb plasmid is populated by several different transposable elements that are not present in the chromosome, suggesting that the plasmid is a recent acquisition. Plasmid pLM80 has a high level of sequence and gene organization similarity to the L.innocua CLIP 11262 plasmid pLI100 (8) and the Bacillus anthracis plasmid pXO2 (23) (Fig. 3). The pXO2 plasmid encodes the genes responsible for regulation and production of the poly-d-glutamic acid capsule, one of the major virulence factors of this pathogen. In comparing these three plasmids, two distinct regions of similarity are evident. Region 1 is specific to Listeria and encodes proteins responsible for the detoxification of arsenate and cadmium. This region also contains six mobile genetic elements, five of which are >80% identical to genes of pLI100 (Fig. 3). The second region of pLM80 is most similar to a region of pXO2, but with a lower similarity level than seen in the Listeria-specific region, suggesting that this region was acquired some time ago, and has diverged substantially from its counterpart in pXO2. It is also possible that L.monocytogenes acquired this portion of the plasmid from a different source. In both pLM80 and pXO2, the genes are transcribed in the same direction, away from the conserved origin of replication, and encode a possible plasmid transfer apparatus. The LMOh7858_pLM80_0022 gene is similar to the TraD/TraG family of proteins that are membrane proteins essential for the assembly of the plasmid transfer apparatus. Additionally, this region encodes a number of proteins that have motifs indicative of proteins involved in the transport of surface-associated proteins. These features may play a role in plasmid transfer; thus far, the mechanism(s) of plasmid transfer in Listeria and many other Gram-positive organisms has not been fully elucidated. In addition to the two regions described above, a set of replication genes (LMOh7858_pLM80_0092–93, pLI0069–70 and BXB0039–40) are shared by pXO2, pLI100 and pLM80. The overall organization of pLM80 suggests that it is a composite plasmid, constructed by gene insertion and deletion events that were aided by Listeria-specific mobile genetic elements. Both plasmids pLM80 and pLI100 probably originated from a similar source, and the acquisition of the pXO2 region may have endowed pLM80 with increased mobility and/or a role in pathogenesis.
Figure 3.
Comparison of pLM80 from L.monocytogenes H7858 (B) with pLI100 from L.innocua CLIP 11262 (C) and pXO-1 from Bacillus anthracis (A). ORF colors are based on function. Matches between plasmid ORFs were based on BLASTP data where pLM80 was the query and the other two plasmids were in a WU-BLAST-formatted database. Only those ORFs with a percentage identity ≥35% and P-value of ≤1 × 10–5 were considered significant. Matches were colored based on percentage identity as follows: blue, 35–49% identity; brown, 50–59% identity; gold, 60–79% identity; green, 80–89% identity; red, 90–100% identity.
Comparative genome analysis
Comparison of the five Listeria genomes at the nucleotide and predicted protein levels revealed a number of differences that could possibly relate to the survival, growth and pathogenicity attributes of these strains and serotypes. A total of 51, 97, 69 and 61 strain-specific genes were identified from strains F2365, F6854, H7858 and EGD-e, respectively (Supplementary table 2). Unique to strain F2365 is a stretch of genes (LMOf2365_0331–LMOf2365_0323) that include a putative type II restriction endonuclease with specificity for GATC sites, a DNA methylase specific for cytosines at GATC sites, and a DNA-binding protein. The strain-specific genes on average are more likely to have atypical composition (Supplementary table 3), suggesting that some of these genes may have been acquired by gene transfer. Some of the strain-specific genes encode putative surface-associated proteins (www.tigr.org/tdb/listeria/) including proteins of the internalin family, and may contribute to the virulence of these strains.
