Abstract
Recent genomic analyses revealed many kinds of tandem repeats of specific sequences. Some of them are related to genetic diseases, but their biological functions and structures are still unknown. Two X-ray structures of a short DNA fragment d(gcGA[G]1Agc) show that four base-intercalated duplexes are assembled to form an octaplex at a low K+ concentration, in which the eight G5 residues form a stacked double G-quartet in the central part. At a higher K+ concentration, however, the octaplex is split into just two halves. These structural features suggest a folding process of eight tandem repeats of d(ccGA[G]4Agg), according to a double Greek-key motif. Such a packaging of the repeats could facilitate slippage of a certain sequence during DNA replication, to induce increase or decrease of the repeats.
INTRODUCTION
The major conformation of DNA is the B-form proposed by Watson and Crick (1), but the A-form (2) at low-humidity (3) and the left-handed Z-DNA (4–6) are also well known. Recently we found a parallel-stranded duplex (7) with homo base pairs. In these structures, however, two DNA strands are associated to form a duplex. In addition, DNA triplexes (8) are considered to be involved in regulation of DNA replication, transcription, recombination and cell development. G-rich repeats in telomeric DNA can form a quadruplex with G-quartet (9). The complementary C-rich repeats can also form a quadruplex with an intercalated cytosine motif (i-motif) (9). It is expected that DNA in a single-stranded state would adopt more complicated structures like functional RNAs. However, our structural knowledge on DNA is still limited to examples cited above.
Recent human genome projects have revealed that the content of coding regions are only a few percent (∼1.5%), whereas the repetitive sequences found in transposon-derived sequences, simple sequence repeats (SSR), segmentally duplicated sequences, telomere and centromere of heterochromatin accounts for >50% of the genome (10). These repeats may have some biological functions with specific structures. To understand the biological significance of such sequences, it is necessary to reveal structural versatility of DNA molecules with specific properties.
Variable number of tandem repeats (VNTR), which is a SSR with a long repetitive unit, are very useful in the analysis of the human genome to identify an individual, like a finger print, because they form a large polymorphism with different lengths, generated by slippage of the repeats during replication (11). Such repetitive regions are dispersed throughout the genomes of all vertebrates (12,13) and are known to be hot spots for meiotic recombination in germ cells (12,14). Recent reports have indicated that some VNTRs with genetic diseases including insulin-dependent diabetes mellitus (15) and myoclonus epilepsy (16) may function as transcriptional or translational regulators, or may modify a protein function when the tandem repetitive sequences invade in the coding region of the gene (17).
A VNTR immediately adjacent to the human pseudoautosomal telomere also shows a high degree of length polymorphism (18). In this region, the G-rich sequence d(ccGA[G]4Agg) is repeated eight times. [Lowercase characters indicate that they can form a Watson–Crick G:C or C:G pair when the fragments are aligned in an anti-parallel fashion for self-assembly or folding.] To reveal the structure of such a repetitive sequence, we started X-ray analysis. For crystallization, the original sequence d(ccGA[G]4Agg) was shortened and modified at both ends as d(ccGA[G]1Agg), d(ggGA[G]1Acc), d(gcGA[G]1Agc) and d(cgGA[G]1Acg). In the previous study (19), we determined the crystal structure of d(gcGA[G]1Agc) with a hexamine–cobalt cation, in which the two fragments are associated to form a duplex as expected. In addition, it was found that three duplexes are further associated to form hexa-assembly, in which two hexamine–cobalt chlorides facilitate the assembly formation. In the present study, we have determined three crystal structures of d(gcGA[G]1Agc) at different potassium concentrations under physiological conditions. In all three crystals, two octamers form a base-intercalated duplex similar to those of the base-intercalated duplexes of d(gcGA[A]1Agc) (20,21) and d(gcGA[T]1Agc) (19). However, surprisingly, it has been found that the four base-intercalated duplexes form an octaplex through double G-quartet formation at lower potassium concentration. This is the largest multiplex, suggesting a folding of the eight repeats of d(ccGA[G]4Agg) with a double Greek-key motif. In addition, it has been found that the octaplex is split into just the two halves at a slightly higher potassium concentration. These structural features suggest a mechanism of the octaplex formation. In this paper, detailed structures of two multiplexes are discussed with their biological significance.
