Abstract
X-ray repair cross-complementing protein-1 (XRCC1)-deficient cells are sensitive to DNA damaging agents and have delayed processing of DNA base lesions. In support of its role in base excision repair, it was found that XRCC1 forms a tight complex with DNA ligase IIIα and also interacts with DNA polymerase β (Pol β) and other base excision repair (BER) proteins. We have isolated wild-type XRCC1–DNA ligase IIIα heterodimer and mutated XRCC1–DNA ligase IIIα complex that does not interact with Pol β and tested their activities in BER reconstituted with human purified proteins. We find that a point mutation in the XRCC1 protein which disrupts functional interaction with Pol β, affected the ligation efficiency of the mutant XRCC1–DNA ligase IIIα heterodimer in reconstituted BER reactions. We also compared sensitivity to hydrogen peroxide between wild-type CHO-9 cells, XRCC1-deficient EM-C11 cells and EM-C11 cells transfected with empty plasmid vector or with plasmid vector carrying wild-type or mutant XRCC1 gene and find that the plasmid encoding XRCC1 protein, that does not interact with Pol β has reduced ability to rescue the hydrogen peroxide sensitivity of XRCC1- deficient cells. These data suggest an important role for the XRCC1–Pol β interaction for coordinating the efficiency of the BER process.
INTRODUCTION
Base excision repair (BER) is the major pathway involved in removal of endogenous and mutagen induced DNA damage. This repair process involves a number of sequential reactions initiated by a damage-specific DNA glycosylase that recognizes and removes the damaged base by cleavage of the N-glycosylic bond connecting the base to the sugar phosphate backbone. The arising apurinic/apyrimidinic site (AP site) is recognized by an apurinic/apyrimidinic endonuclease 1 (APE1) that incises the phosphodiester bond 5′ next to the AP site, followed by addition of one nucleotide to the 3′-OH end of the incised AP site and excision of the base-free sugar phosphate residue by DNA polymerase β (Pol β). Finally, DNA ligase completes repair by sealing the DNA ends (reviewed in 1).
Three major human DNA ligases (DNA ligase I, IIIα and IV) employ the same basic mechanism for DNA ligation, but participate in distinct biological processes namely DNA replication, BER and nonhomologous end joining, respectively (2). The mechanism linking DNA ligases to a particular biological process is unclear. It was proposed that other proteins might support functional diversity of the DNA ligases (3). Indeed, all of the DNA ligases mentioned above were found in tight complexes with other proteins. DNA ligase I forms a complex with PCNA (4), and DNA ligases IIIα and IV with X-ray cross complementing protein 1 (XRCC1) and XRCC4, respectively (5–7). Mutations in a DNA ligase partner protein frequently leads to genetic instability and sensitivity to DNA damaging agents (8–10) and all attempted knockouts of these genes were embryonic lethal (11).
Changes in the XRCC1 gene in characterized mutant cell lines (EM7, EM9, EM-C11 and EM-C12) resulted in undetectable levels of the XRCC1 protein and reduced (3–5 fold) levels of its counterpart protein, DNA ligase IIIα (12,13). These changes affect the mutant cells’ sensitivity to a number of DNA damaging agents including alkylating agents and X-rays, pointing to a deficiency in the BER pathway. Indeed, delayed ligation during BER was demonstrated for XRCC1-deficient cells (14). Since XRCC1 has no established biological activity, it was suggested that it might serve as a scaffold protein recruiting and positioning other components during BER (15,16). This idea was further supported by the multiple interactions of human XRCC1 with other BER proteins including Pol β (15,17), polynucleotide kinase (18), poly(ADP-ribose) polymerase 1 (PARP-1) (17,19), and APE1 (20). Although the physiological role of XRCC1–PARP-1 (21,22) and XRCC1–DNA ligase IIIα (23) interactions have been recently demonstrated, there are no such data for other XRCC1 interactions. In this report, we address the role of the XRCC1–Pol β interaction.
MATERIALS AND METHODS
Materials
Synthetic oligodeoxyribonucleotides purified by high-performance liquid chromatography were obtained from Midland (8-oxoguanine). [γ-32P]ATP (3000 Ci/mmol) was purchased from Perkin-Elmer Life Sciences.
Two-hybrid analysis
Performed as previously described (18).
