Abstract
The timing of events in the cell cycle is of crucial importance, as any error can lead to cell death or cancerous growth. This accurate timing is accomplished through the activation of specific CDC genes. Mutations in the CDC40/PRP17 gene cause cell cycle arrest at the G2/M stage. It was previously found that the CDC40 gene encodes a pre-mRNA splicing factor, which participates in the second step of the splicing reaction. In this paper we dissect the mechanism by which pre-mRNA splicing affects cell cycle progression. We identify ANC1 as the target of CDC40 regulation. Deletion of the ANC1 intron relieves the cell cycle arrest and temperature sensitivity of cdc40 mutants. Furthermore, we identify, through point mutation analysis, specific residues in the ANC1 intron that are important for its splicing dependency on Cdc40p. Our results demonstrate a novel mechanism of cell cycle regulation that relies on the differential splicing of a subset of introns by specific splicing factors.
INTRODUCTION
The orderly progression through the mitotic cycle in eukaryotic cells is ensured by the highly regulated transition from one phase to the next. The accuracy of this process is critical for the high fidelity transmission of genetic information. Accuracy is achieved by the timely activation/inactivation of specific cell division cycle (CDC) genes, and is genetically controlled by surveillance mechanisms termed checkpoints (reviewed in 1).
In this work we have investigated the role of a specific CDC gene, CDC40, in connecting cell cycle progression and pre-mRNA splicing. CDC40 was first identified through the temperature-sensitive mutation cdc40–1. This mutation affects both the mitotic and meiotic cycles (2). At the restrictive temperature, cdc40 strains arrest as large-budded cells with one undivided nucleus containing a duplicated content of DNA, a phenotype characteristic of G2/M arrest (2,3). Besides the role played in the transition from G2 to M, Cdc40p is required for efficient entry into the S phase following release from α-factor-induced G1 arrest (3–5). In addition, cdc40 cells are also slightly sensitive at the restrictive temperature to UV and ionizing irradiation, and show sensitivity to DNA damaging agents such as methyl methane sulfonate (MMS) (6,7). Independently, genetic screens identified PRP17 as a gene involved in the second step of the splicing reaction; mutations in this gene accumulate small amounts of splicing intermediates (8). Sequence comparison revealed that CDC40 and PRP17 are the same gene. Thus the PRP17/CDC40 gene is involved in two seemingly unrelated processes: pre-mRNA splicing and progression through G1/S and G2/M phases of the cell cycle. For simplicity, henceforward we will refer to the gene as CDC40.
Eukaryotic genes are often interrupted by intervening sequences (introns) that must be cleaved out precisely during gene expression. Although introns need to be removed with single-nucleotide precision to avoid introducing catastrophic errors of frame, they contain only very few conserved sequences. Only three short regions are conserved among all yeast introns: the 5′ splice site (GUAUGAGU), the branchpoint (UACUAACA) and the 3′ splice site (PyAG, usually preceded by a pyrimidine-rich tract). These short sequences are highly important for the efficient splicing of pre-mRNA. For example, it has been shown that mutations in the first two nucleotides (GU) of the 5′ splice site or in the AG dinucleotide at the 3′ splice site of Saccharomyces cerevisiae introns block the first or second step of splicing, respectively (9,10).
Pre-mRNA splicing occurs by two transesterification reactions. In the first catalytic step, the 2′ hydroxyl of the conserved intronic adenosine attacks the phosphate at the 5′ splice site, producing a free 5′ exon and a branched species termed the lariat intermediate. In the second catalytic step, the 3′ hydroxyl of the 5′ exon attacks the phosphate at the 3′ splice site, yielding ligated mRNA and a lariat intron. Pre-mRNA splicing requires a large number of trans-acting factors that constitute the spliceosome (11). The splicing reaction is highly dynamic and requires a large number of protein–protein, protein–RNA and RNA–RNA interactions. The spliceosome includes several small nuclear ribonucleoprotein particles (snRNPs) composed of small nuclear RNA molecules (U1, U2, U4, U5 and U6 snRNA) associated with a large number of specific proteins. In addition, a large number of non-snRNP proteins are either transiently associated with, or constitute an integral part of, the spliceosome (11). Briefly, the 5′ splice site is initially recognized through a base-pairing interaction with U1 snRNA (12). The branchpoint sequence is first recognized by the branchpoint binding protein (BBP) in a sequence-specific fashion. This interaction is later replaced by base pairing with U2 snRNA. In the next step, the U4/U5/U6 triple snRNP binds. This binding heralds a large number of RNA:RNA rearrangements. U1 is released from the pre-mRNA, the stable interaction between U4 and U6 is disrupted and U4 is released. This release frees the U6 snRNA, which is now available for base pairing with U2 snRNA (12). Recent findings suggest that the RNA rearrangements are then followed by the recruitment of an additional protein complex (named NTC or CSC) which interacts and modifies the U5 snRNP (13). Having executed these RNA rearrangements, the spliceosome is competent to carry out the two steps of splicing. Although much progress has been made in understanding how the 5′ splice site and branchpoint are recognized, the mechanism that recognizes 3′ splice sites remains ill defined. Following the first transesterification reaction, another rearrangement occurs which aligns the two splicing sites and positions them for the second transesterification reaction (12). Recent findings suggest that all of the above rearrangements may occur within the context of a pre-existing higher-order particle [reviewed in (11)].
Cdc40p plays a role in the second step of the splicing reaction along with Prp8p, Prp16p, Prp18p, Prp22p and Slu7p (8,14). Previously, we implemented a screen to identify mutants that are lethal in the absence of Cdc40p. This screen identified several genes, including three novel genes SYF1–3, which were shown to play a role in both cell cycle progression and splicing (15,16). When the Syf proteins were used as baits in two-hybrid screens, we uncovered a complex network of interactions which suggested the existence of a protein complex (CSC) that plays a role in both splicing and cell cycle progression (15,16). Biochemical analysis in both S.cerevisiae and Schizosaccharomyces pombe has confirmed the existence of this complex (17,18; O. Dahan and M. Kupiec, unpublished results).
In addition to CDC40 and the SYFs, a handful of genes in both S.cerevisiae and S.pombe have also been identified in genetic screens for splicing factors and independently in screens for cell cycle regulators. These include PRP3 (19), PRP8 (20), PRP22 (21) and CEF1 (22) in S.cerevisiae, and prp2 (23), prp5 and prp6 (24), prp8 (25) and prp12 (26) in S.pombe. All of the above strongly indicate that pre-mRNA splicing and cell cycle control are linked in vivo. Despite this wealth of evidence, the exact molecular mechanism underlying the connection between cell cycle progression and splicing is still unclear.
In this paper we examine the role played by CDC40 in splicing and cell cycle regulation. By screening for cDNAs that can overcome the splicing defect in a cdc40 strain we show that the cell cycle arrest phenotype observed in this mutant is due to the inefficient splicing of the ANC1 pre-mRNA. We identify, through point mutation analysis, specific residues in the ANC1 intron that are important for its splicing dependency on Cdc40p. Our results suggest that cell cycle progression can be regulated at the level of splicing and that Cdc40p is important for the efficient splicing of a subset of introns with suboptimal splicing sequences.
