Abstract
Hepatocyte Nuclear Factor 1α (HNF1α) and Hepatocyte Nuclear Factor 4α (HNF4α) are two liver-enriched transcription factors coexpressed in specific tissues where they play a crucial role through their involvement in a complex cross-regulatory network. HNF1α down regulates HNF4α-mediated activation of transcription via a direct protein–protein interaction. Here we show that HNF4α enhances the transcriptional activity of HNF1α in a DNA binding independent manner, thus indicating that it behaves as a HNF1α coactivator. Using mutations in the ligand binding domain (LBD) of HNF4α, we confirmed the involvement of the Activation Function 2 module and demonstrated the requirement of the integrity of the LBD for the interaction with HNF1α. Moreover, we show that HNF4α cooperates with p300 to achieve the highest HNF1α-mediated transcription rates. Our findings highlight a new way by which HNF4α can regulate gene expression and extend our knowledge of the complexity of the transcriptional network involving HNF4α and HNF1α.
INTRODUCTION
Hepatocyte Nuclear Factor 1α (HNF1α) and Hepatocyte Nuclear Factor 4α (HNF4α) are two liver-enriched transcription factors that are also expressed in kidney, intestine and endocrine pancreas (1). They are involved in complex cross-regulatory networks that determine the phenotype of hepatocytes and pancreatic β-cells (1–3). HNF1α is a homeodomain-containing transcription factor (1) whereas HNF4α belongs to the hormone nuclear receptor superfamily (4). Both transcription factors are highly conserved during evolution (4,5). HNF1α and HNF4α directly modulate the expression of a large number of genes (4,6,7). They can also modulate transcription indirectly through the above mentioned transcription factor network, including a HNF1α-mediated negative regulation of genes activated by HNF4α (8,9). The roles of these transcription factors in vivo have been confirmed by defects linked to the invalidation of their genes in mice (10–14). Further underscoring the importance of these transcription factors, mutations in HNF1α and HNF4α genes have been identified in patients with Maturity Onset Diabetes of the Young, MODY3 and MODY1, respectively (15,16). MODY1 and MODY3 mutations result in loss of function of these transcription factors (17). Moreover, it has been reported that expression of a HNF1α dominant negative mutant linked to MODY3 led to an impaired function of pancreatic β-cells (18).
HNF1α contains two domains involved in DNA binding (an atypical homeodomain and a POU-like domain), an N-terminal dimerization domain and a C-terminal transactivation domain (1). Several proteins interact with various HNF1α domains and play crucial roles in HNF1α function. DcoH (Dimerization Cofactor of HNF1), is a small protein, which binds to the HNF1 dimerization domain and is involved in dimer stabilization (19). The ability of various HNF1α domains to interact with multiple coactivators allows formation of a platform for recruitment of a transcriptional complex, leading to a strong enhancement of transcription. CBP/p300 interacts with both the DNA binding domain and the activation domain of HNF1α while P/CAF, SRC-1 and RAC3 interact with the HNF1α activation domain (20,21). Each of these coactivators can independently increase activation of transcription by HNF1α. In addition, they cooperate with each other to further enhance the HNF1α-mediated activation of transcription (21). HNF1α can also interact with GATA 4, GATA 5 and Cdx-2 transcription factors (22). The interactions between HNF1α, GATA5 and Cdx-2 lead to a cooperative enhancement of HNF1α-mediated activation of transcription (22). A synergy between HNF1α and neurogenin 3 was also recently reported (23). HNF1α down regulates HNF4α-mediated activation of transcription via a direct interaction of these transcription factors (8,9).
In this work, we show that HNF4α enhances the transcriptional activity of HNF1α, and that the cooperation between both factors can be further enhanced by p300 recruitment.
MATERIALS AND METHODS
DNA constructs
Plasmids pcDNA3 HNF4α2 and pcDNA3 HNF4α2-E276Q were previously described (24). Plasmid pcDNA3 HNF4α2-D126Y was described by Oxombre et al. (25). Plasmids pcDNA3 HNF4α2-E262A and pGEX2TK HNF4α2 were described by Eeckhoute et al. (26). Plasmids pcDNA3 HNF1α and pcDNA3 HNF1α-P291fsinsC, here named HNF1α-ΔAD, were generous gifts from Drs M. Yaniv and A. Abderrahmani, respectively. Plasmids pcDNA3 HNF4α2-A223F and pcDNA3 HNF4α2-AF2mut were obtained by site-directed mutagenesis using the QuickChange™ kit (Stratagene) to introduce the A223F or the three mutations E363K, L365Q and L366Q, respectively. Plasmid pM3-VP16 was from Clontech and pcDNA3-RXRβ encoding the full-length human RXRβ was a generous gift from Dr R. Polakowska. Plasmid pGEX2TK HNF1α was prepared by a strategy identical to that used for cloning pGEX2TK COUP-TFII (24) by inserting a PCR fragment encompassing the human HNF1α cDNA. Plasmid pCMVβ-NHA p300, PGEX2TK p300(1–595), PGEX2TK p300(340–528) and PGEX2TK p300(1572–2370) were kindly provided by Dr S. R. Grossman. Expression plasmids for VP16, Np300-VP16 and Cp300-VP16 were gifts from Dr D. Hum (27). Plasmid pGL3 (–96/+11) LPK was a generous gift from Dr M. Vasseur-Cognet (28). Plasmid pGL3 (–341/+183) human HNF1α was a gift from Dr G. Bell. Plasmid (GAL4)x5 TATA Luc was described by Chang and Gralla (29). Plasmid pGL3 HNF1α-TATA-Luc was cloned by inserting a double-stranded oligonucleotide encompassing the HNF1α binding site (–56/ –35) of the SRC promoter into the SacI/NheI sites of the pGL3 basic vector in which the TATA box of the adenovirus major late promoter had been previously cloned as described by Suaud et al. (24). All constructs were verified by DNA sequencing.