Proteins
Recombinant human uracil-DNA glycosylase (UDG) was purified as described (24). Histidine-tagged human Pol β, APE1 and DNA ligase IIIα were purified on Ni2+-charged His-Bind Resin (Novagen, Cambridge, MA) as recommended by the manufacturer.
Transfection and survival experiments
The mammalian expression construct pcD2EXH, encoding human XRCC1 protein with a C-terminal decahistidine affinity tag, was described previously (5). A mutant derivative of pcD2EXH containing the V86R mutation was constructed using the ‘QuikChange’ protocol (Stratagene) and confirmed by sequencing. EM-C11 cells were cultured as monolayers in DMEM medium and a plasmid containing wild-type or mutant XRCC1 was introduced into EM-C11 cells using ‘Fugene 6’ transfection reagent (Roche) according to the manufacturer’s protocol. Stable clones were isolated under G418 (1.5 mg/ml) selection. Treatment with hydrogen peroxide was performed in DMEM medium with the indicated concentrations of the mutagen for 15 min. Colonies that appeared after 8–10 days were fixed with methanol, stained with Methylene Blue and counted. Relative colony formation (%) was expressed as colonies per treatment level/colonies that appeared in the control.
Purification of XRCC1–DNA ligase IIIα heterodimer
To purify the XRCC1–DNA ligase IIIα heterodimer, cell extracts prepared from EM-C11 cells by the method of Tanaka et al. (25) as modified by Vodenicharov et al. (26) were brought to 5 mM imidazole and 200 µl of 50% Ni-NTA agarose slurry (Qiagen) was added. After incubation at 4°C for 1 h, beads were washed five times with 0.5 ml of buffer containing 50 mM NaH2PO4 pH 8.0, 0.2 M KCl, 40 mM imidazole and the XRCC1–DNA ligase IIIα heterodimer was eluted with the same buffer containing 250 mM imidazole (3 × 100 µl), dialysed overnight against buffer containing 25 mM HEPES–KOH pH 7.9, 2 mM DTT, 12 mM MgCl2, 0.1 mM EDTA, 17% glycerol and 0.1 M KCl and stored at –80°C.
DNA ligase activity
DNA ligase activity of the purified DNA ligase IIIα and XRCC1–DNA ligase IIIα heterodimer was monitored using a 40mer oligonucleotide duplex containing a 3′-OH and 5′-phosphate nick at position 20. Reactions were carried out in a reaction mixture (10 µl) that contained 45 mM HEPES pH 7.8, 70 mM KCl, 7.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 2 mM ATP, 20 µM each of dATP, dGTP, dCTP and TTP, 0.5 mg/ml BSA, a FAM (6-carboxyfluorescein)-labelled oligonucleotide duplex (∼5 ng, 0.5 pmol) and the indicated amount of DNA ligase IIIα or XRCC1–DNA ligase IIIα heterodimer at 37°C for 20 min. The reaction was stopped by addition of 10 µl of gel loading buffer (95% formamide, 20 mM EDTA, 0.02% Bromophenol Blue and 0.02% xylene cyanole). Following incubation at 90°C for 3 min, the reaction products were separated by electrophoresis on a 20% denaturing polyacrylamide gel. One unit of DNA ligase activity was defined as the amount of enzyme converting 0.1 pmol of the substrate to a full-length product (40mer) at 37°C within 20 min.
Reconstituted repair assays
The reactions were reconstituted in a reaction mixture (10 µl) that contained 45 mM HEPES pH 7.8, 70 mM KCl, 7.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 2 mM ATP, 2 mg/ml BSA, 20 µM each of dATP, dGTP, dCTP and TTP, and a FAM (6-carboxyfluorescein)-labelled 30mer oligonucleotide duplex containing a uracil residue at position 19 (∼5 ng, 0.5 pmol). The reactions were initiated by incubation of the substrate with UDG (∼645 fmol), APE1 (∼300 fmol), Pol β (∼50 fmol) and the indicated amount of DNA ligase IIIα or XRCC1–DNA ligase IIIα heterodimer at 37°C for 20 min. The reaction was stopped by addition of 10 µl of gel loading buffer. Following incubation at 90°C for 3 min, the reaction products were separated by electrophoresis in a 20% denaturing polyacrylamide gel.