MATERIALS AND METHODS
Preparation
A DNA octamer with the sequence d(gcGA[G]1Agc) (hereafter G1) was synthesized on a DNA synthesizer. To resolve the phase problem in X-ray crystallography, the second cytidine residue was modified with 2′-deoxy-5-iodocytidine or 2′-deoxy-5-bromocytidine (hereafter G1-I and G1-Br, respectively). These octamers were purified by HPLC and cation exchange chromatography. Crystallization conditions were surveyed at 277 K by the hanging-drop vapor diffusion method. For the G1-Br, crystals of G1-Br-highK were grown for four days in a droplet prepared by merging 1 µl of 1.5 mM DNA solution and 1 µl of 40 mM sodium cacodylate buffer solution (pH 7.0) that contains 12 mM spermine tetrahydrochloride, 12 mM sodium chloride, 80 mM potassium chloride and 10% (v/v) 2-methyl-2,4-pentanediol (MPD). For the G1-I, two types of crystals were obtained. One (G1-I-highK) was grown up for 1 week under the same conditions as that of G1-Br-highK. The other one (G1-I-lowK) was also obtained for 3 days under the same conditions but DNA concentration was 3.0 mM (two times higher). It is noteworthy that potassium concentration used for crystallizations was within normal physiological levels (22) and that the concentration ratio of potassium ion to DNA was two times higher in G1-I-highK than G1-I-lowK. Fresh crystals were mounted in nylon cryoloops (Hampton Research) with the crystallization solution containing 35% (v/v) MPD as a cryoprotectant and stocked in liquid nitrogen for X-ray experiments.
Data collection
Every X-ray data was taken at 100 K with synchrotron radiation. For multiple-wavelength anomalous dispersion (MAD) phasing using the anomalous scattering of bromine atoms, X-ray data of G1-Br-highK were taken at BL18B of Photon Factory. Three different wavelengths were employed based on XAFS measurements. X-Ray diffractions were recorded on a CCD detector (Quantum 4R) positioned at 170 mm away from the crystal. Diffraction patterns were taken using 2° oscillation with 60 s exposure per frame. Recorded images were processed with the program DPS/MOSFLM (23–26). The diffraction data for G1-I-highK were measured in 1° oscillation frames (24 s each) at BL44XU of SPring-8. Diffraction data, recorded on a CCD detector (PX210) positioned at 200 mm away from the crystal, were processed at a resolution of 1.7 Å with the program d*TREK (27). For G1-I-lowK crystal, the CCD detector (Quantum 4R) was positioned at 150 mm away from the crystal. Diffraction patterns, taken using 1° oscillation with 60 s exposure per frame at BL18B of PF, were processed with the program Crystalclear (Rigaku/MSC). Intensity data of three types of crystals were put on a relative scale and merged into the independent reciprocal space using the program SCALA and TRUNCATE of CCP4 suite (28). All crystals belong to the same space group, but have different unit cell dimensions due to differences of crystallization conditions. Two crystals obtained under higher potassium conditions suggest their isomorphism. On the other hand, G1-I-lowK obtained under lower potassium conditions has different unit cell dimensions. Statistics of the data collection and crystal data are summarized in Table 1.
Table 1. Crystal data, statistics of data collection, and statistics of structure refinement.
| Crystal code | G1-I-lowK | G1-I-highK | G1-Br-highK | ||
|---|---|---|---|---|---|
| Crystal data |
|
|
|
|
|
| Space group |
I222 |
I222 |
I222 |
|
|
| Unit cell (Å) |
a = 36.2, b = 36.4, c = 63.1 |
a = 34.6, b = 42.5, c = 64.1 |
a = 33.8, b = 43.5, c = 64.2 |
|
|
|
Za |
1 |
1 |
1 |
|
|
| Data collection |
|
|
|
|
|
| Wavelength (Å) |
1.00 |
0.900 |
1.00 (remote) |
0.9196 (peak) |
0.9202 (edge) |
| Resolution (Å) |
20–2.3 |
35–1.7 |
32–1.8 |
27–2.2 |
30–2.1 |
| Observed reflections |
9488 |
30 980 |
46 368 |
54 264 |
57 560 |
| Unique reflections |
1980 |
5269 |
4543 |
2587 |
2973 |
| Completeness (%) |
98.4 |
96.5 |
98.1 |
99.9 |
100.0 |
| in the outer shell |
99.0 |
100.0 |
88.6 |
100.0 |
100.0 |
|
Rmergeb (%) |
8.4 |
9.6 |
5.0 |
5.7 |
5.8 |
|
Ranomc (%) |
– |
– |
2.7 |
5.3 |
3.8 |
| Theoretical f′/f′′ |
– |
– |
–2.22/0.59 |
–7.08/3.82 |
–9.33/0.50 |
| Refined f′/f′′ |
– |
– |
–3.15/0.57 |
–7.36/3.83 |
–9.27/2.20 |
| Structure refinement |
|
|
|
|
|
| Resolution range (Å) |
9.0–2.3 |
9.0–1.7 |
9.0–1.8 |
|
|
| Used reflections |
1903 (Fo>3σ) |
5063 (Fo>3σ) |
4227 (Fo>3σ) |
|
|
|
R-factord (%) |
23.4 |
26.2 |
22.5 |
|
|
|
Rfreee |
26.5 |
29.6 |
24.9 |
|
|
| Number of DNA atoms |
332 |
332 |
332 |
|
|
| Number of water molecules |
97 |
87 |
110 |
|
|
| Number of potassium ions |
1/4, 1/2 |
1/2 |
1/2 |
|
|
| R.m.s. deviation |
|
|
|
|
|
| Bond length (Å) |
0.005 |
0.004 |
0.004 |
|
|
| Bond angles (°) |
0.9 |
0.9 |
0.9 |
|
|
| Improper angles (°) | 0.8 | 0.9 | 0.8 |
aNumber of base-intercalated duplexes in the asymmetric unit.