MATERIALS AND METHODS
Media and growth conditions
Yeast cells were grown at various temperatures (in accordance with the required experimental conditions) in either YEPD (1% yeast extract, 2% bactopeptone, 2% dextrose) or SD media (0.67% yeast nitrogen base and 2% dextrose, supplemented with appropriate nutrients). In order to induce expression from the GAL promoter, cells were grown on YEPGal (1% yeast extract, 2% bacto peptone, 2% galactose) or SGal (0.67% yeast nitrogen base and 2% galactose, supplemented with appropriate nutrients) media. Bacto Agar (1.8%) was added for solid media. Selective media lacking one nutrient are designated SD-nutrient (e.g. SD-Ura). Ura– colonies were selected on SD complete medium with uracil (50 mg/l) and 5-fluoroorotic acid (5-FOA) (0.8 g/l). CuSO4 (Merck) was diluted into solid or liquid medium at the concentrations indicated from a stock 1 M solution.
Yeast strains
YR4: MATa ade2-1 his3-RS21 leu2Δ1 lys2-801 trp1Δ1 ura3-52 can1-100. SB37: MATa cdc40::URA3 ade2-1 his3-RS21 leu2Δ1 lys2-801 trp1Δ1 ura3-52 can1-100. MK89: MATa leu2-3,115 trp1-1 ade2-1 ura3-NcoΔ his3-11 can1-100. IM2: MATa cdc40::LEU2 leu2-3,115 trp1-1 ade2-1 ura3-NcoΔ his3-11 can1-100. IM2–M5: MATa cdc40::LEU2 TRP1- PGAL10-TUB1Δi leu2-3,115 trp1-1 ade2-1oc ura3-Nco his3-11 can1-100. YOD6: MATa cdc40::LEU2 TRP1- PGAL10-TUB1Δi ANC1ΔI leu2-3,115 trp1-1 ade2-1oc ura3-NcoΔ his3-11 can1-100. W303: MATa leu2-3,112 trp1Δ1 can1-100 ade2-1 ura3-1 his3-11,15. YOD1-A: MATa anc1::HIS3 leu2-3,112 trp1Δ1 can1-100 ade2-1 ura3-1 his3Δ300. YCL51: MATa ura3 leu2 his3 trp1 lys2 cup1::ura3. SB90: MATα cdcd40::leu2::HIS3 ura3 leu2 his3 trp1 lys2 cup1::ura3.
Plasmids
Saccharomyces cerevisiae cDNA library: yeast cDNA expression library, regulated by the GAL1 promoter cloned into pRS316 vector (URA3-CEN) (kindly provided by Professor D. Kornizer, Technion, Israel). YIplac211: integrative URA3 marked plasmid. pJU83: CUP1-ACT1 reporter plasmid (CEN-LEU2) (27).
The following plasmids contain an ANC1-CUP1 reporter, and were constructed as explained below. pSBY87: ANC1-CUP1 reporter plasmid, based on pJU83 (CEN-LEU2). BOD3: ACT-ACT, contains the ACT1 branchpoint and the ACT1 3′ splice site. BOD6a: ANC-ACT, contains the ANC1 branchpoint and the ACT1 3′ splice site. BOD7: ANC-ANC, contains the ANC1 branchpoint and the ANC1 3′ splice site. BOD8: ACT-ANC, contains the ACT1 branchpoint and the ANC1 3′ splice site. BOD9: ANC T71A-ACT, is BOD6a with a T71A mutation. BOD10: ACT-ANC A97T, is BOD8 with an A97T mutation.
Construction of the CUP1 reporting plasmids
Plasmid pSBY87 was constructed as follows: pJU83 (28) was digested with BamHI followed by partial digestion with Asp718, causing the release of ACT1 intron. A PCR fragment containing the ANC1 intron was produced using the following primers: upper primer: 5′-CGCGGATCCATGGTAGCTGT ATGT-3′; lower primer: 5′-CGGGGTACCTTTTACTGTC TGTCTGTCTGTTG-3′. After digestion of the PCR fragment with both BamHI and Asp718, it was ligated to the linear plasmid to create pSBY87.
The plasmids containing the ACT1-ANC1 chimeric introns were constructed as follows. First, pJU83 was digested with both XhoI and PacI causing the release of the branchpoint and 3′ splice site of the ACT1 intron. PCR fragments containing different combinations of ACT1 and ANC1 branchpoint and 3′ splice site were obtained using the following as templates: pJU83 (in the case of BOD3 and BOD6a), pSBY87 (in the case of BOD7 and BOD8), BOD6a (in the case of BOD9) or BOD8 (in the case of BOD10). The sequence of primers used to construct each of the PCR fragments is available upon request.
All PCR fragments were digested with XhoI and PacI and ligated to the purified linear pJU83 plasmid, yielding the desired clones.
Cell synchronization
Exponentially growing cells were arrested in G1 by incubation in the presence of 4 µM α-factor (Sigma-Aldrich, Israel) for 3 h at 25°C. In order to release cells from G1 arrest, they were washed three times and resuspended in fresh YEPD.
Screening for temperature-sensitivity suppressors
IM2-M5 (cdc40Δ) cells were transformed with a cDNA expression library regulated by the GAL1 promoter on the URA3-CEN vector pRS316. The transformation mixture was plated on SGal-Ura plates at 25°C. After 12 h, cells were shifted to the restrictive temperature (33°C) for 3–4 days until the appearance of colonies. Colonies were then streaked on YEPGal plates and replicated on YEPGal and 5-FOA plates at 25 and 33°C. Colonies that were able to grow on YEPGal plates at 25 and 33°C and on 5-FOA plates at 25 but not at 33°C were further analyzed. Plasmids were extracted, re-checked for their ability to support growth at the restrictive conditions and then their cDNA insert was sequenced.
Creation of a genomic ANC1Δi allele
A PCR fragment containing 785 bp upstream of the ANC1 open reading frame (ORF) and the first 30 bp of the ANC1 cDNA was amplified using the OR1 and OR3 oligonucleotides and the pMW13 plasmid as a template. A second PCR fragment containing the entire ANC1 cDNA was amplified using the OR2 and OR4 oligonucleotides. Both PCR fragments were then used as templates in a third PCR using the OR1 and OR2 oligonucleotides, yielding a 1.6 kb fragment containing 785 bp upstream of the ANC1 ORF fused to the entire ANC1 cDNA. This fragment was cloned into the integrative URA3 marked plasmid YIplac211 and transformed into IM2-M5 cells. 5-FOA-resistant colonies (pop-outs) were checked by PCR in order to identify cells in which the genomic ANC1 was replaced by its cDNA. Primers used are listed below (restriction sites are underlined). OR1: 5′ CGCGGATCCGCTATTACAAGAAGCTTTG 3′. OR2: 5′ CCGGAATTCGCGGTTTTGGAATAAAGTTTAT 3′. OR3: 5′ ACGGATGGTTCTTTTTACTGTAGCTACCATGATTA GTTATCT 3′. OR4: 5′ AGATAACTAATCATGGTAGCT ACAGTAAAAAGAACCATCCGT 3′.
RT–PCR analysis
RNA was extracted from 5 × 107 cells using the RNeasy kit (QIAGEN Inc.) according to the manufacturer’s instructions. Prior to the RT–PCR step, genomic DNA was degraded by RQ1 RNase-free DNase (Promega Inc.). Complete removal of contaminating DNA was verified by negative control PCRs with a specific set of primers. One microgram of total RNA was used as a template for cDNA synthesized using the Expand™ Reverse Transcriptase Kit (Boehringer Mannheim) and 500 ng of oligo (dT)15 (Boehringer Mannheim) as a primer. A quarter of each cDNA preparation was used as a template in a PCR using specific primers spanning the intron of either ANC1 or RPL25.
Copper growth assays
Copper-containing plates were made by diluting CuSO4 into SD agar lacking the relevant nutrients at the concentrations indicated from a stock solution of 1 M. Copper-resistant growth was determined by plating assays. Logarithmic cultures of the strains of interest containing the relevant plasmids were grown to the same cell number (107 cells/ml). Consecutive decimal dilutions were carried out, and drop-out assays were performed on plates with different concentrations of copper. Growth was assayed after 3 days of incubation at either 25 or 30°C.