RESULTS
Purification and characterization of XRCC1–DNA ligase IIIα heterodimer
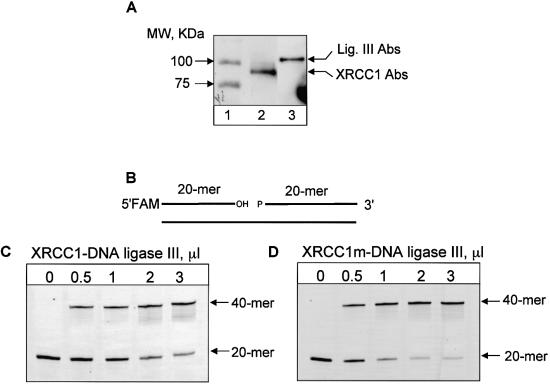
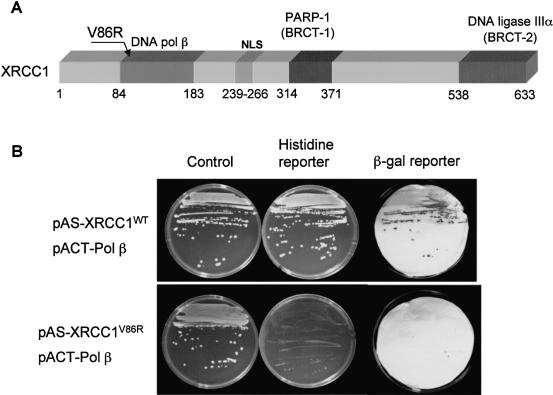
Recombinant XRCC1 and DNA ligase IIIα were shown to form a very tight complex resistant to dissociation at a high salt concentration (12) and it was also demonstrated that the two proteins co-purify over several chromatographic steps (6) and can be co-immunoprecipitated from whole cell extracts (18). To purify XRCC1–DNA ligase IIIα heterodimer, we used EM-C11 cells that contain a G → A substitution at nucleotide 1169 in the XRCC1 gene which leads to a C390 → Y amino acid change in the protein and is located in the putative functional BRCT1 domain (13). This mutation results in an undetectable level of XRCC1 protein in these cells (probably due to instability of the mutated protein) and reduced amounts of DNA ligase IIIα (12,13). We stably transfected these cells with wild-type His-tagged XRCC1 and pulled down the XRCC1–DNA ligase IIIα heterodimer using Ni-NTA agarose. Western blot analysis revealed that the pulled down fraction contained both XRCC1 and DNA ligase IIIα components (Fig. 1A). The complex was also tested and shown to be active in a DNA ligase activity assay with a nicked oligonucleotide (Fig. 1C and D). However, since a Coomassie-stained gel of the heterodimer in addition to XRCC1 and DNA ligase IIIα also contained several background protein bands characteristic for non-specific binding to Ni-NTA agarose, the complex was tested for enzymatic activities that may interfere with BER. We did not find any substantial activity of DNA polymerase, UDG, AP endonuclease or any other nucleases in a reconstituted repair assay (data not shown). In control experiments, no ligase activity could be pulled-down from XRCC1-deficient cells (EM-C11) or HeLa cells containing untagged XRCC1 (data not shown). Marintchev et al. (27) have recently characterized a number of XRCC1 point mutations specifically disrupting the interaction of Pol β with the N-terminal domain of XRCC1. To test whether this interaction is important for the activity of the XRCC1–DNA ligase IIIα heterodimer in the BER reaction, we used site-directed mutagenesis to generate a V86R mutation in a cloned human XRCC1 cDNA (Fig. 2A). Yeast 2-hybrid analysis confirmed that the V86R mutation greatly reduced or ablated the interaction with Pol β in vivo (Fig. 2B). This was indicated by the ability of the wild-type protein, but not mutated protein, to activate the histidine and β-galactosidase reporter genes in yeast expressing Pol β. The mutation in XRCC1 did not affect the interaction of XRCC1 with DNA ligase IIIα, however (data not shown).
Figure 1.