bRmerge = 100 × Σhklj|Ihklj – <Ihklj>| / Σhklj<Ihklj>.
cRanom = 100 × Σhklj|Ihklj(+) – Ihklj(–)| / Σhklj[Ihklj(+) + Ihklj(–)].
dR-factor = 100 × Σ||Fo| – |Fc|| / Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
eCalculated using a random set containing 10% of observations that were not included throughout refinement (36).
Structure determination and refinement
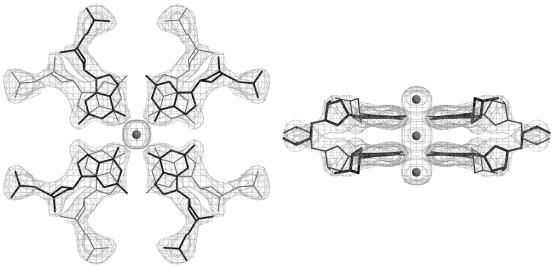
From the three data sets of G1-Br-highK with different wavelengths, initial phases were estimated by the MAD method using the program SOLVE (29). Two bromine atoms were found from the anomalous difference Patterson map, from which the initial phases were estimated with the figure-of-merit of 0.6. Electron density, modified by a solvent flattening technique (solvent content 43.9%) with the program CNS (30), showed clearly the phosphate–ribose backbone with the individual bases. The molecular structure was constructed on a graphic workstation with the program QUANTA (Accelrys Inc.). Phases of G1-I-lowK and G1-I-highK were derived by molecular replacement technique with the program AMoRe (31) using a part of the G1-Br-highK structure as a probe. The molecular structure was constructed and modified on a graphic workstation by inspecting |Fo| – |Fc| omit maps at every nucleotide residue with the same program described above. The atomic parameters of the three crystal structures were refined with the program CNS through a combination of rigid-body, simulated-annealing, crystallographic conjugate gradient minimization refinement and B-factor refinements, followed by interpretation of an omit map at every nucleotide residue. In the initial stage of refinements, conformational restraints were applied to all ribose rings to be a C2′-endo pucker, but in the later stages, these restraints were released. No restraints were applied to base-pair geometry. In G1-I-lowK, two potassium ions, found at a cross-center of two 2-fold axes (K1) and on the 2-fold axis along c (K2), and 97 water molecules were included in the final refinement. Similarly, a potassium ion on the 2-fold axis and 87 water molecules in G1-I-highK, and a potassium ion on the 2-fold axis and 110 water molecules in G1-Br-highK, were included in the final respective refinements. Statistics of the structure refinements are summarized in Table 1. All local helical parameters including torsion angles and pseudorotation phase angles of ribose rings were calculated using the program 3DNA (32). Figure 1 shows a 2|Fo| – |Fc| map of the central double G-quartets found in the G1-I-lowK crystal, which was drawn with the program O (33). Figures 2–5 were drawn with the program RASMOL (34).
Figure 1.
Local 2|Fo| – |Fc| maps for the two stacked G-quartets at the center of the octaplex of G1-I-lowK. Density is contoured at 1σ level. Gray spheres represent potassium cations.
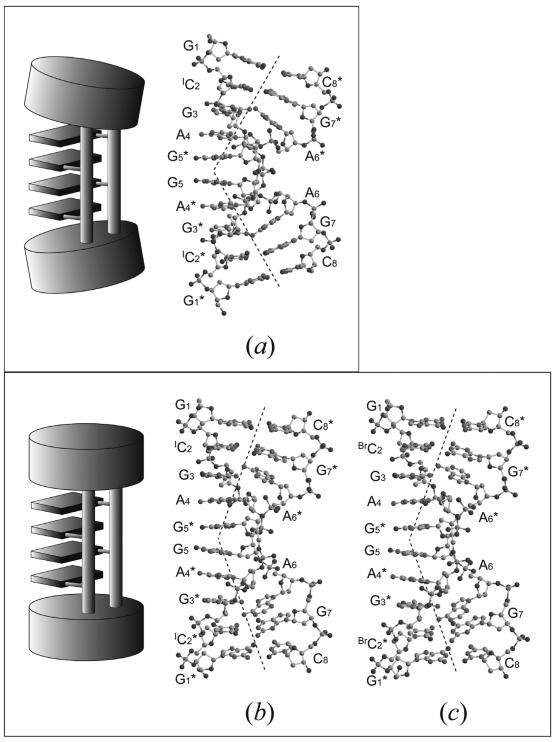
Figure 2.