Eighty-three genes were restricted to the serotype 1/2a strains (genomic division I), and 51 genes were restricted to the serotype 4b strains (genomic division II). Thirty-seven (44%) of the serotype 1/2a-specific and 33 (65%) of the serotype 4b-specific genes are hypothetical proteins for which there is no biochemical information (Supplementary table 2). Among the serotype 1/2a-specific genes are three clusters that encode pathways for the transport and metabolism of carbohydrates including ribose, and an unidentified pentose sugar. The serotype 1/2a-specific genes also include an operon that encodes the biosynthetic pathway for the antigenic rhamnose substituents that decorate the cell wall-associated teichoic acid polymer in serogroup 1/2a strains (3), five glycosyl transferases and an adenine-specific DNA methyltransferase.
A total of 8605 and 105 050 high quality SNPs were found in the genomes of L.monocytogenes H7858 and L.monocytogenes F6854, respectively, when compared with strain F2365. Of these high quality SNPs, 1984 (23%) and 16 811 (16%) resulted in a non-synonymous (NS) change in amino acid sequence in strains H7858 and F6854, respectively (Table 3; Supplementary figure 1A and B). When grouped by role category, there are a higher number of NS-SNPs in cell envelope and cellular processes (which includes pathogenesis and toxin production), as well as energy metabolism and transport, than in other role categories (Supplementary figure 2). Although strain F6854 contained many more SNPs than strain H7858, the SNP distribution across the role categories is largely conserved (Supplementary figure 2). The higher mutation rate of genes involved in energy metabolism and transport most probably reflects varying abilities to withstand adverse environments and to colonize different environmental niches. Variations in the genes involved in cell wall metabolism and in genes encoding cell wall-anchored proteins is likely to reflect the ability to interact with and infect various cell types and tissues.
Table 3. High quality SNPs in L.monocytogenes 4b H7858 and L.monocytogenes 1/2a F6854 when compared with L.monocytogenes 4b F2365.
| L.monocytogenes 4b H7858 | L.monocytogenes 1/2a F6854 | |
|---|---|---|
| Total high quality SNPs |
8605 |
105 050 |
| Intergenic SNPs |
653 |
6780 |
| Synonymous SNPs |
5968 |
81 459 |
| Codon position 1 |
233 |
3736 |
| Codon position 2 |
3a |
17a |
| Codon position 3 |
5732 |
77 706 |
| Non-synonymous SNPs |
1984 |
16 811 |
| Codon position 1 |
915 |
7328 |
| Codon position 2 |
687 |
5121 |
| Codon position 3 |
732 |
4362 |
| Transition rate |
70.1% |
68.3 |
| Transversion rate | 39.9% | 31.7% |
aSynonymous SNPs at the second position of a stop codon TGA→TAA or synonymous SNPs at both the first and second position of a serine codon.
Interestingly, the pleiotropic regulatory activator prfA (LMOf2365_0211 in strain F2365) and the four genes comprising the agr locus (agrA–D; LMOf2365_0057–60 in strain F2365) are completely conserved across all four L.monocytogenes strains. The PrfA regulon controls the major virulence genes (hly, plcA, plcB, mpl, actA, inlA and inlB) that are critical for virulence of Listeria. Recently, Autret et al. (24) demonstrated a role for the agr locus in bacterial virulence and in the secretion of proteins such as LLO, also under the control of PrfA. The fact that these regulatory systems interact to modulate the virulence network of Listeria, and that they are conserved across the different strains, suggests that they are under selective pressure to be maintained without mutations. The same agr genes are also present in the non-pathogenic L.innocua CLIP 11262, but with NS changes in the homologs of LMOf2365_0057, LMOf2365_0058 and LMOf2365_0059.