Isolation and characterization of XRCC1–DNA ligase IIIα heterodimer. (A) XRCC1–DNA ligase IIIα heterodimer pulled down by Ni-NTA agarose from EM-C11 cells transfected with His-tagged human XRCC1 was subjected to electrophoresis on a 10% polyacrylamide gel, transferred onto PVDF membrane, and immunoblotted against XRCC1 (lane 2) or DNA ligase IIIα (lane 3) antibodies. Molecular weight markers are shown on lane 1. (B) Schematic representation of nick-containing oligonucleotide substrate 5′-labelled with FAM. (C and D) DNA ligase activity of XRCC1–DNA ligase IIIα heterodimer. The indicated amount of wild-type (C) or mutated (D) XRCC1–DNA ligase IIIα complex was incubated with nick-containing substrate. Following incubation for 20 min at 37°C, reactions were stopped by addition of an equal volume of gel loading buffer and after incubation at 90°C for 3 min, the reaction products were separated by electrophoresis on a 20% denaturing polyacrylamide gel.
Figure 2.
Mutated XRCC1 (V86R) does not interact with Pol β in a yeast two-hybrid system. (A) Schematic representation of human XRCC1. Sites of interaction with other BER proteins, the nuclear localization signal (NLS) and site-directed point mutation (V86R) affecting XRCC1-Pol β interaction are indicated. Adapted from Thompson and West (28). (B) Wild-type and mutant human XRCC1 proteins were examined for interaction with human Pol β by yeast two-hybrid analysis. Yeast Y190 cells harbouring pACT-Pol β and either pAS-XRCC1WT or pAS-XRCC1V86R were examined for activation of the histidine and β-galactosidase reporter genes that are indicative of a protein–protein interaction. For this purpose, Y190 cells harbouring these constructs were plated onto minimal media lacking tryptophan and leucine, to select for the pAS and pACT constructs (‘Control’), and also on the above plates additionally lacking histidine and containing 50 mM 3-aminotriazole to test for protein–protein interaction (‘Histidine reporter’). Colonies from the control plates above were also examined for β-galactosidase activity by colony filter assays (‘β-gal reporter’).
Then we stably transfected EM-C11 cells with mutated His-tagged XRCC1 (XRCC1m) pulled down the XRCC1m–DNA ligase IIIα heterodimer using Ni-NTA agarose and tested its DNA ligase activity. The V86R mutation did not affect DNA ligase activity and the complex proved to be highly active (Fig. 1D).
Interaction of XRCC1–DNA ligase IIIα with Pol β is required for efficient ligation in reconstituted BER
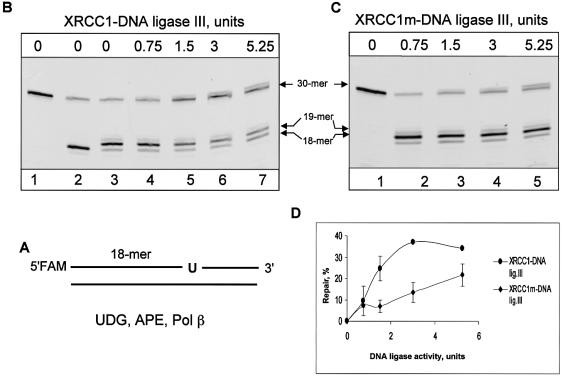
The potential function of XRCC1 as a scaffold protein has been discussed in a number of papers and the interaction of XRCC1 with Pol β was suggested as a possible molecular basis for such a role (reviewed in 16,18,28), however this model has not been verified experimentally. To demonstrate that the V86R mutation in XRCC1 protein affects the ability of the mutated XRCC1m–DNA ligase IIIα heterodimer to function in BER, we tested its DNA ligase activity and its ability to support BER in a reconstituted repair assay. We reconstituted short-patch repair of uracil in DNA with purified human recombinant proteins including UDG, APE1, Pol β and XRCC1–DNA ligase IIIα or mutated XRCC1m–DNA ligase IIIα heterodimer. The DNA end-joining activity of the XRCC1–DNA ligase IIIα heterodimer and XRCC1m–DNA ligase IIIα was initially measured using a single nick-containing oligonucleotide (Fig. 1C and D, correspondingly) and an equal amount of activity units was subsequently used in the reconstituted reaction (one unit of DNA ligase activity was defined as the amount of enzyme converting 0.1 pmol of the nicked substrate to full-length product at 37°C within 20 min).