Schematic diagrams and molecular structures of three base-intercalated duplexes, found in G1-I-lowK (a), in G1-I-highK (b) and in G1-Br-highK (c). The two ends of the duplex are largely bent in (a) to form G-quartets in the central part, indicating that the four base-intercalated duplexes are tightly bound to form an octaplex. Those of the split quadruplex seem to be relaxed, as compared with (a).
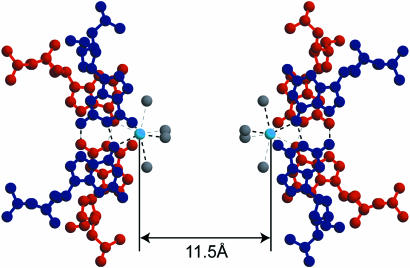
Figure 5.
Two potassium cations bound to four guanine bases in the central parts of the split quadruplexes. Four water molecules cover the outer side of each cation. The G-duets are colored in red or blue. The value indicates the K…K distance in G1-I-highK. Light blue and gray spheres represent potassium cations and water oxygen atoms, respectively. Broken lines indicate eight coordination bonds around the respective potassium cations and possible hydrogen bonds.
RESULTS
Two types of crystal structures of d(gcGA[G]1Agc)
The G1 octamer was crystallized under two different conditions, in which the starting concentrations of DNA were 1.5 and 3.0 mM at the constant potassium concentration of 80 mM, the concentration ratio of potassium ion to DNA being two times higher for the G1-Br-highK and G1-I-highK crystals (80/1.5 mM) than for the G1-I-lowK crystal (80/3.0 mM). Therefore, the former two are isomorphous to each other with similar unit cell dimensions. Although, G1-I-lowK belongs to the same space group I222, the unit cell dimensions are different suggesting a different crystal packing or molecular structure.
Base-intercalated duplex formation as a basic unit
In any crystal structures, the two octamers are in an anti-parallel orientation and form a base-intercalated duplex, as shown in Figure 2 (note the numbering systems of nucleotide residues). At both ends of the duplex, two Watson–Crick G:C base pairs are formed. Halogenations at the cytidine residues have no significant effect on the base-pair formations. The subsequent G3 residue forms a pair in a sheared-type geometry with the A6 residue of the counter strand, the G3:A6* and A6:G3* pairs being formed through the two hydrogen bonds, N3(G)…H-N6(A) and N2-H(G)…N7(A). On the other hand, the central A4 and G5 residues are not involved in any base-pair formations. Their base moieties are respectively stacked on those of the other strand so that A4 is intercalated between G3 and G5*, and G5 is intercalated between G5* and A4*. These four bases, A4, G5*, G5 and A4*, expose their Watson–Crick and major-groove sites into the solvent region and interact with the neighboring molecules to form multiplexes, as described below. These structural features are similar to those of the base-intercalated duplexes of d(gcGA[A]1Agc) (20,21) and d(gcGA[T]1Agc) (19) (hereafter A1 and T1).
Octaplex formation with I-motif of G-quartet under a relatively lower potassium concentration
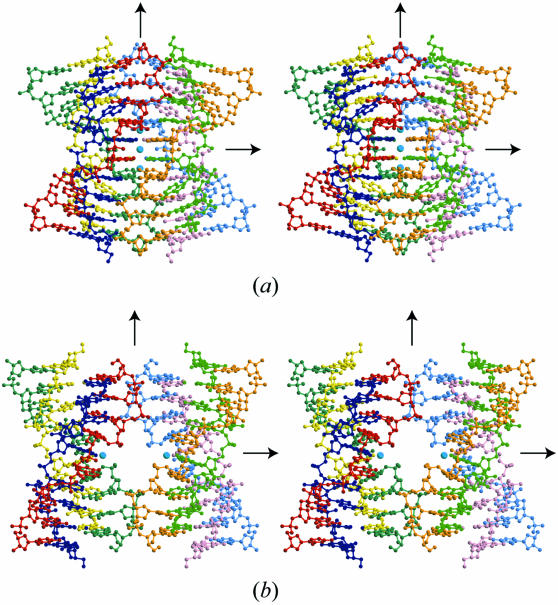
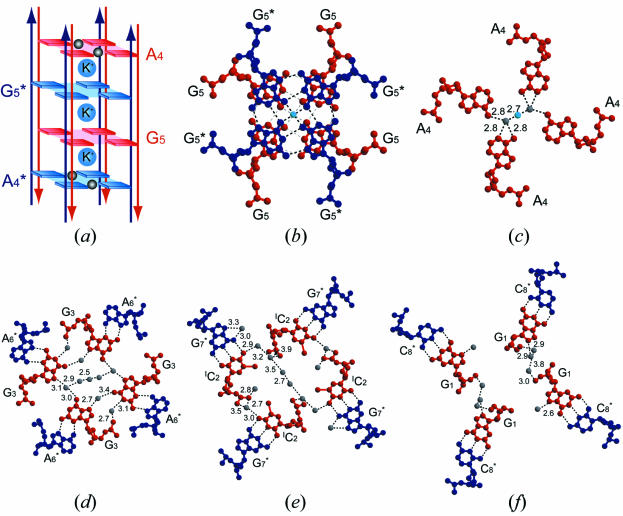
In the G1-I-lowK crystal obtained under conditions containing relatively lower potassium concentration, the four base-intercalated duplexes are assembled according to the crystallographic 222-symmetry to form an octaplex at the central part (Fig. 3a). This is the largest multiplex found in nucleic acid crystals so far.