Mutagenesis of ANC1 3′ splice site
Two complementary oligonucleotides, each containing the ANC1 3′ splice site, were synthesized (named DEG5 and DEG3). Random nucleotides were incorporated at five different sites in DEG5 and DEG3 (corresponding to positions 94, 95, 97, 99 and 101 in the ANC1 intron) as follows: DEG5, 5′ CTATTCTCATCATTANNTNANCNA CAGACGTTAAAG 3′; DEG3, 5′ CTTTAACGTCTGTNG NTNANNTAATGATGAGAATAG 3′.
Each degenerate oligonucleotide was then used to generate a PCR fragment using BOD8 as a template in combination with oligonucleotides CUP5 or CUP3 which are complementary to upstream and downstream sequences from BOD8: CUP5, 5′ CTTTTAGATTTTTCACGC 3′; CUP3, 5′ GACT ATTCGTTTCATTTC 3′.
The two PCR fragments were then used as templates in a new PCR using the CUP5 and CUP3 primers, yielding PCR fragments containing random sequences at the desired positions.
Screening for mutated clones that confer copper resistance
The mutated PCR fragments were co-transformed into SB90 cells along with a linear BOD8 plasmid (digested with XhoI and PacI). The cells were then plated on SD-Leu plates containing 0.2 mM copper and incubated at 25°C for 2–3 days until the appearance of colonies. This copper concentration was chosen because it was just above the maximal copper concentration at which BOD8 allowed growth. After rechecking the ability of the colonies to grow at high copper concentrations, the colonies were streaked on YEPD plates to allow plasmid loss and then replica plated on SD plates with or without 0.2 mM copper. Colonies that were unable to grow on plates containing copper without the plasmid were further analyzed. Plasmids were extracted, rechecked for their ability to support growth at high copper concentration and subjected to sequence analysis.
RESULTS
The cell cycle arrest phenotype of cdc40Δ cells can be suppressed by ANC1 cDNA
Since splicing constitutes a key cellular mechanism, genes encoding splicing factors are usually essential. Many temperature-sensitive splicing mutants have been isolated; most of them arrest at the restrictive temperature without any defined cell cycle morphology (16). However, as has been shown previously, cdc40 mutant cells are temperature sensitive, arresting at the restrictive temperature with a large bud and one undivided nucleus (2). FACS analysis and β-tubulin staining showed that cdc40 mutant cells held at the restrictive temperature contain 2C DNA content and a short spindle indicating that cells are arrested at the G2/M phase of the cell cycle (2,3). We reasoned that inefficient processing of a small number of intron-containing genes particularly dependent on Cdc40p for their pre-mRNA splicing might cause the cell cycle specific arrest of cdc40 mutant cells. If this were the case, we predicted that introduction of the relevant target gene(s) in the form of cDNA should be sufficient to bypass the need for Cdc40p and should allow growth at the restrictive temperature.
To test this hypothesis, we conducted a screen to isolate cDNAs able to suppress the temperature-sensitive cell cycle arrest of cdc40Δ cells. A cdc40Δ strain was transformed with a yeast cDNA expression library and screened for cDNAs that support cell growth at 33°C, a restrictive temperature for cdc40 cells. About 100 000 colonies were screened; in addition to the CDC40 gene itself, we isolated five independent clones, all carrying the ANC1 cDNA.
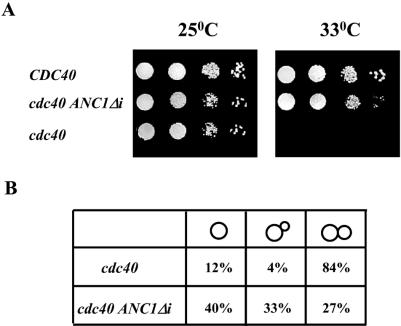
In order to ensure that the suppression by ANC1 cDNA results from the ability to bypass the cdc40Δ splicing defect, and not simply from over-expression of ANC1, we constructed a cdc40Δ strain in which the chromosomal ANC1 gene was replaced by its cDNA under the control of its endogenous promoter (designated ANC1Δi). As shown in Figure 1A, the ANC1Δi allele was able to support growth of cdc40Δ cells at the restrictive temperature of 33°C. However, no growth was observed at higher temperatures. Microscopic examination of cultures held at 37°C revealed that in contrast with cdc40Δ mutants, cdc40Δ anc1Δi cells do not exhibit a cell cycle specific arrest (Fig. 1B). The fact that ANC1 cDNA was unable to support full vigorous growth of cdc40Δ cells at 37°C (data not shown), suggests that other genes, in addition to ANC1, are dependent on Cdc40 for splicing. These genes, however, are not required for a specific cell cycle stage.
Figure 1.
ANC1 cDNA suppresses the temperature-sensitive phenotype of a cdc40Δ strain. (A) A wild-type strain (MK89), a cdc40Δ strain containing ANC1 cDNA under its endogenous promoter (ANC1Δi, YOD6) and a cdc40Δ strain (IM2) were diluted, spotted on YEPD plates and assayed for viability at either permissive (25°C) or restrictive (33°C) temperatures. (B) IM2 (cdc40Δ) and YOD6 (ANC1Δi) were synchronized at G1 with α-factor and transferred to 37°C. After 6 h incubation cells were analyzed by microscopic observation and classified as being in the G1 (unbudded), S (small-budded cells) or G2/M (large-budded cells) cell cycle stage. More than 300 cells were classified in this manner.
ANC1 pre-mRNA splicing is not cell cycle regulated
ANC1 was first isolated in a screen for mutants that fail to complement a temperature-sensitive allele of ACT1 in the heterozygous state. anc1 mutant cells grow slowly at 30°C and are unable to grow at 37°C (29). In addition, anc1 mutants exhibit defects in actin cytoskeleton organization and several morphological aberrations which correlate with cytoskeletal imperfections such as unusual cell morphology, sensitivity to high osmolarity and low mating (29). However, Anc1p resides exclusively in the nucleus, which is devoid of actin, suggesting that Anc1p is not involved in the actin cytockeleton function per se. Subsequently, it was shown that Anc1p is a component of several transcription complexes, including the Mediator (30), the TFIID and TFIIF basal transcription complexes (31), the SWI/SNF chromatin remodeling complex (32) and the NuA3 histone aceytelation complex (33).
It should be noted that Anc1 is not a ‘sticky’ protein that is co-purified artifactually with all yeast complexes. It has been identified only in a selective minority of yeast complexes that affect transcription/chromatin, and always in stoichiometric quantities. Anc1p is remarkable in that it is an abundant protein present in several regulatory complexes, and yet, unlike the other members of these complexes, it is not essential for cell viability. The role of ANC1 remains unclear; it may play a role in regulating transcriptional regulators or it may act as a mediator between several complexes, linking different processes in the cell.
Since the expression of ANC1 cDNA was able to suppress the cdc40Δ cell cycle arrest phenotype, we assumed that Anc1p is required for cell cycle progression through the G2/M transition at high temperatures. A search in the current databases of DNA microarray experiments shows that the steady state RNA level of ANC1 is very low and its transcript levels remain constant and are not cell cycle regulated. Therefore ANC1 constitutes a good candidate for a gene whose expression is regulated by splicing. In an attempt to find out whether the splicing pattern of ANC1 pre-mRNA varies at different cell cycle stages, we measured its expression by RT–PCR. Wild-type cells were synchronized at the G1 stage of the cell cycle with α-factor and then released into fresh YEPD medium at 30°C. At different time points, samples were taken, RNA was extracted and RT–PCR analysis using primers spanning the ANC1 intron was performed. In addition, the cell cycle distribution of the cell population was determined. As shown in Figure 2, ANC1 pre-mRNA splicing in wild-type cells remains efficient and constant through all cell cycle stages and does not seem to be regulated during cell cycle progression.