For repair reactions, a 30mer oligonucleotide duplex containing a single uracil at position 19 was used as a substrate (Fig. 3A). Incubation of the 30mer substrate (Fig. 3B, lane 1) with UDG and APE1 leads to generation of the 18mer labelled product resulting from removal of uracil and incision of the AP site (Fig. 3B, lane 2). Pol β, when added to the reaction mixture, continues repair by adding one nucleotide to the 3′-end of the incised AP site and simultaneously removing the 5′-sugar phosphate, thus generating a substrate for DNA ligase (Fig. 3B, lane 3). The XRCC1–DNA ligase IIIα heterodimer stimulated ligation (observed as the 30mer full-length product) and stimulation depended on the amount of the heterodimer added (Fig. 3B and D). In contrast, the equivalent amount (in units of DNA ligase activity) of XRCC1m–DNA ligase IIIα was about 2–3 fold less efficient in reconstituted BER (Fig. 3C and D). We thus concluded that interaction of the XRCC1–DNA ligase IIIα complex with Pol β is required for efficient DNA ligation.
Figure 3.
Interaction of XRCC1–DNA ligase IIIα with Pol β is required for efficient ligation in reconstituted BER. (A) Schematic representation of the FAM-5′-labelled oligonucleotide substrate containing uracil (U). A 33 bp duplex oligonucleotide (5 ng, 500 fmol) containing a uracil residue at position 19 was incubated in reaction buffer containing magnesium, dNTPs, 20 ng (645 fmol) of UDG, 10 ng (300 fmol) of APE1, 2 ng (50 fmol) of Pol β and the indicated amount of wild-type (B) or mutated (C) XRCC1–DNA ligase IIIα complex. Untreated substrate was loaded on lane 1 of (B) and (C), and the reaction loaded on lane 2 of (B) contained only UDG and APE1. Following incubation for 20 min at 37°C, reactions were stopped by addition of an equal volume of gel loading buffer and after incubation at 90°C for 2 min, the reaction products were separated by electrophoresis on a 20% denaturing polyacrylamide gel. (D) Graphical representation of three independent experiments.
Disruption of XRCC1–DNA ligase IIIα–Pol β interaction increases cell sensitivity to hydrogen peroxide
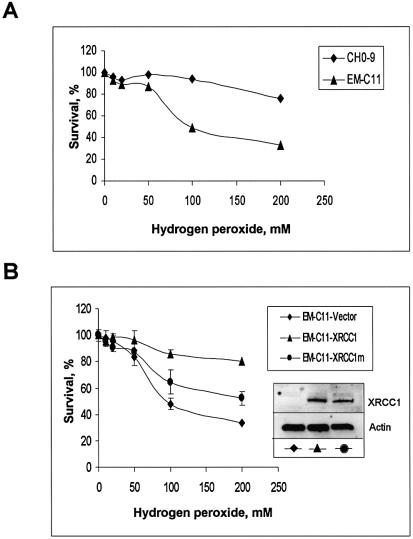
Finally, to address the biological role of this mutation, we compared sensitivity to hydrogen peroxide between wild-type CHO-9 cells, EM-C11 cells and EM-C11 cells transfected with empty plasmid vector and with plasmid vector carrying wild-type or mutant XRCC1 gene. Western blot analysis revealed that both clones transfected with the plasmid vector containing wild-type or mutated XRCC1 express similar levels of XRCC1, and no XRCC1 was detected in the EM-C11 cells transfected with the empty vector (Fig. 4B). In a clonogenic survival assay, we found that EM-C11 cells were approximately 2-fold more sensitive to killing by hydrogen peroxide than CHO-9 cells (Fig. 4A) and transfection of the EM-C11 cells with the vector carrying wild-type XRCC1, but not empty vector, completely rescued the EM-C11 cells (Fig. 4B). However, cells carrying mutated XRCC1 remained sensitive to hydrogen peroxide, although not as much as the EM-C11 cells (Fig. 4B).
Figure 4.