GST pull-down assays
GST pull-down assays were performed as described previously (24) using [35S]methionine-labelled in vitro synthesized HNF1α or HNF4α and bacterially expressed GST-fusion proteins indicated in legends to figures.
Cell culture and transient transfection assays
HeLa cells (5.5 × 104 cells per 24-well dish) were grown and transfected as described by Eeckhoute et al. (30) with plasmid amounts indicated in figure legends. Luciferase activities were measured using the Bright-Glo Luciferase assay system (Promega).
Western blotting
Western blotting performed from whole-cell extracts and using the α455 HNF4α antiserum was carried out as indicated by Eeckhoute et al. (26).
Data analysis
Statistical analyses were based on Student’s t-test for unpaired data using Prism software. Statistical significance was set at ***P < 0.001, **P < 0.01 and *P < 0.05.
RESULTS
Enhancement of HNF1α-mediated activation of transcription by HNF4α
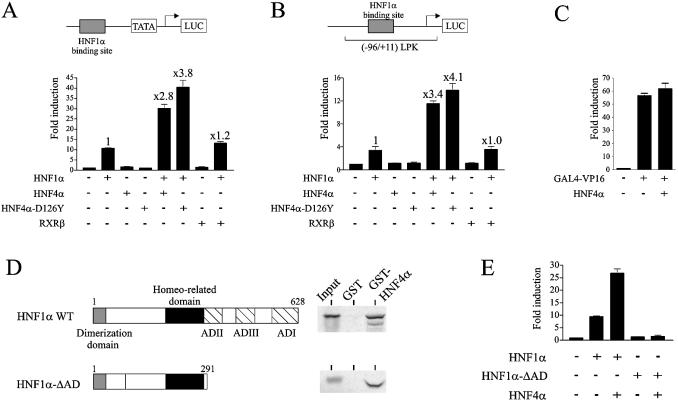
HNF1α and HNF4α can positively regulate one another’s expression in cell types that endogenously express these two factors (3,6). To avoid any interference with the endogenous proteins, the effect of HNF4α on the transactivation activity of HNF1α was analysed in HeLa cells. The experiments were performed on a synthetic promoter consisting of one HNF1α binding site located upstream of the TATA box (HNF1α-TATA promoter) and on the natural liver pyruvate kinase promoter (–96/+11) LPK containing one HNF1α binding site (site L1, position –96/–72) but lacking the HNF4α response element (28). As expected, both promoters were activated by HNF1α but not by HNF4α (Fig. 1A and B). Cotransfection of HNF1α and HNF4α resulted in a marked increase in the HNF1α-mediated activation of these promoters (2.8- and 3.4-fold activation of the synthetic and LPK promoters, respectively) (Fig. 1A and B). The enhancement of the HNF1α activity was not impaired when introducing the D126Y mutation in HNF4α, which significantly decreases its DNA binding and transactivation activities (25) (data not shown), thus confirming that the synergy between HNF1α and HNF4α does not require the DNA binding-dependent activities of HNF4α (Fig. 1A and B). This induction was not an artefact linked to expression of a protein since cotransfection of an unrelated factor failed to enhance the HNF1α-mediated activation of transcription (compare the second and last columns in Fig. 1A and B).
Figure 1.
HNF4α enhanced the HNF1α-dependent activation of transcription. HeLa cells were transiently transfected with 250 ng of the HNF1α-TATA promoter (A) or the (–96/+11) LPK promoter (B), 15 ng of HNF1α and 20 ng of HNF4α or RXRβ expression plasmids, as indicated. (C) HNF4α was unable to cooperate with the VP16 activation domain. HeLa cells were transiently transfected with 250 ng of the (GAL4)x5 TATA promoter, 5 ng of GAL4-VP16 and 20 ng of HNF4α expression vectors, as indicated. In (A), (B) and (C), the total amounts of transfected DNA were equalized with the empty expression plasmids (–). Fold induction refers to the basal activities of promoters (left columns). Results are means ± S.E. of three experiments performed in triplicate. The extent of HNF4α-mediated enhancement of HNF1α-dependent activation of transcription is indicated (values above bars). (D) In vitro interaction between HNF1α and HNF4α analysed by pull-down assays. A schematic representation of the HNF1α and HNF1α-ΔAD proteins is presented, AD denotes activation domain. [35S]methionine-labelled HNF1α or HNF1α-ΔAD was incubated with immobilized GST or GST-HNF4α. Bound proteins were analysed by SDS–PAGE and PhosphorImager (Molecular Dynamics). Inputs correspond to 10% of amounts of labelled proteins used in the assays. (E) Requirement of the activation domains of HNF1α for the cooperation between HNF4α and HNF1α. HeLa cells were transfected as in (A) with the indicated HNF1α constructs. Fold induction refers to the basal activity of the promoter (left column). Results are means ± S.E. of three experiments performed in triplicate.
To control the possibility that HNF4α does not act as a transcriptional partner of another transcription factor, we analysed the effect of HNF4α on the VP16 activation domain fused to the GAL4 DBD (construct GAL4-VP16). HNF4α was unable to enhance the VP16-mediated activation of transcription (Fig. 1C), thus indicating the selectivity of the cooperation between HNF4α and HNF1α.
As predicted from the results of Ktistaki and Talianidis (9), we observed that full-length HNF1α and HNF4α physically interact (Fig. 1D). Interestingly, pull-down experiments also showed that HNF1α lacking its activation domain (HNF1α-ΔAD) efficiently interacted with HNF4α (Fig. 1D). This result indicates that the sequence 1–291 of HNF1α, containing the dimerization and DNA binding domains, is able to interact with HNF4α. This prompted us to investigate whether coexpression of HNF4α and HNF1α-ΔAD could activate the HNF1α-TATA promoter. Despite their efficient interaction, HNF1α-ΔAD and HNF4α were unable to activate transcription (Fig. 1E), thus indicating that cooperation between HNF1α and HNF4α requires the HNF1α activation domain.