Environmental aspects and metabolism
The primary reservoir for L.monocytogenes is likely to be the natural environment. Environmental adaptations of the organism include the ability to grow at temperatures from 0 to 44°C, to utilize a wide variety of carbohydrate substrates, to grow in the presence of oxygen and under microaerophilic and anaerobic conditions, and to grow between pH 4.4 and 9.6. The bacterium can also grow in environments containing 10% (w/v) NaCl, and can survive at even higher salt concentrations. Profiling the metabolic capabilities encoded by the genome of strain F2365, sequenced to completion, revealed a range of substrate utilization and transporter abilities, traits that were also present in strains F6854, H7858 and EGD-e. The genomes have intact glycolysis and pentose phosphate pathways, and have genes for transport and utilization of a number of simple and complex sugars including fructose, rhamnulose, rhamnose, glucose, mannose, chitin, sucrose, cellulose, pullulan, trehalose and tagatose. These sugars are largely associated with the environments where L.monocytogenes is harbored, and conservation of genes for substrate utilization provides subtle clues regarding the survival and growth of the organism. In addition, the mevalonate biosynthesis pathway is intact, and the amino acids alanine, arginine, aspartate, glutamate, glycine, lysine, serine and threonine are probably substrates for growth. Listeria monocytogenes strain EGD-e possessed a substantial number of sugar phosphotransferase system (PTS) and ABC transporters comprising 4% of the genome (8); the additional L.monocytogenes strains display a similar overall array of sugar transporters. The overall similarities in metabolism and transport between the different L.monocytogenes serotypes suggest that substrate utilization patterns are not critical to the ability of the different serotypes to successfully colonize different environmental niches.
Virulence factor differences among strains
Differences in virulence among the different serotypes of Listeria, or even in strains belonging to the same serotype, remain undefined. Since major virulence determinants, such as the internalins InlA and InlB (internalization), listeriolysin LLO and phospholipases PlcA and PlcB (escape from the host vacuole), ActA (movement within the host cell cytoplasm) or the master regulator of virulence PrfA, are conserved in virulent and in less virulent strains of Listeria, the presence of these genes alone is not enough to explain the differences in virulence of any one particular strain. Through whole genome comparisons, we have identified a number of previously unknown protein sequences that have motifs characteristic of putative and known virulence factors (Supplementary tables 4 and 5). Among these newly identified putative virulence factors are cell wall-associated LPXTG proteins, choline-binding proteins, lipoproteins and internalins, all of which are variable in number across the four strains of L.monocytogenes that were compared. In addition, strain-specific genes associated with cell wall and teichoic acid biosynthesis, as well as glycosyl transferases, are probably related to differences in somatic antigens and may be involved in virulence and immunogenicity of listeriae.
The most noticeable example of a virulence-associated protein family with pronounced diversity among the sequenced genomes was the internalin family, members of which (internalin A and B) mediate bacterial internalization by non-professional phagocytes. Internalins are characterized by the presence of leucine-rich repeat (LRR) domains, consisting of tandem repeats of an amino acid sequence motif with leucine residues in fixed positions. The different internalins identified in the present study indicate an unusually high rate of mutation in these genes (Supplementary figure 3). Searches in the genome of L.monocytogenes strain F2365 revealed 25 internalins, 16 of which were associated with the cell wall, and nine secreted. The other serotype 4b strain, H7858, had the same total number of internalins, with 16 associated with the cell wall and nine secreted. The serotype 1/2a strain F6854 had 26 internalins, 17 associated with the cell wall, nine secreted, and three of which were unique to that strain. As for strain EGD-e, our analysis revealed 24 internalins, two of which were unique to this strain (Supplementary table 5). Phylogenetic analysis (Supplementary figure 3) across the strains shows that some internalins from the laboratory strain EGD-e, such as lmo0263, lmo2396 and lmo2445, cluster separately from the internalins of the newly sequenced outbreak strains, whether of serotype 4b (F2365 and H7858) or 1/2a (F6854) (Supplementary figure 3). This could reflect the fact that this strain was isolated from animal illness and is not a human outbreak isolate.