Figure 2.
ANC1 pre-mRNA splicing is not cell cycle regulated. Wild-type cells (strain W303) were synchronized in G1 with α-factor and then released into fresh YEPD media at 30°C. At timely intervals aliquots were collected and samples were analyzed by microscope for cell cycle distribution (A), or were subject to RT–PCR analysis using ANC1 specific primers (B). Unbudded cells were classified as being in G1, cells with small buds (less than half the size of the mother cell) were considered to be in S phase and cells with large buds were classified as being in G2/M. As a control an RT–PCR was carried out with a cdc40Δ strain incubated for 2 h at 37°C.
Cdc40p is required for the efficient splicing of ANC1 pre-mRNA
The ability of ANC1 cDNA to suppress the cell cycle arrest of cdc40Δ cells in restrictive conditions supports the notion that Cdc40p is required for the efficient splicing of ANC1 pre-mRNA. To test this possibility, we carried out RT–PCR analysis. Total RNA from cdc40Δ and wild-type strains was isolated before and after the cells were shifted to the non-permissive temperature. RNA was subjected to RT–PCR analysis using primers spanning either the ANC1 intron or, as a control, the intron-containing ribosomal gene RPL25. As shown in Figure 3, when cells deleted for CDC40 were incubated at the non-permissive temperature, an accumulation of unspliced ANC1 was observed. Such an accumulation was not seen in cells incubated at the permissive temperature, or in wild-type cells at either temperature. No pre-mRNA accumulation was seen using primers specific to the ribosomal gene RPL25 (Fig. 3) or to six additional intron-containing genes (data not shown). These results indicate that the splicing of ANC1 pre-mRNA is highly dependent on the Cdc40 protein, and that the requirement for Cdc40p as a splicing factor is restricted to a subset of intron-containing genes.
Figure 3.
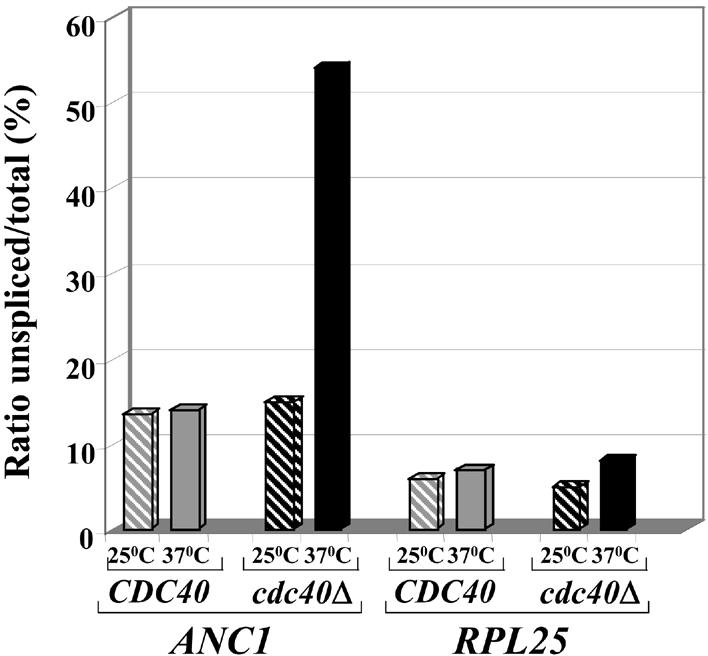
Cdc40 protein is required for efficient splicing of the ANC1 intron. cdc40Δ (IM2) and wild-type (MK89) strains were grown exponentially at 25°C and then transferred to either permissive (25°C) or restrictive (37°C) temperatures for 3 h. RNA was extracted and analyzed by RT–PCR using ANC1 or RPL25 specific oligonucleotides spanning the relevant introns. The level of products for spliced and unspliced transcripts of ANC1 and RPL25 was quantified after gel electrophoresis. For each strain and temperature, the ratio of unspliced to total is shown. Similar results were obtained after 1.5 and 6 h incubation at 37°C.
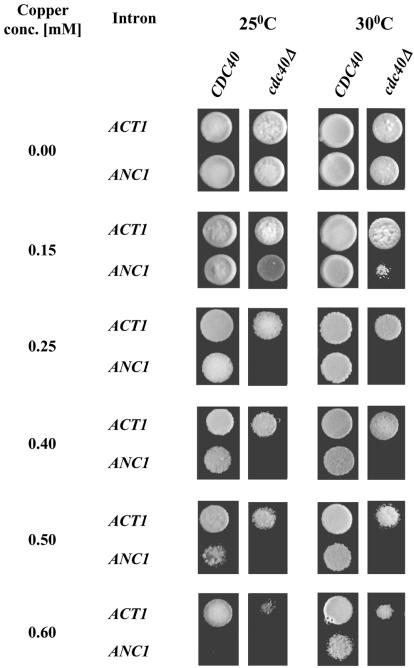
Additionally, in order to monitor pre-mRNA splicing in vivo and further examine the ANC1 splicing dependency on Cdc40p, we have developed a reporting assay based on the ACT1-CUP1 system (28). CUP1 is a non-essential gene that encodes a 61 amino acid metallothionein protein. Cup1p protects cells from copper toxicity by chelating the metal in a dosage-dependent manner, therefore allowing cells to grow in the presence of otherwise lethal concentrations of copper. CUP1 thus provides a sensitive assay for studying splicing (28).
In the splicing reporting system, the CUP1 gene was cloned downstream to an intron-containing fragment of the ACT1 gene. The ACT1 promoter was replaced by the strong constitutive GPD promoter (28). In order to use this reporter system for the analysis of ANC1 splicing, we replaced the ACT1 intron by the intervening sequence of ANC1. The reporter-containing plasmids (carrying either the ACT1 or the ANC1 intron) were introduced into either cup1Δ cdc40Δ or cup1Δ CDC40 strains. The transformants were then spotted on copper-containing plates. Since the ability to grow in the presence of different copper concentrations depends on the amount of correctly spliced CUP1 pre-mRNA, one can estimate the splicing efficiency of the ANC1 intron by assaying the level of copper resistance. The results are summarized in Figure 4. cup1Δ CDC40 cells were able to grow at high copper concentrations (0.5 mM) in the presence of either ACT1 or ANC1 introns. Note that splicing of the ANC1 intron at 25°C is slightly less efficient in these cells than splicing of the ACT1 intron. Similarly, cup1Δ cdc40Δ cells containing the ACT1 intron were able to grow at high copper concentrations (0.6 mM). However, when the same cells were transformed with the ANC1-intron-containing plasmid, they exhibited a marked decrease of copper tolerance (≤0.15 mM copper). These results prove that the splicing efficiency of the ANC1 intron (but not that of ACT1) is highly reduced in the absence of Cdc40p.
Figure 4.
Cdc40 protein is required for efficient splicing of the ANC1 intron. cup1Δ cdc40Δ (SB90) and cup1Δ CDC40 (YCL51) strains were transformed with plasmids bearing ACT1-CUP1 or ANC1-CUP1 reporter gene fusions. The transformants were diluted, spotted on minimal selective plates and tested for the ability to grow in the presence of increasing copper concentrations.