Reduced survival after hydrogen peroxide treatment of EM-C11 cells transformed with plasmid expressing mutated XRCC1. (A) Survival of wild-type (CHO-9) cells and XRCC1-deficient (EM-C11) cells after exposure to hydrogen peroxide. (B) Survival of EM-C11 cells transfected with expression plasmid harbouring wild-type XRCC1, V86R mutant XRCC1 or an empty vector after exposure to hydrogen peroxide. Expression of the XRCC1 gene in the transfected cell lines used for survival experiments was verified by running 30 µg whole cell extract proteins on a 10% SDS–polyacrylamide gel followed by transfer onto a PVDF membrane, and immunoblot analysis with the indicated antibodies.
DISCUSSION
Unless BER enzymes are organized in a preassembled DNA repair complex, BER should be a highly coordinated process to avoid accumulation of repair intermediates, since some of them, such as single strand breaks introduced by APE1 during the course of repair, are a major threat to genome integrity and should be repaired promptly. Recently, several groups proposed the so-called ‘passing the baton’ model for BER (1,29,30). According to this model, the complex of a repair protein with damaged DNA formed at one step of the repair is recognized by the enzyme involved in the next step. Thus, a DNA repair intermediate is ‘handed’ from one repair protein to another without exposure to a cellular milieu. In this model, interactions of XRCC1 protein with other BER proteins were suggested to play an important role. XRCC1 interacts with several enzymes implicated in BER including Pol β (15,17), PARP-1 (17,19) and DNA ligase IIIα (5–7). While the in vivo effects of disrupting the interactions with PARP-1 and DNA ligase IIIα have been characterized previously (21–23), the biological importance of the Pol β interaction had not been assessed until now. XRCC1 interacts with the palm-thumb domain of Pol β via its N-terminal domain (NTD) and this interaction is therefore distinct from the sites of interaction with either PARP-1 or DNA ligase IIIα (31). Recently, NMR chemical shift mapping of the XRCC1 NTD–Pol β interaction interface has identified several key residues involved in this protein–protein interaction and their importance has been confirmed by site-directed mutagenesis (27,32). We selected one of the XRCC1 mutants (V86R) for further study since it has been shown to completely abolish binding of the XRCC1 NTD to Pol β (27). The mutation is located in the five-stranded β-sheet of the core β-sandwich (33) and disrupts a key hydrophobic interaction with the Pol β thumb loop without, crucially, affecting the folding of the NTD (27).
In this study, we provide direct evidence of a biological role for the interaction of XRCC1–DNA ligase IIIα with Pol β by analysing the relative efficiency of wild-type and non-interacting mutant XRCC1–DNA ligase IIIα heterodimer in BER reactions reconstituted with purified human proteins. We first demonstrated that the V86R mutation does not affect the DNA ligase activity of the mutated XRCC1m–DNA ligase IIIα heterodimer as measured with a nicked oligonucleotide duplex substrate (Fig. 1). We then used similar amounts of DNA ligase activity in reconstituted BER reactions and found that the XRCC1–DNA ligase IIIα heterodimer, which does not interact with Pol β, has reduced ability to support BER (Fig. 3). We further demonstrated that this reduced ability of mutated heterodimer to function as a DNA ligase in BER is correlated with a reduced ability of the mutated XRCC1 gene to rescue the DNA damage-sensitive phenotype of XRCC1-deficient cells (Fig. 4). Taken together, our data provide the first evidence that interaction of Pol β with XRCC1–DNA ligase IIIα heterodimer plays an important role in cellular defence against DNA damage by supporting efficient BER.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr R. Robson for fruitful discussions and Dr M. Zdzienicka for providing EM-C11 cells. This work was supported in part by EC grant FIGH-CT 2002–0027.