Figure 3.
Stereo-pair drawings of the octaplex at low [K+] (a) and the split quadruplexes at high [K+] (b). Light blue spheres represent potassium cations.
In the central part, the eight G5 residues form two G-quartets, which are stacked on each other, as shown in Figure 4a and b. Another interesting feature is that the four DNA strands with a G-quartet are aligned in a parallel fashion. The other four strands with another G-quartet are in the opposite direction to deny the strand polarity (5′–3′) of the octaplex (Fig. 4a). The four A4 residues of the parallel strands are stacked above or below the double G-quartets. Therefore, the eight strands seem to be assembled by intercalation of the two G-quartets. So we designate this type of octaplex formation as I-motif of G-quartet. Between the two G-quartets, a potassium cation occupies the center so that the eight O6 atoms of the guanine bases surround the potassium cation. In addition, two other potassium cations are bound to the four O6 atoms above and below the double G-quartet, respectively. Furthermore, on the other side of these two cations, two water molecules are bound. Subsequently, these water molecules are bridged to the four A4 residues, as shown in Figure 4c. At each end of the octaplex, four stems of the base-intercalated duplexes are assembled by mediation of water molecules, as shown in Figure 4d–f. These hydrogen-bonded networks are indispensable in stabilizing the octaplex.
Figure 4.
The interior structures along the central axis of the octaplex (G1-I-lowK). A schematic diagram of the I-motif of G-quartet (a), and its local structures, the stacked double G5-quartets (b), water-mediated A4-assembly (c), four G3 residues paired with the A6 residues of the anti-parallel strands (d), four IC2 residues paired with the G7 residues of the anti-parallel strands (e) and four G1 residues paired with the C8 residues of the anti-parallel strands (f). At both ends of the octaplex, phospahte groups are gathered around the center. Arrows colored in red and dark blue represent 5′–3′ directions of DNA strands. Light blue and gray spheres represent potassium cations and water oxygen atoms, respectively. Broken lines indicate possible hydrogen bonds.
Splitting of the octaplex into two identical quadruplexes under a relatively higher potassium concentration
Another interesting structural feature of the octaplex is found in the G1-I-high crystal obtained under a condition containing a higher potassium concentration. As shown in Figure 3b, the octaplex is split into just two halves of quadruplexes, keeping the original 222-symmetry. In each half, a potassium cation is bound on the split surface (Fig. 5). These two potassium cations separate the split quadruplexes at a distance of 11.5 Å. The structure of the bromo-derivative in G1-Br-highK is also split into two halves, the potassium–potassium distance being 11.7 Å. The two structures are isomorphous with the following structural features.
In the central part of each quadruplex, the four G5 residues form two G-duets. As shown in Figure 5, they are stacked between the two upper Gs and the two lower Gs. The two upper Gs belonging to the parallel strands form a pair through the two hydrogen bonds (N1-H…O6 and N2-H…N7). The two lower Gs also pair in a similar way, but their strand polarity is reversed. These four Gs are bound to the central potassium cation. The four O6 atoms and four water molecules are coordinated to the potassium cation. At the fourth residue, the two adenine bases form a pair through the direct hydrogen bond between N6-H and N1. This type of pair is known to occur only between the parallel strands (7), but the geometry is deformed from a trans position. At each end of the split quadruplex, the two stems of the base-intercalated duplexes are assembled by mediation of water molecules, which is similar to that of the octaplex.