Taken together, the RT–PCR and ACT1-CUP1 reporting system results show that the splicing of ANC1 is extremely dependent on Cdc40p.
Both the branchpoint and the 3′ splice site of ANC1 pre-mRNA cause splicing dependency on the Cdc40 protein
The results presented so far support the possibility that splicing factors such as Cdc40p are differentially required for the efficient processing of a subset of intron-containing transcripts. This dependency could be due to unique intron features. For example, it has been shown that the requirement for Slu7p during the second step of splicing increases with the distance between the branchpoint and the 3′ splice site. Slu7p is necessary only for efficient splicing of introns in which the distance between the 3′ splice site and the branchpoint is >7 nt (34). Since Cdc40p is required for the splicing of only a subset of genes (including ANC1), we reasoned that Cdc40p target genes might contain some unique features that make their splicing highly dependent on Cdc40p.
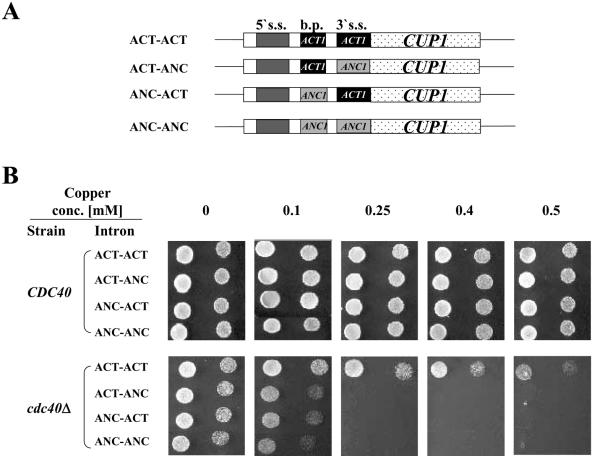
In order to identify sites in the ANC1 intron which are responsible for its splicing dependency on Cdc40p, we compared the ANC1 intron splice sites (5′ splice site, branchpoint and 3′ splice site) with the yeast consensus sites (35). This comparison reveals that while the ANC1 5′ splice site is identical to the consensus sequence, unusual bases in conserved positions are found at both the branchpoint and the 3′ splice site. ANC1 splicing dependency on Cdc40p could be due to the unusual ANC1 branchpoint, the 3′ splice site or both. In order to examine this hypothesis and localize the feature in the ANC1 intron that confers its splicing dependency on Cdc40p, we used the reporting systems based on ANC1 and ACT1 introns described. These two introns are differently affected by the absence of Cdc40p (Fig. 4). By shuffling the branchpoint and the 3′ splice site of the ANC1 and ACT1 introns, we created a series of chimeric introns fused to the CUP1 reporting gene (Fig. 5A). All the constructs analyzed shared the same (ACT1) intron sequences upstream of the branchpoint. Plasmids carrying these fusions were introduced into cup1Δ CDC40 and cup1Δ cdc40Δ cells and examined for their ability to allow growth at different copper concentrations. As can be seen in Figure 5B, cdc40Δ cells transformed with a clone containing the ACT1 branchpoint and 3′ splice site (ACT-ACT) were able to grow at high copper concentrations (>0.5 mM). In contrast, all the other chimeric clones, which contain either the ANC1 branchpoint (ANC-ACT), the ANC1 3′ splice site (ACT-ANC) or both (ANC-ANC), were defective in their splicing ability and allowed cup1Δ cdc40Δ cells to grow only in the presence of low copper concentrations (≤0.1 mM). However, these clones were spliced normally in the presence of Cdc40p, and supported growth of CDC40 cells at high copper concentrations (Fig. 5B). The fact that both ANC-ACT (ANC1 branchpoint and ACT1 3′ splice site) and ACT-ANC (ACT1 branchpoint and ANC1 3′ splice site) clones were barely spliced in the absence of Cdc40p indicates that both the branchpoint and the 3′ splice site of ANC1 pre-mRNA participate in the dependency on CDC40 for efficient splicing.
Figure 5.
Both the branchpoint and the 3′ splice site of the ANC1 intron contribute to the dependency on Cdc40p for splicing. (A) A series of chimeric plasmids containing different combinations of ANC1 and ACT1 splice sites fused to the CUP1 reporting gene were constructed. (B) cdc40Δ cup1Δ (SB90) and cup1Δ CDC40 (YCL51) strains were transformed with each of the chimeric clones. Serial dilutions of each transformant were spotted on minimal selective media at 30°C and tested for the ability to grow in the presence of different copper concentrations.
Characterization of the ANC1 intron
In order to identify the specific residues in each site (branchpoint and 3′ splice site) that make ANC1 splicing highly dependent on Cdc40p, we carried out site-directed mutagenesis experiments.
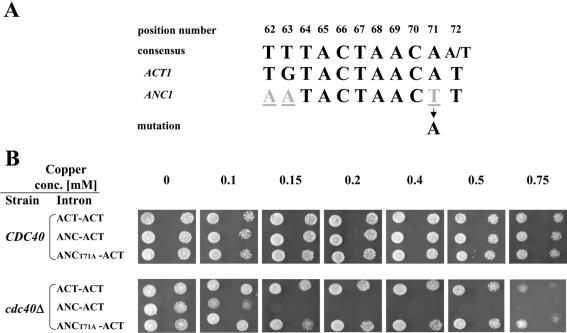
The ANC1 branchpoint differs from the consensus at three positions (62, 63 and 71). Position 71 is the most conserved among yeast introns. A mutation was introduced replacing the unusual thymidine at position 71 by the conserved adenine in the ANC-ACT clone (Fig. 6A). The mutated T71A clone was transformed into cup1Δ CDC40 and cup1Δ cdc40Δ cells and its ability to support growth at different copper concentrations was examined. As shown in Figure 6B, whereas cup1Δ cdc40Δ cells containing wild-type ANC1 branchpoint (ANC-ACT clone) grew only at a low copper concentration (0.1 mM), the same strain containing the mutant clone (ANC T71A-ACT) was extremely resistant to copper and grew at high copper concentrations, similar to cells containing the ACT1 branchpoint (ACT-ACT). These results demonstrate that a single nucleotide is responsible for the ANC1 branchpoint reliance on Cdc40p for splicing, and mutating this nucleotide is sufficient to alleviate this dependency.
Figure 6.
Site-directed mutagenesis of the ANC1 intron branchpoint. (A) The branchpoint sequence of both ACT1 and ANC1 introns compared with the consensus sequence. The position number of each residue of the ANC1 branchpoint (numbering starting from the first nucleotide of the ANC1 intron) is indicated. Non-canonical nucleotides are underlined. (B) cup1Δ cdc40Δ (SB90) and cup1Δ CDC40 (YCL51) strains were transformed with wild-type and mutated chimeric clones. The transformants were diluted and tested at 30°C for the ability to grow in the presence of different copper concentrations.