DISCUSSION
Listeriosis remains a significant public health problem, and whole genome comparison of major outbreak strains is providing important insights into the genetic complement that defines the survival, growth and pathogenicity characteristics of L.monocytogenes. The genomes presented here included representatives of the two major genomic divisions of this species as represented by strains of serotypes 1/2a and 4b. Strain F2365 was implicated in the California outbreak of 1985 (25) and is a representative of epidemic clone I, which includes a number of genetically closely related serotype 4b strains implicated in numerous geographically and temporally unrelated outbreaks (3). These outbreaks have involved diverse food vehicles, including soft cheeses, coleslaw and specialty meats, suggesting that this is a clonal group prevalent among processed foods and clearly capable of causing invasive illness in humans. The other serotype 4b strain (H7858) represents a different epidemic-associated clonal group (epidemic clone II) implicated in the 1998–1999 multistate outbreak in the USA, and possibly in a subsequent multistate outbreak in 2002. Both outbreaks involved contaminated processed meats.
Strain F6854 was implicated in human illness in 1988, and traced to contaminated turkey frankfurters from a specific processing plant. The same processing plant was implicated in a multistate outbreak in 2001, and the genetic fingerprint of the implicated bacteria was indistinguishable from that of strain F6854 by pulsed-field gel electrophoresis (3). For these reasons, this isolate serves as a model for serotype 1/2a strains capable of persistence in the processing plant, product contamination and invasive human illness. The genomic comparisons between strains EGD-e (also of serotype 1/2a) and F6854 have revealed sequences unique to each strain. This provides important information in light of the documented genetic diversity and plethora of strain subtypes in serotype 1/2a (3). In addition, the analysis revealed serogroup-specific genes that are shared by both serotype 1/2a strains, but are absent from the serotype 4b strains. The sequence of L.monocytogenes strain F6854 may prove more representative of serotype1/2a strains linked to human illness compared with the EGD-e strain, which was isolated from animal illness cases in 1924 (26).
Although strain-specific and serotype-specific genes were identified, the genomes of all four L.monocytogenes strains were remarkably similar in gene content and organization. Whole genome analysis has revealed that the L.mono cytogenes genomes are syntenic, with the majority of genomic differences consisting of phage insertions, transposable elements, scattered unique genes, and islands encoding proteins of mostly unknown function, as well as SNPs in many genes associated with virulence functions. With the exception of prophage sequences, genes found in L.innocua CLIP 11262 that were absent from strain EGD-e were typically absent from the genomes of the other three L.monocytogenes strains, suggesting that gene loss from a lineage ancestral to L.monocytogenes and L.innocua preceded the genomic diversification of L.monocytogenes into genomic divisions I and II. Conceivably, such gene loss may have contributed to genomic streamlining of L.monocytogenes, perhaps conferring higher fitness to the organism.
Considering the differences in epidemiologic background, genomic division and serotype of the strains, the high degree of similarity in the genomes is surprising. These findings suggest that L.monocytogenes strains prevalent in human and animal illness have surprisingly high genomic stability, and rely on a relatively small number of unique regions for antigenic diversity and epidemiologically relevant attributes. Such findings serve to direct the focus of research efforts to a relatively small number of specific genomic regions, to elucidate their possible involvement in virulence and adaptive physiology attributes of epidemic-associated bacteria. The relatively small number of unique genes and gene clusters suggests significant roles for these genes in virulence and/or the ecology of listeriae.