DISCUSSION
Stability of the base-intercalated duplex
The base-intercalated duplex is the basic unit of two multiplexes, the octaplex and split quadruplexes. Even when the fifth residue is replaced with other nucleotides, the unit duplex adopts a structure similar to those found in A1 octamers (20,21) and a T1 octamer (19). It is, therefore, concluded that the generalized sequences d(gcGA[X]1Agc) always form the base-intercalated duplex. Furthermore, it may be possible to extend the central region with the sequence d(gcGA[X]nAgc). As discussed in a previous paper (19), the sheared G:A base-pair formations in both stems of the duplexes bring the two phosphate backbones closer to facilitate the A-(X*-X)n-A* intercalated base–base stacking.
Conformational changes between the two multiplexes
When the two duplexes, G1-I-lowK and G1-I-highK, are superimposed, the differences we have outlined are found in the duplex conformation, as shown in Figure 2. In the octaplex, the bending of the duplex becomes larger at the sheared G3:A6 base pairs, indicating that the base-intercalated duplex is flexible at these points, like two elbows. As a result, the unit-cell of G1-I-highK is expanded by 1 Å along the c-axis. (At the same time, the b-length is also increased for splitting of the octaplex along the b-axis.) The two ends of the duplex seem to be relaxed in the quadruplex. For these movements, the torsion angles around the P-O5′ (α) and C5′-C4′ (γ) bonds of the A6 residue are changed from +gauche and –gauche in G1-Br-highK and G1-I-highK to –gauche and +gauche in G1-I-lowK. The guanine moieties of the G5 residues of G1-I-lowK protrude largely to form direct interactions between the four guanine bases. These flexibilities of the base-intercalated duplex thus allow conformational changes between the two multiplexes under different potassium concentrations.
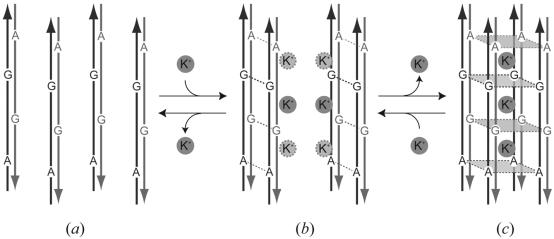
Structural equilibrium between the two multiplexes
From a simple difference with high similarity between the two structures, the octaplex and the split quadruplexes, it is plausible to speculate that at least these two states equilibrate in solution depending on potassium concentration. These structural features make it possible to simulate a dynamic formation of an octaplex. Firstly, two DNA fragments are aligned in an anti-parallel fashion and associated to form a base-intercalated duplex (Fig. 6a). Secondly, two duplexes are assembled to form a quadruplex through potassium-ion mediation (Fig. 6b). Finally, two quadruplexes are further associated to make an octaplex by releasing some potassium cations (Fig. 6c). These dynamic changes would be reversible. In the G1-I-highK and G1-Br-highK crystals, only one potassium ion is visible at a certain position. However, other ions may be disordered to cover the surface of the quadruplex. As mentioned previously, the potassium concentrations used are within the normal physiological conditions. Therefore, it is expected that the two multiplexes may occur in some biological processes, when DNA is unwound to perform a function.
Figure 6.
Schematic diagram of the dynamic transition to form an octaplex. Arrows in gray and black represent 5′–3′ directions of DNA strands. Potassium cations are circled with gray. (a) Two octamers are aligned in an anti-parallel fashion and associate to form a base-intercalated duplex. (b) Two duplexes are associated to form a quadruplex through potassium-ion mediation. (c) Two quadruplexes assemble to make an octaplex by releasing some potassium cations.
A possibility of octaplex formation in VNTRs
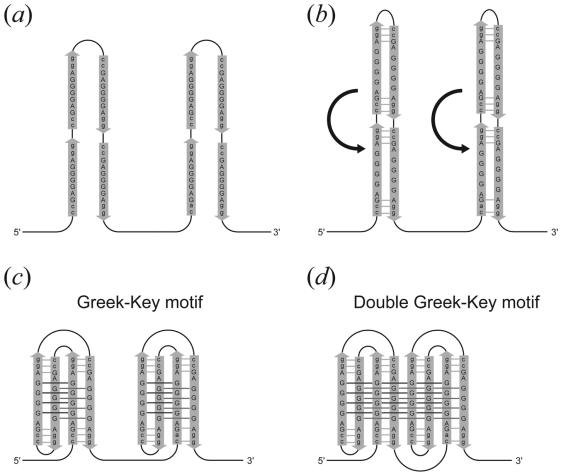
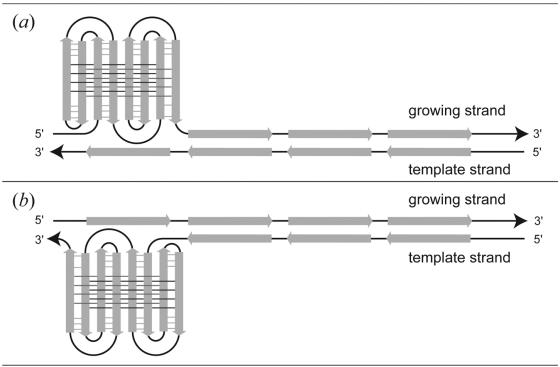
The present sequence d(gcGA[G]1Agc) can extend to the sequence d(ccGA[G]4Agg). The latter sequence is repeated eight times in a VNTR with a high degree of length polymorphism, which is immediately adjacent to the human pseudoautosomal telomere (18). These eight tandem repeats can fold into an octaplex. We suggest that the octaplex in vivo could be formed exactly as a Greek-key motif does in proteins, as shown in Figure 7. At first, the long chain is folded in the two parts between the second and third repeats and between the sixth and seventh repeats (Fig. 7a). Then base-intercalated duplexes are formed in each part (Fig. 7b). In the next step, the two parts are further folded; one occurs between