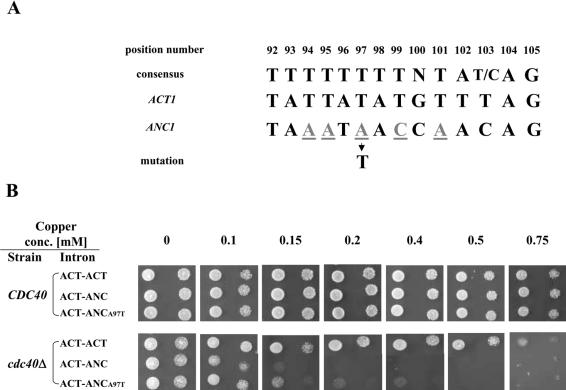
The ANC1 3′ splice site contains several nucleotides that differ from the consensus (Fig. 7A). The ninth nucleotide from 3′ end of the intron (position 97 of the ANC1 intron) is occupied by a thymidine in 86% of all the yeast introns (35). However, in the ANC1 intron there is an adenine at this position. To test the possible role of this nucleotide, we introduced the A97T transversion in the ACT-ANC clone. The mutated clone was transformed into cup1Δ CDC40 and cup1Δ cdc40Δ cells and its ability to support growth at different copper concentrations was examined. Unlike the results obtained by mutating the ANC1 branchpoint, the single-nucleotide exchange only partially increased ANC1 splicing efficiency (as indicated by the ability of cup1Δ cdc40Δ carrying ACT-ANC A97T clone to grow in the presence of 0.4 mM copper) (Fig. 7B). However, the splicing efficiency of cells containing the mutant clone was lower than that of the ACT-ACT clone, indicating that this nucleotide is not solely responsible for the dependency of the splicing of the ANC1 3′ splice site on the Cdc40 protein.
Figure 7.
Site-directed mutagenesis of the ANC1 intron 3′ splice site. (A) The sequences of the 3′ splice site of both ACT1 and ANC1 introns are compared with each other and with the consensus sequence. The position number of each residue of the ANC1 3′ splice site is indicated. Non-canonical nucleotides are underlined. (B) cup1Δ cdc40Δ (SB90) and cup1Δ CDC40 (YCL51) strains were transformed with wild-type and mutated chimeric clones. The transformants were diluted and tested at 30°C for the ability to grow in the presence of different copper concentrations.
In an attempt to identify other residues in the ANC1 3′ splice site that are necessary for its efficient splicing in the absence of Cdc40p, we adopted a random mutagenesis strategy. Five non-canonical positions at the ANC1 3′ splice site (positions 94, 95, 97, 99 and 101) in the ACT-ANC clone were mutagenized using degenerate primers. A thymidine (T) is present at all of these positions in the consensus sequence, as well as in the efficient ACT1 3′ splice site, whereas the ANC1 intron carries either adenine (A) or cytosine (C). The mutated clones were then transformed into cup1Δ cdc40Δ cells and screened for their ability to support growth in the presence of restrictive copper concentrations. Clones that supported growth were extracted and further checked in both cup1Δ CDC40 and cup1Δ cdc40Δ cells, and the nature of mutations was determined by sequencing. The results are summarized in Table 1. Unlike the results obtained for the branchpoint, our mutation analysis of the ANC1 3′ splice site failed to identify a single position crucial for the ANC1 splicing dependency on Cdc40p. However, several points could be inferred from our experiments. The nucleotides at positions 94 and 101 do not seem to play an important role, since all four nucleotides were found at similar frequencies at these positions. At position 99, there seems to be a bias against the cytosine present at the ANC1 intron, suggesting that C at this position lowers splicing efficiency. Positions 95 and 97 clearly have the strongest impact on splicing efficiency. Out of 15 copper-resistant clones, 8 and 11 carried thymidines at positions 95 and 97, respectively. Thus our results indicate that thymidines are important and cannot be replaced by the other pyrimidine, cytosine, which seems to be under-represented in the mutant clones (especially at positions 95 and 97). A genome-wide search revealed a total of 12 introns carrying adenine residues at positions 95 and 97. We are presently investigating whether these introns are also controlled by a differential requirement of Cdc40p activity.
Table 1. Sequence of mutagenized clones conferring resistance to high levels of copper.
| Clone | Position 94 | Position 95 | Position 97 | Position 99 | Position 101 |
|---|---|---|---|---|---|
| ACT-ACT |
T |
T |
T |
T |
T |
| ACT-ANC |
A |
A |
A |
C |
A |
| |
|
|
|
|
|
| P47 |
T |
T |
T |
T |
C |
| P22 |
T |
T |
T |
Δ |
Δ |
| P53 |
A |
T |
T |
G |
T |
| P83 |
A |
T |
T |
G |
T |
| P89 |
A |
T |
T |
G |
T |
| P105 |
T |
T |
T |
T |
C |
| P65 |
C |
A |
T |
T |
G |
| P49 |
A |
A |
T |
G |
C |
| P81 |
T |
A |
T |
T |
T |
| P35 |
T |
G |
T |
C |
A |
| P1 |
A |
A |
T |
C |
A |
| P75 |
C |
T |
A |
G |
A |
| P31 |
A |
T |
G |
A |
C |
| P4 |
G |
G |
A |
T |
T |
| P28 |
C |
G |
G |
T |
G |
| |
|
|
|
|
|
| T |
5 |
8 |
11 |
6 |
5 |
| A |
6 |
4 |
2 |
1 |
3 |
| G |
1 |
3 |
2 |
5 |
2 |
| C | 3 | 0 | 0 | 2 | 4 |
In summary, our analysis was able to identify specific residues in both the branchpoint and the 3′ splice site that are important for ANC1 splicing dependency on Cdc40p.
DISCUSSION
Cell cycle progression and pre-mRNA splicing are two apparently independent processes in eukaryotic cells. Despite this, a growing body of evidence has recently accumulated indicating that these processes may be connected. However, understanding the molecular basis underlying this connection has remained a challenge. Our results provide a molecular explanation for this connection.
Mutations in most splicing genes do not cause a cell cycle arrest. Furthermore, in some cases, different alleles of splicing genes behave differently. For example, not all splicing-defective alleles of PRP8 cause a cell cycle stage specific arrest (20). One possible explanation for the association between pre-mRNA splicing and cell cycle progression may be the need for specific splicing factors for the efficient splicing of subsets of intron-containing genes (‘target’ genes). According to this model, a small number of genes that are essential for the different transitions through the phases of the cell cycle contain introns with unique features. These introns are differentially dependent on specific splicing factors. Failure to accurately splice these genes prevents the synthesis of the specific protein(s) and leads to cell cycle arrest. By transforming cdc40Δ cells with a cDNA library we have isolated the ANC1 cDNA as a suppressor of cell cycle arrest of cdc40Δ cells. By deleting the intron in the genomic copy of ANC1, we were able to show that this suppression is truly due to bypass of the need for ANC1 pre-mRNA splicing, rather than to ANC1 over-expression.
Anc1p was found to be associated with several protein complexes in the cell (30–33). The precise role of Anc1p in all these complexes is presently unclear. A common feature of these complexes, however, is that they are all involved in transcription and chromatin remodeling, and thus the expression of Anc1p is likely to be important for the proper transcription of many genes, including cell cycle specific genes. Two human homologs of ANC1 were identified. These genes, ANL and AF9, were shown to be the target of reciprocal translocations that are common in myeloid leukemias. New evidence suggests that the resulting fusion proteins affect transcription partly by interacting with a repressor of transcription, BcoO. The negative effect on transcription deregulates cell cycle progression and thus increases susceptibility to leukemia (36).
The steady state level of ANC1 RNA is very low; its transcription is not cell cycle regulated and remains constant in most experimental conditions. We have shown that elimination of the ANC1 intron is sufficient to allow suppression of the cdc40Δ cell cycle arrest phenotype. Using both RT–PCR analysis and the CUP1 reporting system, we have proved that the efficient splicing of ANC1 pre-mRNA requires Cdc40p. Thus regulation of the ANC1 gene occurs not at the level of transcription, but at the level of splicing. The dependence of ANC1 on Cdc40p is not universal, since other intron-containing genes are less dependent on Cdc40p for splicing. Using microarray experiments specially designed to detect splice junctions and introns, the Ares group has recently examined the genome-wide response to the loss of individual splicing factors. They showed that intron-containing genes depend on different splicing factors to different extents. These findings are in accordance with our target gene hypothesis. Furthermore, in the same study, ANC1 splicing was shown to be extremely dependent on Cdc40/Prp17p (as well as on Prp18p) (37) in agreement with our RT–PCR and CUP1 reporting analysis.