Finally, the comparison of the four sequenced L.monocytogenes genomes has provided valuable insight into defining the core genetic complement of the organism. This core complement forms the basic genetic underpinnings that define the organism’s ability to survive and grow in the many habitats it populates. Such information is especially important since measures to control the organism in the natural environment, in foods and in cases of human infection will probably exploit the products of these core genetic targets.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Michael Heaney, Michael Holmes, Vadim Sapiro, Dr Robert Strausberg and Michael Brown at The Institute for Genomic Research, for support with various aspects of this project. The authors also thank Dr Bala Swaminathan and colleagues at the Centers for Disease Control and Prevention for providing strains. This research was funded by the USDA, Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Kathariou S. (2003) Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes. In Torrence,M.E. and Isaacson,R.E. (eds), Microbial Food Safety in Animal Agriculture. Iowa State University Press, Ames, IA. [Google Scholar]
- 2.Mead P.S., Slutsker,L., Dietz,V., McCaig,L.F., Bresee,J.S., Shapiro,C., Griffin,P.M. and Tauxe,R.V. (1999) Food-related illness and death in the United States. Emerg. Infect. Dis., 5, 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathariou S. (2002) Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot., 65, 1811–1829. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization Joint FAO/WHO Food Standards. (2001) Programme, Proposed Draft Guidelines for the Control of Listeria monocytogenes in Foods. Technical Report No. Agenda Item 6. Codex Alimentarius Commission. [Google Scholar]
- 5.Bibb W.F., Gellin,B.G., Weaver,R., Schwartz,B., Plikaytis,B.D., Reeves,M.W., Pinner,R.W. and Broome,C.V. (1990) Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol., 56, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piffaretti J.C., Kressebuch,H., Aeschbacher,M., Bille,J., Bannerman,E., Musser,J.M., Selander,R.K. and Rocourt,J. (1989) Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl Acad. Sci. USA, 86, 3818–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosch R., Chen,J. and Luchansky,J.B. (1994) Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol., 60, 2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser P., Frangeul,L., Buchrieser,C., Rusniok,C., Amend,A., Baquero,F., Berche,P., Bloecker,H., Brandt,P., Chakraborty,T. et al. (2001) Comparative genomics of Listeria species. Science, 294, 849–852. [DOI] [PubMed] [Google Scholar]
- 9.Mascola L., Lieb,L., Chiu,J., Fannin,S.L. and Linnan,M.J. (1988) Listeriosis: an uncommon opportunistic infection in patients with acquired immunodeficiency syndrome. A report of five cases and a review of the literature. Am. J. Med., 84, 162–164. [DOI] [PubMed] [Google Scholar]
- 10.(1989) Epidemiological notes and reports listeriosis associated with consumption of Turkey Franks. MMWR Weekly, 38, 267–268. [PubMed] [Google Scholar]
- 11.(1998) Multistate outbreak of Listeriosis—United States, 1998. MMWR Weekly, 47, 1085–1086. [PubMed] [Google Scholar]
- 12.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 13.Delcher A.L., Phillippy,A., Carlton,J. and Salzberg,S.L. (2002) Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res., 30, 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 18.Engdahl H.M., Hjalt,T.A. and Wagner,E.G. (1997) A two unit antisense RNA cassette test system for silencing of target genes. Nucleic Acids Res., 25, 3218–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefan A., Reggiani,L., Cianchetta,S., Radeghieri,A., Gonzalez Vara y Rodriguez,A. and Hochkoeppler,A. (2003) Silencing of the gene coding for the epsilon subunit of DNA polymerase III slows down the growth rate of Escherichia coli populations. FEBS Lett., 546, 295–299. [DOI] [PubMed] [Google Scholar]
- 20.Jansen R., Embden,J.D., Gaastra,W. and Schouls,L.M. (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol., 43, 1565–1575. [DOI] [PubMed] [Google Scholar]
- 21.Jansen R., van Embden,J.D., Gaastra,W. and Schouls,L.M. (2002) Identification of a novel family of sequence repeats among prokaryotes. Omics, 6, 23–33. [DOI] [PubMed] [Google Scholar]
- 22.Loessner M.J., Inman,R.B., Lauer,P. and Calendar,R. (2000) Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol., 35, 324–340. [DOI] [PubMed] [Google Scholar]
- 23.Read T.D., Salzberg,S.L., Pop,M., Shumway,M., Umayam,L., Jiang,L., Holtzapple,E., Busch,J.D., Smith,K.L., Schupp,J.M. et al. (2002) Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science, 296, 2028–2033. [DOI] [PubMed] [Google Scholar]
- 24.Autret N., Raynaud,C., Dubail,I., Berche,P. and Charbit,A. (2003) Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun., 71, 4463–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuchat A., Swaminathan,B. and Broome,C.V. (1991) Listeria monocytogenes CAMP reaction. Clin. Microbiol. Rev., 4, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray E.G. (1953) The story of Listeria. Trans. R. Soc. Can., 47, 15–21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.