the first and second repeats and between the third and fourth repeats, and the other occurs between the fifth and sixth repeats and between the seventh and eighth repeats, to form a quadruplex in each part (Fig. 7c). This folding pattern is similar to a Greek-key motif, which is well known in protein folding. Finally the two quadruplexes are associated to form an octaplex with a double Greek-key motif (Fig. 7d). A similar repetitive sequence of d(tgGA[G]3Aca) is also found in human 3′-flanking mini-satellite pMS51 (35). Packaging of these repeats through the present mechanism might induce slippage of the repeats during DNA replication. As illustrated in Figure 8, there will be an increase, or a decrease, of the number of repeats depending on whether slippage occurs on the growing, or on the template strand. In fact, it was reported that such slippages will frequently occur to regulate transcription and translation of genes according to its number of repeats (17). Moreover, these are known to be hot spots for meiotic recombination in germ cells (12,14). Therefore, the octaplex structure may be an indicator of such a sequence undergoing recombination. To confirm the validity of the above hypotheses, more extensive and intensive investigations are required. In addition, it is necessary to reveal the structures of longer octaplexes with I-motif of G-quartets.
Figure 7.
A proposed folding mechanism of eight tandem repeats of d(ccGA[G]4Agg). (a) A long chain is folded at the two linkers between the second and third repeats and between the sixth and seventh repeats. (b) Base-intercalated duplexes are formed between the anti-parallel strands. (c) The two parts are further folded; one is between the first and second repeats and between the third and fourth repeats, and the other is between the fifth and sixth repeats and between the seventh and eighth repeats, to form a quadruplex in each part. This folding pattern is similar to a Greek-key motif, which is well known in protein folding. (d) Finally the two quadruplexes are associated to form an octaplex with a double Greek-key motif.
Figure 8.
Mechanisms of repeat increase (a) and decrease (b). Slippage occurs in the growing strand for repeat increase (a). On the other hand, slippage occurs in the template strand for repeat reduction (b).
Data bank accession codes
The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the ID codes 1V3P for G1-I-lowK, 1V3O for G1-I-highK and 1V3N for G1-Br-highK.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Suzuki, N. Igarashi and A. Nakagawa for facilities and help during data collection, T. Higashi for data processing of G1-I-lowK, and T. Simonson for proofreading of the original manuscript. This work was supported in part by Grants-in-Aid for Scientific Research (No.12480177 and 14035217) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
PDB ID codes 1V3P, 1V3O and 1V3N
REFERENCES
- 1.Watson J.D. and Crick,F.H. (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 2.Fuller W., Wilkins,M.H.F., Wilson,H.R. and Hamilton,L.D. (1965) The molecular configuration of deoxyribonucleic acid. IV. X-ray diffraction study of the A form. J. Mol. Biol., 12, 60–80. [DOI] [PubMed] [Google Scholar]
- 3.Leslie A.G.W., Arnott,S., Chandrasekaran,R. and Ratliff,R.L. (1980) Polymorphism of DNA double helices. J. Mol. Biol., 143, 49–72. [DOI] [PubMed] [Google Scholar]
- 4.Wang A.H.-J., Quigley,G.J., Kolpak,F.J., Crawford,J.L., van Boom,J.H., van der Marel,G. and Rich,A. (1979) Molecular structure of a left-handed double helix DNA fragment at atomic resolution. Nature, 282, 680–686. [DOI] [PubMed] [Google Scholar]
- 5.Drew H., Takano,T., Tanaka,S., Itakura,K. and Dickerson,R.E. (1980) High-salt d(CpGpCpG): a left-handed double helix. Nature, 286, 567–573. [DOI] [PubMed] [Google Scholar]
- 6.Brown B.A. 2nd and Rich,A. (2001) The left-handed double helical nucleic acids. Acta Biochim. Pol., 48, 295–312. [PubMed] [Google Scholar]
- 7.Sunami T., Kondo,J., Kobuna,T., Hirao,I., Watanabe,K., Miura,K. and Takénaka,A. (2002) Crystal structure of d(GCGAAAGCT) containing a parallel-stranded duplex with homo base pairs and an anti-parallel duplex with Watson–Crick base pairs. Nucleic Acids Res., 30, 5253–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang E. and Feigon,J. (1999) Structures of nucleic acid triplexes. In Neidle,S. (ed.), Nucleic Acid Structure. Oxford University Press, Oxford, UK, pp. 355–388. [Google Scholar]