AF2 is not the only sequence within the HNF4α LBD required for interaction and cooperation with HNF1α
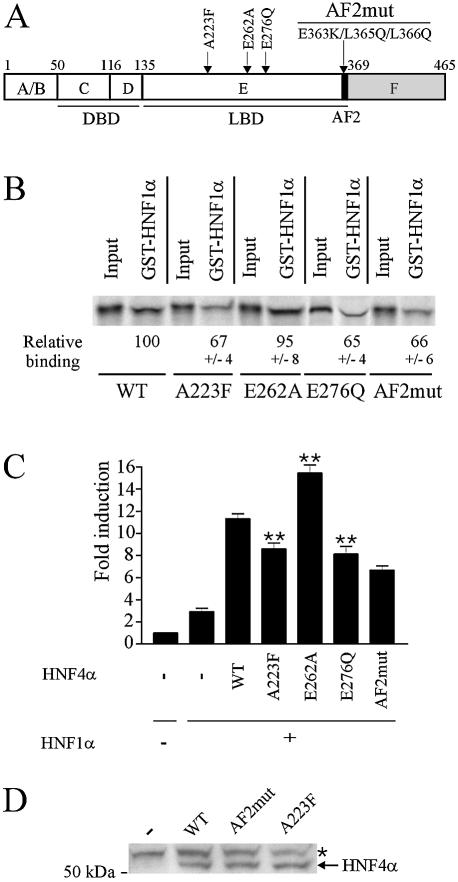
The repressive action of HNF1α on HNF4α was unambiguously shown to require the HNF4α LBD (9). However, using truncated fragments of HNF4α, Ktistaki and Talianidis mapped the sequence involved in the interaction with HNF1α between residues 337 and 368, leading to the conclusion that the HNF4α AF2 activation function is necessary and sufficient for interaction with HNF1α (9). In fact, this sequence does not contain solely the AF2 (sequence 358–366 corresponding to helix 12 of the LBD), but also a fragment forming part of the large helix termed H10–H11, according to conventional nomenclature (31). Indeed, H10 and H11 are contiguous in HNF4α and form a helix that plays a key role in HNF4α function (31). Furthermore, the integrity of this helix is crucial for protein conformation (32). These recent findings begged a re-evaluation of the HNF4α sequence involved in interaction with HNF1α. Using full-length HNF4α, we analysed the effects of point mutations located in the HNF4α LBD (Fig. 2A) on the interaction and cooperation with HNF1α. The E262A mutation was shown to affect HNF4α dimerization (26). The E276Q mutation does not alter the intrinsic HNF4α transcriptional activity but impairs recruitment of HNF4α transcriptional partners (24,33). Owing to the bulky side chain of phenylalanine, the A223F mutation was hypothesized to affect occupancy of the ligand binding pocket by fatty acids that act as structural cofactors rather than as conventional ligands (31). Accord ingly, A223F mutation mildly impaired HNF4α transcriptional activity and interaction with coactivators (data not shown). In the AF2 module, the acidic group of the amino acid residue side chain at position 363 and the hydrophobic nature of the amino acid residue side chain at position 365 are strictly conserved and play a key role in nuclear receptor function (34). Residue L366 is also important since the L366Q mutation alters HNF4α transcriptional activity (35). Pull-down assays showed that introducing three mutations E363K, L365Q and L366Q in the AF2 (HNF4α-AF2mut construct) altered interaction between HNF4α and HNF1α (Fig. 2B). Note however that the decrease in interaction was mild. This result confirms the involvement of the AF2 in the interaction between HNF1α and HNF4α but indicates that this module is not the unique sequence required for interaction between these proteins. Interaction with HNF1α was also significantly decreased by the A223F and E276Q mutations while mutation E262A had no effect (Fig. 2B). Consistent with data from pull-down assays, the HNF4α-mediated enhancement of HNF1α-dependent transcriptional activation of the (–96/+11) LPK promoter was not impaired by the E262A mutation but was significantly decreased with the mutants A223F, E276Q and HNF4α-AF2mut (Fig. 2C). We controlled so that mutant and wild-type HNF4α were expressed at a similar level in transfected cells as shown in Figure 2D for mutants A223F and HNF4α-AF2mut and in (24,26) for the mutant E276Q and E262A, respectively. It appears therefore that the interaction and cooperation between HNF4α and HNF1α depend on both the AF2 module (helix 12) and the integrity of the HNF4α LBD.
Figure 2.
The AF2 is not the only sequence within the HNF4α LBD required for interaction and cooperation with HNF1α. (A) Positions of mutations used in this study. A scheme of HNF4α structure with the various domains is given: DBD, DNA binding domain; LBD, ligand binding domain; AF2, activation function 2 module. (B) Effects of HNF4α mutations on the physical interaction with HNF1α. [35S]methionine-labelled wild-type or mutated HNF4α were incubated with immobilized GST-HNF1α. Bound proteins were analysed by SDS–PAGE and PhosphorImager. Values under photographs indicate binding of HNF4α mutants relative to that of wild-type HNF4α from three independent experiments. Inputs, corresponding to 5% of amounts of labelled proteins used in the assays, were taken into account for binding quantifications. (C) Effects of HNF4α mutations on the cooperation between HNF4α and HNF1α on the (–96/+11) LPK promoter. HeLa cells were transfected as in Figure 1B. Fold induction refers to the basal activity of the promoter (left column). Results are means ± S.E. of three experiments performed in triplicate. Statistical significance of differences with values obtained with wild-type HNF4α2 is indicated by stars (P < 0.01). (D) Western blotting of wild-type (WT), A223F and AF2mut HNF4α expressed in HeLa cells using the α455 HNF4α antiserum (43). Position of HNF4α is indicated. The star denotes a non-specific band obtained from HeLa cells transfected with the empty vector (–). The number on the left indicates the molecular mass of a marker size in kDa.