The ANC1 intron sequence differs from most other yeast introns at several positions. By creating chimeric introns bearing different combinations of the ACT1 and ANC1 branchpoint and 3′ splice sites, we have shown that both the branchpoint and the 3′ splice site of ANC1 are essential for its splicing dependency on Cdc40p. Each of the two splice sites seems to contain unique features that generate dependency on Cdc40p. The degree of nucleotide conservation at each position of the intron splice sites is highly correlated with its importance in the splicing reaction. For instance, mutations in the first and fifth positions of the 5′ splice site (invariant Gs for all naturally occurring yeast introns) adversely affect both the efficiency of splicing and the specificity of the first catalytic step, whereas mutations at other positions have less severe effects (10). Similarly, branchpoint mutations at positions not found to vary naturally are the most detrimental to splicing efficiency, in contrast with the weak effect of mutations in less conserved residues (38). The existence of natural introns bearing non-standard signals indicates that these residues may possess some regulatory role. Comparison of the ANC1 splice site with the consensus sequence (35) revealed the existence of several unorthodox residues at highly conserved positions in both the branchpoint and the 3′ splice site (Figs 6 and 7). We have shown that in the case of the ANC1 branchpoint, replacing the unusual nucleotide (T) at position 71 with the most conserved nucleotide (A) completely abolished the splicing dependency of the ANC-ACT clone on Cdc40p. Similarly, replacement of the adenine at position 97 of the ANC1 intron with the consensus (T) residue increased splicing efficiency in the absence of CDC40. These results indicate that positions 71 and 97 are important for regulating the ANC1 pre-mRNA splicing involving Cdc40p. Sequence comparison of the ANC1 intron with its orthologs from other related yeast species (Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces bayanus) (39) revealed that both unorthodox nucleotides at position 71 in the ANC1 branchpoint and position 97 in the ANC1 3′ splice site are conserved in all the yeast species examined. This strengthens the notion that these residues may play an important role in controlling ANC1 splicing.
Position 71 in ANC1 is located near the highly conserved branchpoint residues that are engaged in base pairing with U2 snRNPs during the splicing reaction (40). We propose that Cdc40p is important for stabilizing the interaction between U2 snRNA and branchpoint sequences containing residues different from the consensus (as in the ANC1 intron). In the absence of Cdc40p this interaction is disrupted, especially at higher temperatures, resulting in a reduction in splicing efficiency. This hypothesis is supported by the ability of a compensatory mutation in U2 snRNA (T33A) that restores the stable base paring at position 71 of the ANC1 intron to support cell growth in the absence of Cdc40p (data not shown) presumably by allowing efficient splicing of the ANC1 branchpoint even in the absence of Cdc40p.
As for the 3′ splice site, although there is no indication for base pairing between the 3′ splice site and any known snRNA, the 3′ splice site region has been shown to interact with several splicing proteins such as Prp8, Prp16, Prp22 and Slu7 (41,42). Recently, McPheeters and Muhlenkamp (43) monitored, by cross-linking analysis, the RNA–protein interactions involving the 3′ splice site during yeast pre-mRNA splicing. According to their results, extensive associations of the U2-snRNP-associated splicing factor Hsh155p with the branchpoint and 3′ splice site region are established upon pre-spliceosome formation. Following the first catalytic step of splicing, Prp8p, and later Prp16p, interacts with the 3′ end of the intron, leading to binding of Prp22p (43). Our results indicate that Cdc40p cooperates with these factors, directly or indirectly interacting with the 3′ splice site.
Previously, extensive genetic interactions between CDC40 and PRP8 were described. In the absence of Cdc40p, mutations in PRP8 can either suppress the temperature-sensitive phenotype or confer synthetic lethality (44). In addition, a strong correlation between the ability of prp8 alleles to suppress the temperature-sensitive phenotype of cdc40Δ cells and their ability to recognize mutant 3′ splice sites was observed (44). Similar interactions were also found between prp8 alleles and mutants of the CSC (data not shown). Since we have shown that Cdc40p plays a role in governing the splicing of the ANC1 intron through its branchpoint and 3′ splice site, one can propose the following model. Prp8p plays a central role in the second splicing reaction, presumably by helping in the recognition of the 3′ splice site. However, the efficient splicing of some introns (presumably those with non-canonical splice sites) requires the assistance of Cdc40p (or other factors such as the CSC). Cdc40p, in addition to its role in stabilizing the base pairing between the branchpoint of some introns and the U2 snRNA, may help in positioning Prp8p (and probably other proteins as well) at the correct site on the pre-mRNA. Alternatively, Cdc40p may help stabilize interactions within the spliceosome. In the absence of the Cdc40p, at the restrictive temperature, the interactions become unstable and splicing of introns with suboptimal sequences such as ANC1 is disrupted, causing cell cycle arrest.
The relationship between Cdc40p and the CSC is still not clear. The CSC was originally discovered in a screen for mutants that depend on Cdc40p for survival (15). This dependency hints at an overlapping role for CSC and Cdc40p. Subsequently, it was found that mutations in components of the CSC, such as cef1 or isy1Δ syf2Δ, also cause arrest at the G2/M phase of the cell cycle. The arrest is due to inefficient splicing of the TUB1 and TUB3 genes encoding α-tubulin (45,46). We have also shown that this cell cycle arrest is mediated by the spindle checkpoint pathway (46). However, introduction of TUB1 or TUB3 cDNA does not suppress the temperature-sensitive phenotype of cdc40Δ cells (5, data not shown). These results are supported by a DNA microarray analysis of cdc40Δ cells in which no increase in TUB1 or TUB3 unspliced signals were observed (37). Thus, although the CSC and Cdc40p seem to have somewhat overlapping functions, their specificity may differ. Different subgroups of introns in essential genes may require Cdc40p, the CSC or both, explaining both the synthetic lethality and the specificity observed.
Budding yeast cells have a relatively small number of intron-containing genes. Although several essential yeast genes have intervening sequences, there is a clear evolutionary trend of discarding introns, perhaps by a cDNA-directed homologous recombination mechanism as proposed by Fink (47). It is conceivable that those introns that remained may serve in the regulation of cellular processes. A similar mechanism, for example, is used for the regulation of meiosis. A number of genes that are essential for meiotic functions are transcribed in both vegetative and meiotic cells. Their expression, however, is meiosis specific because they are spliced only in meiotic cells. The meiosis-specific splicing depends on MER1, a meiosis-specific U1-associated splicing factor that is required for efficient splicing of introns containing non-canonical 5′ splice sites (48). Mutation in the 5′ splice site of one target gene (MER2) that recreates the consensus bypasses the Mer1p requirement for splicing (49). Thus the meiosis-specific splicing factors, together with the unique 5′ splice site, ensure efficient splicing of introns only in the right circumstances.
Similarly, we have shown that Cdc40p serves as an auxiliary splicing factor that is dispensable for most yeast introns but has become essential for splicing of the ANC1 gene. We have provided a mechanism for this dependence which relies on the presence of non-canonical sequences at the branchpoint and 3′ splice site. The evolutionary conservation of Cdc40p (50), as well as that of the non-canonical residues in the ANC1 gene, suggests that the mechanism of splicing-dependent cell cycle regulation uncovered here may be conserved in other organisms. Current work is aimed at analyzing the dependence on Cdc40p of additional introns with non-canonical sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We thank all members of the Kupiec group for support and ideas, especially Dr Sigal Ben Yehuda and Dr Orna Landman. We also thank D. Kornitzer, M. Ares, C. Guthrie and D. Drubin for reagents. This work was supported by grants to M.K. by AICR, ICRF and the Israeli Cancer Association.