REFERENCES
- 1.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 2.Tomkinson A.E. and Mackey,Z.B. (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. [DOI] [PubMed] [Google Scholar]
- 3.Tomkinson A.E., Chen,L., Dong,Z., Leppard,J.B., Levin,D.S., Mackey,Z.B. and Motycka,T.A. (2001) Completion of base excision repair by mammalian DNA ligases. Prog. Nucleic Acid Res. Mol. Biol., 68, 151–164. [DOI] [PubMed] [Google Scholar]
- 4.Levin D.S., Bai,W., Yao,N., O’Donnell,M. and Tomkinson,A.E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: implication for Okazaki fragment synthesis and joining. Proc. Natl Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldecott K.W., McKeown,C.K., Tucker,J.D., Ljungquist,S. and Thompson,L.H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 7.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 8.Thompson L.H., Brookman,K.W., Jones,J.J., Allen,S.A. and Carrano,A.V. (1990) Molecular cloning of the human XRCC1 gene, which corrects defective DNA repair and sister chromatid exchange. Mol. Cell. Biol., 10, 6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zdzienicka M.Z., van der Schans,G.P., Natarajan,A.T., Thompson,L.H., Neuteboom,I. and Simons,J.W. (1992) A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis, 7, 265–269. [DOI] [PubMed] [Google Scholar]
- 10.Stamato T.D., Weinstein,R., Giaccia,A. and Mackenzie,L. (1983) Isolation of cell cycle-dependent gamma ray-sensitive Chinese hamster ovary cell. Somatic Cell Genet., 9, 165–173. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E.C. and Meira,L.B. (2003) Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage. Version 5. DNA Repair (Amst), 2, 501–530. [DOI] [PubMed] [Google Scholar]
- 12.Caldecott K.W., Tucker,J.D., Stanker,L.H. and Thompson,L.H. (1995) Characterization of the XRCC1–DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen M.R., Zdzienicka,M.Z., Mohrenweiser,H., Thompson,L.H. and Thelen,M.P. (1998) Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Res., 26, 1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cappelli E., Taylor,R., Cevasco,M., Abbondandolo,A., Caldecott,K. and Frosina,G. (1997) Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem., 272, 23970–23975. [DOI] [PubMed] [Google Scholar]
- 15.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 16.Caldecott K.W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 23, 447–455. [DOI] [PubMed] [Google Scholar]
- 17.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehouse C.J., Taylor,R.M., Thistlethwaite,A., Zhang,H., Karimi-Busheri,F., Lasko,D.D., Weinfeld,M. and Caldecott,K.W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell, 104, 107–117. [DOI] [PubMed] [Google Scholar]
- 19.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidal A.E., Boiteux,S., Hickson,I.D. and Radicella,J.P. (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein–protein interactions. EMBO J., 20, 6530–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano S., Lan,L., Caldecott,K.W., Mori,T. and Yasui,A. (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol., 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Khamisy S.F., Matsutani,M., Suzuki,H. and Caldecott,K. (2003) A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res., 31, 5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore D.J., Taylor,R.M., Clements,P. and Caldecott,K.W. (2000) Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl Acad. Sci. USA, 97, 13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slupphaug G., Eftedal,I., Kavli,B., Bharati,S., Helle,N.M., Haug,T., Levine,D.W. and Krokan,H.E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry, 34, 128–138. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M., Lai,L.S. and Herr,W. (1992) Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell, 68, 755–767. [DOI] [PubMed] [Google Scholar]
- 26.Vodenicharov M.D., Sallmann,F.R., Satoh,M.S. and Poirier,G.G. (2000) Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res., 28, 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marintchev A., Gryk,M.R. and Mullen,G.P. (2003) Site-directed mutagenesis analysis of the structural interaction of the single-strand-break repair protein, X-ray cross-complementing group 1, with DNA polymerase beta. Nucleic Acids Res., 31, 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson L.H. and West,M.G. (2000) XRCC1 keeps DNA from getting stranded. Mutat. Res., 459, 1–18. [DOI] [PubMed] [Google Scholar]
- 29.Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature, 403, 451–454. [DOI] [PubMed] [Google Scholar]
- 30.Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nat. Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- 31.Marintchev A., Robertson,A., Dimitriadis,E.K., Prasad,R., Wilson,S.H. and Mullen,G.P. (2000) Domain specific interaction in the XRCC1–DNA polymerase beta complex. Nucleic Acids Res., 28, 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gryk M.R., Marintchev,A., Maciejewski,M.W., Robertson,A., Wilson,S.H. and Mullen,G.P. (2002) Mapping of the interaction interface of DNA polymerase beta with XRCC1. Structure (Camb), 10, 1709–1720. [DOI] [PubMed] [Google Scholar]
- 33.Marintchev A., Maciejewski,M.W. and Mullen,G.P. (1999) 1H, 15N and 13C resonance assignments for the N-terminal 20 kDa domain of the DNA single-strand break repair protein XRCC1. J. Biomol. NMR, 13, 393–394. [DOI] [PubMed] [Google Scholar]