HNF4α cooperates with the coactivator p300 to enhance HNF1α transcriptional activity
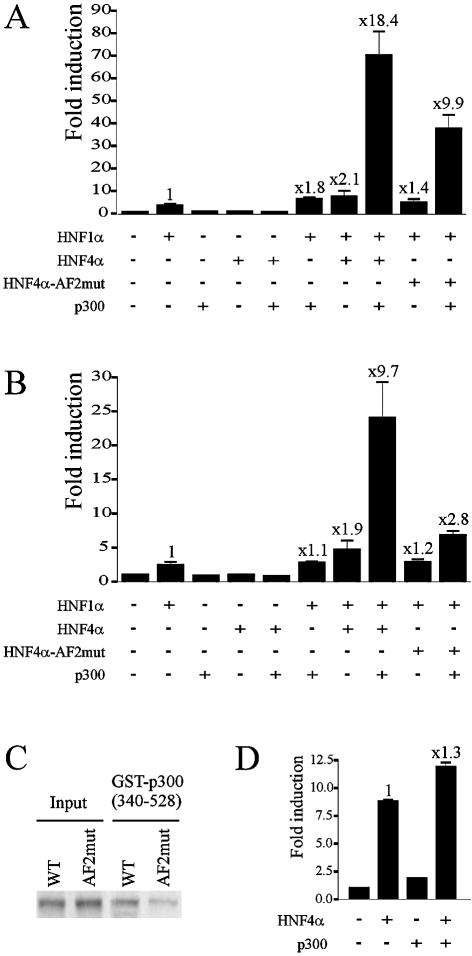
Because CBP/p300 are key coactivators of HNF1α, we hypothesized that cooperation between HNF4α and HNF1α could involve p300. In the absence of HNF1α, the coactivator p300 alone or coexpressed with HNF4α was unable to activate the HNF1α-TATA promoter (Fig. 3A). On this promoter, p300 slightly enhanced HNF1α transcriptional activity (Fig. 3A). Note that this moderate increase is due at least in part to the low ratio of p300/HNF1α expression vectors used in our experiments (3:1) compared to that used in other studies performed on other promoters [e.g. 10:1 in Soutoglou et al. (21)]. We have verified that by using a higher ratio, we could obtain a stronger enhancement of HNF1α activity by p300 alone (data not shown). Interestingly, in the presence of HNF4α, p300 synergistically enhanced HNF1α-mediated activation of transcription: the activity of the promoter was increased 18.4-fold, which is 4.7-fold higher than the additive value for the induction of HNF1α activity by p300 and HNF4α alone (1.8 and 2.1, respectively). The marked p300-mediated enhancement of cooperation between HNF1α and HNF4α could also be observed on the (–96/+11) LPK promoter (Fig. 3B). On both promoters, the enhanced cooperation between HNF1α and HNF4α was significantly impaired by mutations in the HNF4α AF2 module (Fig. 3A and B). These results strongly argue for the involvement of HNF4α in the p300-mediated activation of these promoters despite the absence of HNF4α response element in their sequences. The result obtained with the HNF4α-AF2mut led us to investigate whether mutations in AF2 affect p300 binding. Pull-down experiments clearly showed a much weaker interaction of p300 with HNF4α-AF2mut than with wild-type HNF4α (Fig. 3C). This result is in line with our previous observation that deletion of the AF2 resulted in a decrease in interaction (about 70%) between HNF4α and p300 (33). Therefore, we cannot exclude that the impaired cooperativity of HNF4α-AF2mut was also due to reduced p300 recruitment. Because it has been documented that HNF4α and CBP/p300 form a transcriptionally active complex (36), we verified whether in our experimental conditions this complex was sufficient to achieve the strong cooperation. This question was addressed by the use of human HNF1α promoter containing a HNF4α response element but lacking a HNF1α binding site. Using the same p300/HNF4α expression vector amounts used in Figure 3A and B, cotransfection of p300 failed to strongly enhance the HNF4α-mediated activation of transcription (Fig. 3D). It appears therefore that the strong activation of transcription observed in Figure 3A and B requires a ternary complex including HNF1α, HNF4α and p300.
Figure 3.
Involvement of p300 in the cooperation between HNF1α and HNF4α. The cooperation was analysed on the HNF1α-TATA and (–96/+11) LPK promoters, (A) and (B), respectively. HeLa cells were transiently transfected with 250 ng of promoter plasmids, 10 ng of HNF1α, 15 ng of HNF4α or HNF4α-AF2mut and 30 ng of p300 expression plasmids, as indicated. Fold induction refers to the basal activity of the promoter (left column). Values above bars indicate the enhancement of HNF1α- mediated activation of transcription by HNF4α or p300 alone or in cooperation. Results are means ± S.E. of three experiments performed in triplicate. (C) Effect of the mutations in the AF2 of HNF4α on its interaction with p300 analysed by pull-down assay. [35S]Methionine-labelled HNF4α or HNF4α-AF2mut were incubated with immobilized GST-p300(340–528). Bound proteins were analysed by SDS–PAGE and PhosphorImager. (D) HeLa cells were transiently transfected with expression vector amounts indicated in (A) and 250 ng of the human HNF1α promoter. Fold induction, calculated from three experiments performed in triplicate refers to the basal activity of the promoter.