- 9.Patel D.J., Bouaziz,A., Kettani,A. and Wang,Y. (1999) Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric and triplet repeat disease DNA sequences. In Neidle,S. (ed.), Nucleic Acid Structure. Oxford University Press, Oxford, UK, pp. 389–453. [Google Scholar]
- 10.International Human Genome Sequencing Consortium. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 11.Jeffreys A.J., Royle,N.J., Wilson,V. and Wong,Z. (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature, 332, 278–281. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys A.J., Wilson,V. and Thein,S.L. (1985) Hypervariable ‘minisatellite’ regions in human DNA. Nature, 314, 67–73. [DOI] [PubMed] [Google Scholar]
- 13.Jeffreys A.J., Wilson,V. and Thein,S.L. (1985) Individual-specific ‘fingerprints’ of human DNA. Nature, 316, 76–79. [DOI] [PubMed] [Google Scholar]
- 14.Wahls W.P., Wallace,L.J. and Moore,P.D. (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell, 60, 95–103. [DOI] [PubMed] [Google Scholar]
- 15.Lucassen A.M., Julier,C., Beressi,J.P., Boitard,C., Froguel,P., Lathrop,M. and Bell,J.I. (1993) Susceptibility to insulin dependent diabetes mellitus maps to a 4.1 kb segment of DNA spanning the insulin gene and associated VNTR. Nature Genet., 4, 305–310. [DOI] [PubMed] [Google Scholar]
- 16.Virtaneva K., D’Amato,E., Miao,J., Koskiniemi,M., Norio,R., Avanzini,G., Franceschetti,S., Michelucci,R., Tassinari,C.A., Omer,S., Pennacchio,L.A., Myers,R.M., Dieguez-Lucena,J.L., Krahe,R., de la Chapelle,A. and Lehesjoki,A.E. (1997) Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nature Genet., 15, 393–396. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura Y., Koyama,K. and Matsushima,M. (1998) VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J. Hum. Genet., 43, 149–152. [DOI] [PubMed] [Google Scholar]
- 18.Inglehearn C.F. and Cooke,H.J. (1990) A VNTR immediately adjacent to the human pseudoautosomal telomere. Nucleic Acids Res., 18, 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo J., Umeda,U., Fujita,K., Sunami,T. and Takénaka,A. (2004) X-Ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue. J. Synchrotron Rad., 11, 117–120. [DOI] [PubMed] [Google Scholar]
- 20.Sunami T., Kondo,J., Hirao,I., Watanabe,K., Miura,K. and Takénaka,A. (2004) Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure. Acta Crystallogr. D, 60, 90–96. [DOI] [PubMed] [Google Scholar]
- 21.Sunami T., Kondo,J., Hirao,I., Watanabe,K., Miura,K. and Takénaka,A. (2004) Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G·A pairs to form a functional G·A×A·G crossing. Acta Crystallogr. D, 60, 422–431. [DOI] [PubMed] [Google Scholar]
- 22.Wandergem R. (1982) Intracellular potassium activity during liver regeneration. In McKeehan,W., Boynton,A. and Whitfield,J. (eds), Ions, Cell Proliferation and Cancer. Academic Press, New York, pp. 175–186. [Google Scholar]
- 23.Leslie A.G.W. (1992) Molecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystallography Computing 5, From Chemistry to Biology. Oxford University Press, Oxford, UK, pp. 50–61. [Google Scholar]
- 24.Steller I., Bolotovsky,R. and Rossmann,M.G. (1997) An algorithm for automatic indexing of oscillation images using Fourier analysis. J. Appl. Crystallogr., 30, 1036–1040. [Google Scholar]
- 25.Rossmann M.G. and van Beek,C.G. (1999) Data processing. Acta Crystallogr. D, 55, 1631–1640. [DOI] [PubMed] [Google Scholar]
- 26.Powell H.R. (1999) The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D, 55, 1690–1695. [DOI] [PubMed] [Google Scholar]
- 27.Pflugrath J.W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D, 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P. and Grosse-Kunstleve,R.W. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 31.Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- 32.Lu X.J. and Olson,W.K. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res., 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 34.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 35.Armour J.A., Wong,Z., Wilson,V., Royle,N.J. and Jeffreys,A.J. (1989) Sequences flanking the repeat arrays of human minisatellites: association with tandem and dispersed repeat elements. Nucleic Acids Res., 17, 4925–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structure. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]