REFERENCES
- 1.Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- 2.Kassir Y. and Simchen,G. (1978) Meiotic recombination and DNA synthesis in a new cell cycle mutant of Saccharomyces cerevisiae. Genetics, 90, 49–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaisman N., Tsouladze,A., Robzyk,K., Ben-Yehuda,S., Kupiec,M. and Kassir,Y. (1995) The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol. Gen. Genet., 247, 123–136. [DOI] [PubMed] [Google Scholar]
- 4.Boger-Nadjar E., Vaisman,N., Ben-Yehuda,S., Kassir,Y. and Kupiec,M. (1998) Efficient initiation of S-phase in yeast requires Cdc40p, a protein involved in pre-mRNA splicing. Mol. Gen. Genet., 260, 232–241. [DOI] [PubMed] [Google Scholar]
- 5.Chawla G., Sapra,A.K., Surana,U. and Vijayraghavan,U. (2003) Dependence of pre-mRNA introns on PRP17, a non-essential splicing factor: implications for efficient progression through cell cycle transitions. Nucleic Acids Res., 31, 2333–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassir Y., Kupiec,M., Shalom,A. and Simchen,G. (1985) Cloning and mapping of CDC40, a Saccharomyces cerevisiae gene with a role in DNA repair. Curr. Genet., 9, 253–257. [DOI] [PubMed] [Google Scholar]
- 7.Kupiec M. and Simchen,G. (1986) DNA-repair characterization of cdc40–1, a cell-cycle mutant of Saccharomyces cerevisiae. Mutat. Res., 162, 33–40. [DOI] [PubMed] [Google Scholar]
- 8.Vijayraghavan U., Company,M. and Abelson,J. (1989) Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev., 3, 1206–1216. [DOI] [PubMed] [Google Scholar]
- 9.Vijayraghavan U., Parker,R., Tamm,J., Iimura,Y., Rossi,J., Abelson,J. and Guthrie,C. (1986) Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J., 5, 1683–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquier A., Rodriguez,J.R. and Rosbash,M. (1985) A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell, 43, 423–430. [DOI] [PubMed] [Google Scholar]
- 11.Jurica M.S. and Moore,M.J. (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol. Cells, 12, 5–14. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen T.W. (1998) RNA/RNA interactions in nuclear pre-mRNA splicing. In Simons,R. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 79–307. [Google Scholar]
- 13.Makarov E.M., Makarova,O.V., Urlaub,H., Gentzel,M., Will,C.L., Wilm,M. and Luhrmann,R. (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science, 298, 2205–2208. [DOI] [PubMed] [Google Scholar]
- 14.Schwer B. and Gross,C.H. (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J., 17, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Yehuda S., Dix,I., Russell,C.S., McGarvey,M., Beggs,J.D. and Kupiec,M. (2000) Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics, 156, 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell C.S., Ben-Yehuda,S., Dix,I., Kupiec,M. and Beggs,J.D. (2000) Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA, 6, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.H., Yu,W.C., Tsao,T.Y., Wang,L.Y., Chen,H.R., Lin,J.Y., Tsai,W.Y. and Cheng,S.C. (2002) Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res., 30, 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston L.H. and Thomas,A.P. (1982) The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet., 186, 439–444. [DOI] [PubMed] [Google Scholar]
- 20.Shea J.E., Toyn,J.H. and Johnston,L.H. (1994) The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res., 22, 5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang L.H. and Murray,A.W. (1997) A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell, 8, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K., Yamada,H. and Yanagida,M. (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell, 5, 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potashkin J., Kim,D., Fons,M., Humphrey,T. and Frendewey,D. (1998) Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet., 34, 153–163. [DOI] [PubMed] [Google Scholar]
- 25.Lundgren K., Allan,S., Urushiyama,S., Tani,T., Ohshima,Y., Frendewey,D. and Beach,D. (1996) A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell, 7, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habara Y., Urushiyama,S., Shibuya,T., Ohshima,Y. and Tani,T. (2001) Mutation in the prp12+ gene encoding a homolog of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe. RNA, 7, 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umen J.G. and Guthrie,C. (1996) Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics, 143, 723–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesser C.F. and Guthrie,C. (1993) Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics, 133, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch M.D. and Drubin,D.G. (1994) A nuclear protein with sequence similarity to proteins implicated in human acute leukemias is important for cellular morphogenesis and actin cytoskeletal function in Saccharomyces cerevisiae. Mol. Biol. Cell, 5, 617–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry N.L., Campbell,A.M., Feaver,W.J., Poon,D., Weil,P.A. and Kornberg,R.D. (1994) TFIIF-TAF-RNA polymerase II connection. Genes Dev., 8, 2868–2878. [DOI] [PubMed] [Google Scholar]
- 31.Poon D., Bai,Y., Campbell,A.M., Bjorklund,S., Kim,Y.J., Zhou,S., Kornberg,R.D. and Weil,P.A. (1995) Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 92, 8224–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cairns B.R., Henry,N.L. and Kornberg,R.D. (1996) TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol., 16, 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John S., Howe,L., Tafrov,S.T., Grant,P.A., Sternglanz,R. and Workman,J.L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev., 14, 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 34.Brys A. and Schwer,B. (1996) Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA, 2, 707–717. [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez P.J. and Seraphin,B. (1999) Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA, 5, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srinivasan R.S., de Erkenez,A.C. and Hemenway,C.S. (2003) The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene, 22, 3395–3406. [DOI] [PubMed] [Google Scholar]
- 37.Clark T.A., Sugnet,C.W. and Ares,M., Jr (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science, 296, 907–910. [DOI] [PubMed] [Google Scholar]
- 38.Jacquier A. and Rosbash,M. (1986) RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc. Natl Acad. Sci. USA, 83, 5835–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 40.Parker R., Siliciano,P.G. and Guthrie,C. (1987) Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell, 49, 229–239. [DOI] [PubMed] [Google Scholar]
- 41.Teigelkamp S., Newman,A.J. and Beggs,J.D. (1995a) Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J., 14, 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McPheeters D.S., Schwer,B. and Muhlenkamp,P. (2000) Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res., 28, 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McPheeters D.S. and Muhlenkamp,P. (2003) Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol., 23, 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Yehuda S., Russell,C.S., Dix,I., Beggs,J.D. and Kupiec,M. (2000) Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics, 154, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burns C.G., Ohi,R., Mehta,S., O’Toole,E.T., Winey,M., Clark,T.A., Sugnet,C.W., Ares,M.,Jr and Gould,K.L. (2002) Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahan O. and Kupiec,M. (2002) Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res., 30, 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fink G.R. (1987) Pseudogenes in yeast? Cell, 49, 5–6. [DOI] [PubMed] [Google Scholar]
- 48.Spingola M. and Ares,M.,Jr (2000) A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cells, 6, 329–338. [DOI] [PubMed] [Google Scholar]
- 49.Nandabalan K. and Roeder,G.S. (1995) Binding of a cell-type-specific RNA splicing factor to its target regulatory sequence. Mol. Cell. Biol., 15, 1953–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.BenYehuda S., Dix,I., Russell,C.S., Levy,S., Beggs,J.D. and Kupiec,M. (1998) Identification and functional analysis of hPRP17, the human homolog of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA, 4, 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]