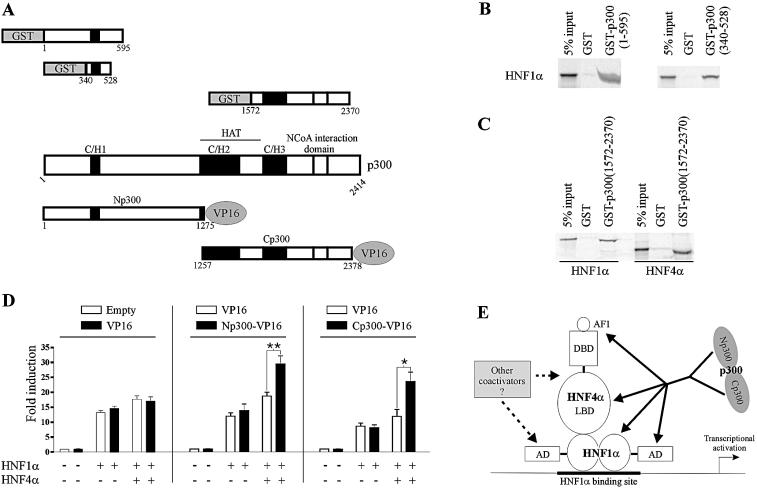
With the aim to investigate the mechanism underlying the p300-mediated enhancement of the cooperation between HNF1α and HNF4α, we surveyed the p300 domains involved in interactions with these transcription factors. Using a double hybrid approach, it was previously shown that the sequences 180–662 and 1818–2079 of p300 interact with the HNF1α transactivation domain (20). Using GST pull-down assays, we mapped more precisely the amino-terminal sequence of p300 involved in interaction with full-length HNF1α between residues 340 and 528 (Fig. 4B). We also confirmed that full-length HNF1α interacts with the carboxy-terminal region of p300 (Fig. 4C). We have previously shown the ability of HNF4α to interact with the amino-terminal domain of p300 (33). HNF4α also interacted with the p300 carboxy-terminal region as shown in Figure 4C. The presence of multiple interaction surfaces between these proteins led us to analyse the effect of HNF4α on the recruitment of the amino- and carboxy-terminal regions of p300 to HNF1α. The strategy consisted of determining whether HNF4α affects the cooperation between HNF1α and the amino- or carboxy-terminal fragments of p300 fused to the VP16 activation domain named Np300-VP16 and Cp300-VP16, respectively (Fig. 4A). Expression of the VP16 activation domain affected neither the basal activity of the promoter nor its activity in the presence of expression vectors for HNF1α alone or together with HNF4α (Fig. 4D, left part). Expression of Np300-VP16 failed to affect the basal activity of the promoter and the HNF1α-mediated activation of this promoter (Fig. 4D, middle). Similar results were obtained with expression of Cp300-VP16 (Fig. 4D, right). Conversely, in the presence of both HNF1α and HNF4α, Np300-VP16 and Cp300-VP16 markedly enhanced the promoter activity (Fig. 4D). These results indicate that, in a cellular context, p300 recruitment by the HNF1α–HNF4α complex can be mediated through both its amino- and carboxy-terminal regions and reinforce data obtained in Figure 3 suggesting that HNF4α improves p300 recruitment to a promoter containing a HNF1α binding site (Fig. 4E).
Figure 4.
HNF4α enhanced the cooperation between HNF1α and p300 through interactions with both the amino- and carboxy-terminal sequences of the coactivator. (A) Schematic representation of p300 fragments fused to either GST or the VP16 activation domain used in interaction studies with HNF1α and HNF4α. (B) Interaction of HNF1α with the amino-terminal domain of p300 analysed by pull-down assays. [35S]Methionine-labelled HNF1α was incubated with immobilized GST, GST-p300(1–595) or GST-p300(340–528). (C) Interaction of HNF1α and HNF4α with the carboxy-terminal domain of p300 analysed by pull-down assays. [35S]Methionine-labelled HNF1α or HNF4α were incubated with immobilized GST or GST-p300(1572–2370). In (B) and (C), bound proteins were analysed by SDS–PAGE and PhosphorImager. (D) HNF4α potentiates enhancement of HNF1α activity by Np300- and Cp300-VP16. HeLa cells were transiently transfected with 250 ng of HNF1α-TATA promoter, 10 ng of HNF1α, 15 ng of HNF4α and 60 ng of VP16 or its empty vector (left), VP16 or Np300-VP16 (middle) and VP16 or Cp300-VP16 (right) expression plasmids. In each case, the total amount of transfected DNA was kept constant using empty pcDNA3. Fold induction refers to the basal activity of the promoter (left columns). Results are means ± S.E. of at least three experiments performed in triplicate. (E) Mechanisms by which HNF4α could enhance HNF1α transcriptional activity. The amino- and/or carboxy-terminal domains of p300 can be involved in p300-mediated activation of transcription. HNF4α could enhance p300 recruitment through direct binding to these p300 fragments and/or through an indirect effect on the HNF1α–p300 interactions. HNF4α could also help recruitment of other coactivators to the transcriptional complex.
DISCUSSION
It has been previously shown that HNF1α acts as a repressor of HNF4α-mediated activation of transcription (8,9). In this study, we show that HNF4α can serve as a coactivator for HNF1α since HNF4α is able to enhance HNF1α activity in a DNA-binding independent manner. A synergy between HNF1α and HNF4α can be inferred from data obtained in undifferentiated Caco-2 cells on a promoter containing binding sites for these two HNFs. Since mutation of the HNF4α binding site did not affect this synergy, the authors suggested that this effect probably did not involve HNF4α DNA binding (37). In light of our results, these data can now most probably be explained by the ability of HNF4α to directly act as a HNF1α coactivator. Analysis of the physiological role of this HNF4α coactivator function in cell types that endogenously express HNF1α and HNF4α cannot be realized by conventional targeting of HNF4α expression since HNF4α also positively regulates HNF1α expression. Therefore, this concern will first require development of an experimental model that permits to distinguish between HNF4α effects linked to regulation of HNF1α expression on one hand and to regulation of HNF1α transcriptional activity on the other hand (Fig. 5).
Figure 5.
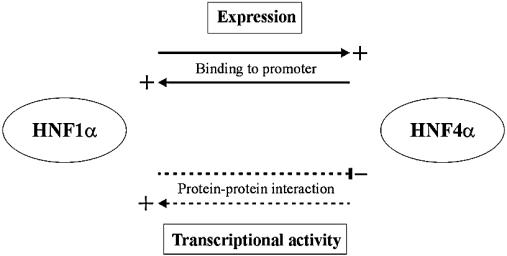
Cross-talk between HNF1α and HNF4α. The cross-talk involves both expression activation, through binding to promoters (solid lines), and modulation of the transcriptional potential, through protein–protein interactions (dotted lines), of the other HNF transcription factor.
Interplays between two transcription factors in both activation and repression of transcription have already been documented. Indeed, cross-talk between HNF6 and HNF3β, two other liver-enriched transcription factors, also results in opposite effects on the activities of these proteins (38).
Despite their efficient interaction, HNF4α and HNF1α lacking its activation function (HNF1α-ΔAD) were unable to cooperate. Such a behaviour is reminiscent of that of CBP and P/CAF with HNF1α (1–280) (21). Only the simultaneous overexpression of CBP and P/CAF together with that of the truncated HNF1α could activate transcription (21). This points to a critical role for the HNF1α activation domain, which could be the recruitment of other required coactivators and/or stabilization of the transcriptional complex recruited by HNF1α.
Our results indicate that the HNF4α AF2 is involved in interaction with HNF1α, but also that other regions of the HNF4α LBD are required for full interaction between these proteins. Ktistaki and Talianidis previously stated that the HNF4α AF2 is sufficient for interaction with HNF1α (9). The discrepancy between their conclusion and our results is likely explained by the fact that these authors used deletion of a fragment (residues 337–368), which includes not only the AF2 but also part of the large helix H10–H11, the integrity of which has meanwhile been shown to be crucial for the protein conformation (32). We observed that two-point mutations in the HNF4α LBD, A223F and E276Q, significantly decreased interaction and cooperation with HNF1α thus demonstrating that the integrity of the HNF4α LBD is required for the synergy with HNF1α.
The similarity of phenotypes exhibited by diabetic patients carrying MODY1 and MODY3 mutations has been ascribed to the mutual control of transcription of HNF1α and HNF4α. The ability of HNF4α to serve as an HNF1α coactivator could provide an additional explanation to the closely related phenotypes of these MODY forms of diabetes. Impairment of interaction and of enhancement of HNF1α transcriptional activity by the HNF4α E276Q mutation may be physiologically relevant since this mutation is correlated with MODY1. Unfortunately, the low transfection efficiency and the presence of large amounts of endogenous HNF4γ in pancreatic β-cells prevented us from analysing the effects of this mutation on the synergy between HNF4α and HNF1α in these cells.
Our results support a model that involves the combined action of HNF4α and the coactivator p300 to achieve the highest rate of transcription mediated by HNF1α. Our results also strongly suggest that HNF4α improves p300 recruitment. The synergy between HNF4α and p300 may be mediated in two ways. The first involves simultaneous interactions of HNF4α and p300 with HNF1α (Fig. 4E). In this case, HNF4α could indirectly improve HNF1α–p300 interaction through induction of a HNF1α conformational change. Such a mechanism is reminiscent of that of the CBP-mediated improvement of the interaction between HNF1α and P/CAF (21). Similarly, upon docking to PPARγ, PGC-1 undergoes a conformational change that permits binding of SRC-1 and p300 (39). The second involves docking of p300 by HNF4α, which is bound to HNF1α (Fig. 4E). The recruitment of p300 to the HNF1α–HNF4α complex could be mediated by its amino- and/or carboxy-terminal regions, which contain the C/H1 and C/H3 domains, respectively. These domains are known to be involved in protein–protein interactions (40). The presence of multiple interaction surfaces in HNF1α, HNF4α and p300 most probably facilitates formation of a ternary complex formed with these proteins (Fig. 4E). The remaining synergy between HNF1α and HNF4α after mutations in the HNF4α AF2 (Fig. 3) may be accounted for by the ability of p300 to interact and activate the HNF4α AF1 (33,41) and by the fact that the two above mentioned mechanisms may not be mutually exclusive.
In conclusion, our results highlight a new way by which HNF4α can regulate gene expression. HNF4α not only directly binds to promoters but also, through interaction with other transcription factors already bound to DNA, can facilitate coactivator recruitment to further enhance transcription. Recently, it has been shown that HNF4α also serves as coactivator for Sterol Regulatory Element-Binding protein-2 (42). Furthermore, our results yield insights into a higher complexity of the transcriptional network and on the primordial relationship between HNF1α and HNF4α (Fig. 5). In the regulatory loop between HNF1α and HNF4α, HNF4α can activate both HNF1α expression and transcriptional activity. In contrast to these additive processes, the negative effect of HNF1α on HNF4α transcriptional activity can attenuate the HNF1α-mediated activation of expression. These regulatory feedback mechanisms will have to be taken into account considering that HNF1α and HNF4α are involved in a large diversity of pathways controlling function of multiple organs, notably liver and endocrine pancreas.
Acknowledgments
ACKNOWLEDGEMENTS
Drs M. Yaniv, A. Abderrahmani, M. Vasseur-Cognet, R. Polakowska, S. Grossman, D. W. Hum and G. Bell are acknowledged for generous gifts of plasmids. We acknowledge Dr P. Sacchetti for helpful discussions and I. Briche for skillful technical assistance.
REFERENCES
- 1.Cereghini S. (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J., 10, 267–282. [PubMed] [Google Scholar]
- 2.Duncan S.A., Navas,M.A., Dufort,D., Rossant,J. and Stoffel,M. (1998) Regulation of a transcription factor network required for differentiation and metabolism. Science, 281, 692–695. [DOI] [PubMed] [Google Scholar]
- 3.Ferrer J. (2002) A genetic switch in pancreatic beta-cells: implications for differentiation and haploinsufficiency. Diabetes, 51, 2355–2362. [DOI] [PubMed] [Google Scholar]
- 4.Sladek F.M. and Seidel,S.D. (2001) Hepatocyte Nuclear Factor 4alpha. In Burris,T.P. and McCabe,E. (eds), Nuclear Receptors and Genetic Disease. Academic Press, San Francisco, CA, pp. 309–361. [Google Scholar]
- 5.Bartkowski S., Zapp,D., Weber,H., Eberle,G., Zoidl,C., Senkel,S., Klein-Hitpass,L. and Ryffel,G.U. (1993) Developmental regulation and tissue distribution of the liver transcription factor LFB1 (HNF1) in Xenopus laevis. Mol. Cell. Biol., 13, 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odom D.T., Zizlsperger,N., Gordon,D.B., Bell,G.W., Rinaldi,N.J., Murray,H.L., Volkert,T.L., Schreiber,J., Rolfe,P.A., Gifford,D.K. et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science, 303, 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tronche F., Ringeisen,F., Blumenfeld,M., Yaniv,M. and Pontoglio,M. (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J. Mol. Biol., 266, 231–245. [DOI] [PubMed] [Google Scholar]
- 8.Kritis A.A., Ktistaki,E., Barda,D., Zannis,V.I. and Talianidis,I. (1993) An indirect negative autoregulatory mechanism involved in hepatocyte nuclear factor-1 gene expression. Nucleic Acids Res., 21, 5882–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ktistaki E. and Talianidis,I. (1997) Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science, 277, 109–112. [DOI] [PubMed] [Google Scholar]
- 10.Pontoglio M., Barra,J., Hadchouel,M., Doyen,A., Kress,C., Bach,J.P., Babinet,C. and Yaniv,M. (1996) Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell, 84, 575–585. [DOI] [PubMed] [Google Scholar]
- 11.Pontoglio M., Sreenan,S., Roe,M., Pugh,W., Ostrega,D., Doyen,A., Pick,A.J., Baldwin,A., Velho,G., Froguel,P. et al. (1998) Defective insulin secretion in hepatocyte nuclear factor 1 alpha-deficient mice. J. Clin. Invest., 101, 2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y.-H., Sauer,B. and Gonzalez,F.J. (1998) Laron Dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α Knockout mouse. Mol. Cell. Biol., 18, 3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayhurst G.P., Lee,Y.H., Lambert,G., Ward,J.M. and Gonzalez,F.J. (2001) Hepatocyte Nuclear Factor 4alpha (Nuclear Receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol., 21, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee J., Inoue,Y., Yoon,J.C., Puigserver,P., Fan,M., Gonzalez,F.J. and Spiegelman,B.M. (2003) Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc. Natl Acad. Sci. USA, 100, 4012–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagata K., Oda,N., Kaisaki,P.J., Menzel,S., Furuta,H., Vaxillaire,M., Southam,L., Cox,R.D., Lathrop,G.M., Boriraj,V.V. et al. (1996) Mutations in the hepatocyte nuclear factor-1 alpha gene in maturity-onset diabetes of the young (MODY3). Nature, 384, 455–458. [DOI] [PubMed] [Google Scholar]
- 16.Yamagata K., Furuta,H., Oda,N., Kaisaki,P.J., Menzel,S., Cox,N.J., Fajans,S.S., Signorini,S., Stoffel,M. and Bell,G.I. (1996) Mutations in the hepatocyte nuclear factor-4 alpha gene in maturity-onset diabetes of the young (MODY1). Nature, 384, 458–460. [DOI] [PubMed] [Google Scholar]
- 17.Ryffel G.U. (2001) Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol., 27, 11–29. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Hagenfeldt-Johansson,K., Otten,L.A., Gauthier,B.R., Herrera,P.L. and Wollheim,C.B. (2002) Experimental models of transcription factor-associated maturity-onset diabetes of the young. Diabetes, 51 (Suppl. 3), S333–S342. [DOI] [PubMed] [Google Scholar]
- 19.Mendel D.B., Khavari,P.A., Conley,P.B., Graves,M.K., Hansen,L.P., Admon,A. and Crabtree,G.R. (1991) Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science, 254, 1762–1767. [DOI] [PubMed] [Google Scholar]
- 20.Ban N., Yamada,Y., Someya,Y., Miyawaki,K., Ihara,Y., Hosokawa,M., Toyokuni,S., Tsuda,K. and Seino,Y. (2002) Hepatocyte nuclear factor-1alpha recruits the transcriptional co-activator p300 on the GLUT2 gene promoter. Diabetes, 51, 1409–1418. [DOI] [PubMed] [Google Scholar]
- 21.Soutoglou E., Papafotiou,G., Katrakili,N. and Talianidis,I. (2000) Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem., 275, 12515–12520. [DOI] [PubMed] [Google Scholar]
- 22.Mitchelmore C., Troelsen,J.T., Spodsberg,N., Sjostrom,H. and Noren,O. (2000) Interaction between the homeodomain proteins Cdx2 and HNF1alpha mediates expression of the lactase-phlorizin hydrolase gene. Biochem. J., 346, 529–535. [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S.B., Gasa,R., Watada,H., Wang,J., Griffen,S.C. and German,M.S. (2003) Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J. Biol. Chem., 278, 38254–38259. [DOI] [PubMed] [Google Scholar]
- 24.Suaud L., Hemimou,Y., Formstecher,P. and Laine,B. (1999) Functional study of the E276Q mutant Hepatocyte Nuclear Factor-4α found in type 1 Maturity-Onset Diabetes of the Young, impaired synergy with Chicken Ovalbumin Upstream Promoter Transcription Factor II on the Hepatocyte Nuclear factor-1 promoter. Diabetes, 48, 1162–1167. [DOI] [PubMed] [Google Scholar]
- 25.Oxombre B., Moerman,E., Eeckhoute,J., Formstecher,P. and Laine,B. (2002) Mutations in hepatocyte nuclear factor 4alpha gene associated with diabetes result in greater loss of HNF4alpha function in pancreatic beta-cells than in nonpancreatic beta-cells and in reduced activation of the apolipoprotein CIII promoter in hepatic cells. J. Mol. Med., 80, 423–430. [DOI] [PubMed] [Google Scholar]
- 26.Eeckhoute J., Oxombre,B., Formstecher,P., Lefebvre,P. and Laine,B. (2003) Critical role of charged residues in helix 7 of the ligand binding domain in hepatocyte nuclear factor 4 alpha. Nucleic Acids Res., 31, 6640–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monte D., DeWitte,F. and Hum,D.W. (1998) Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem., 273, 4585–4591. [DOI] [PubMed] [Google Scholar]
- 28.Cognet M., Bergot,M.O. and Kahn,A. (1991) cis-acting DNA elements regulating expression of the liver pyruvate kinase gene in hepatocytes and hepatoma cells. Evidence for tissue-specific activators and extinguisher. J. Biol. Chem., 266, 7368–7375. [PubMed] [Google Scholar]
- 29.Chang C. and Gralla,J.D. (1993) Properties of initiator-associated transcription mediated by GAL4-VP16. Mol. Cell. Biol., 13, 7469–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eeckhoute J., Moerman,E., Bouckenooghe,T., Lukoviak,B., Pattou,F., Formstecher,P., Kerr-Conte,J., Vandewalle,B. and Laine,B. (2003) Hepatocyte Nuclear Factor 4alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology, 144, 1686–1694. [DOI] [PubMed] [Google Scholar]
- 31.Dhe-Paganon S., Duda,K., Iwamoto,M., Chi,Y.I. and Shoelson,S.E. (2002) Crystal structure of the HNF4alpha ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem., 277, 37973–37976. [DOI] [PubMed] [Google Scholar]
- 32.Bogan A.A., Dallas-Yang,Q., Ruse,M.D.,Jr, Maeda,Y., Jiang,G., Nepomuceno,L., Scanlan,T.S., Cohen,F.E. and Sladek,F.M. (2000) Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J. Mol. Biol., 302, 831–851. [DOI] [PubMed] [Google Scholar]
- 33.Eeckhoute J., Formstecher,P. and Laine,B. (2001) MODY1 diabetes-associated mutations R154X and E276Q in HNF4alpha gene impair recruitment of p300, a key transcriptional coactivator. Mol. Endocrinol., 15, 1200–1210. [DOI] [PubMed] [Google Scholar]
- 34.Hadzopoulou-Cladaras M., Kistanova,E., Evagelopoulou,C., Zeng,S., Cladaras,C. and Ladias,J.A.A. (1997) Functional domains of the nuclear receptor Hepatocyte Nuclear Factor 4. J. Biol. Chem., 272, 539–550. [DOI] [PubMed] [Google Scholar]
- 35.Iyemere V.P., Davies,N.H. and Brownlee,G.G. (1998) The activation function 2 domain of hepatic nuclear factor 4 is regulated by a short C-terminal proline-rich repressor domain. Nucleic Acids Res., 26, 2098–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soutoglou E., Katrakili,N. and Talianidis,I. (2000) Acetylation regulates transcription factor activity at multiple levels. Mol. Cell, 5, 745–751. [DOI] [PubMed] [Google Scholar]
- 37.Hu C. and Perlmutter,D.H. (1999) Regulation of alpha1-antitrypsin gene expression in human intestinal epithelial cell line Caco-2 by HNF-1alpha and HNF-4. Am. J. Physiol., 276, 1181–1194. [DOI] [PubMed] [Google Scholar]
- 38.Rausa F.M., Tan,Y. and Costa,R.H. (2003) Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol., 23, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puigserver P., Adelmant,G., Wu,Z., Fan,M., Xu,J., O’Malley,B. and Spiegelman,B.M. (1999) Activation of PPARgamma coactivator-1 through transcription factor docking. Science, 286, 1368–1371. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian C., Hasan,S., Rowe,M., Hottiger,M., Orre,R. and Robertson,E.S. (2002) Epstein–Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol., 76, 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Padilla M.E., Sladek,F.M. and Weiss,M.C. (2002) Developmentally regulated N-terminal variants of the nuclear receptor HNF4alpha mediate multiple interactions through coactivator and corepressor/HDAC complexes. J. Biol. Chem., 277, 44677–44687. [DOI] [PubMed] [Google Scholar]
- 42.Misawa K., Horiba,T., Arimura,N., Hirano,Y., Inoue,J., Emoto,N., Shimano,H., Shimizu,M. and Sato,R. (2003) Sterol regulatory element-binding protein-2 interacts with hepatocyte nuclear factor-4 to enhance sterol isomerase gene expression in hepatocytes. J. Biol. Chem., 278, 36176–36182. [DOI] [PubMed] [Google Scholar]
- 43.Sladek F.M., Zhong,W., Lai,E. and Darnell,J.E.,Jr (1990) Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev., 4, 2353–2365. [DOI] [PubMed] [Google Scholar]