Abstract
We have determined the stability of intramolecular DNA quadruplexes in which the four G3-tracts are connected by non-nucleosidic linkers containing propanediol, octanediol or hexaethylene glycol, replacing the TTA loops in the human telomeric repeat sequence. We find that these sequences all fold to form intramolecular complexes, which are stabilized by lithium < sodium < potassium. Quadruplex stability increases in the order propanediol < hexaethylene glycol < octanediol. The shallower shape of the melting profile with propanediol linkers and its lower dependency on potassium concentration suggests that this complex contains fewer stacks of G-quartets. The sequence with octanediol linkers displays a biphasic melting profile, suggesting that it can adopt more than one stable structure. All these complexes display melting temperatures above 310 K in the presence of 10 mM lithium, without added potassium, in contrast to the telomeric repeat sequence. These complexes also fold much faster than the telomeric repeat and there is little or no hysteresis between their melting and annealing profiles. In contrast, the human telomeric repeat sequence and a complex containing two hexaethylene glycol groups in each loop, are less stable and fold more slowly. The melting and annealing profiles for the latter sequence show significant differences, even when heated at 0.2°C min–1. CD spectra for the oligonucleotides containing non-nucleosidic linkers show positive maxima at 264 nm, with negative minima ∼244 nm, which are characteristic of parallel quadruplex structures. These results show that the structure and stability of intramolecular quadruplexes is profoundly influenced by the length and composition of the loops.
INTRODUCTION
Guanine-rich nucleic acid sequences can associate into four-stranded structures that contain stacks of G-quartets (1–4) (Fig. 1A). Intermolecular complexes are generated by the association of four separate DNA molecules (5–7) or by dimerization of molecules that contain two G-tracts (8,9), while intramolecular quadruplexes (as shown in Fig. 1B) are formed by folding a single strand of DNA (10–14). The formation of these structures requires the presence of monovalent cations (especially potassium) (15–17) while stable folding is inhibited by lithium (18). The cations fit within the central core of guanine carbonyls and can lie between or within the plane of each quartet.
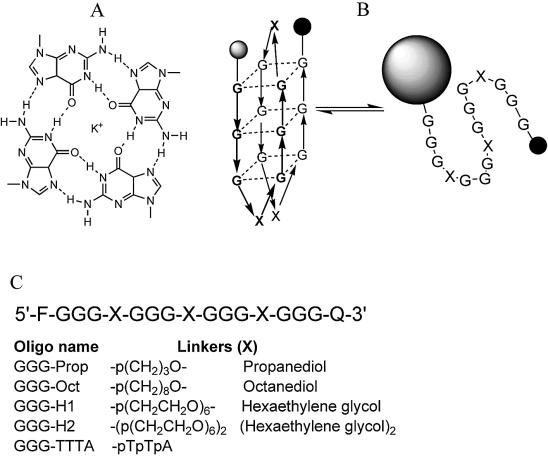
Figure 1.
(A) Chemical structures of the G-quartet. (B) Schematic representation of the folding and melting of the intramolecular quadruplexes. The fluorophore and quencher are represented by the shaded and filled circles respectively. Although several different folded structures are possible, for simplicity only one antiparallel structure is shown. (C) Sequences of the oligonucleotides used in this work.
G-rich sequences with the ability to form quadruplex structures are found in telomeric regions of DNA, which contain highly repeated sequences such as (GGGTTA)n in humans and most other higher eukaryotes (19–21), (GGG GTT)n in Tetrahymena and (GGGGTTTT)n in Oxytricha and Stylonchia (22). Similar sequences are also found in the promoters of several genes including c-myc (23,24), the immunoglobulin switch region (16), insulin regulatory sequences (25) and in fragile X syndrome (26). Quadruplex formation may also be involved in the dimerization of HIV RNA (27), and this structure is found in the core of a number of aptamers including the thrombin binding aptamer (11,28,29) and an inhibitor of HIV integrase (30–32). Quadruplex-forming oligonucleotides have also been suggested as potassium sensing agents (33), and as nanowires for molecular nanoelectronics (34).
High resolution X-ray (7,13,35) and NMR (9,11,12,14) structures have been determined for several DNA quadruplexes and reveal that these complexes are polymorphic. Intermolecular quadruplexes adopt a structure in which all the strands are parallel, while the arrangement in intramolecular complexes can be parallel, antiparallel or a combination of both (3). In the usual representation of the intramolecular quadruplex (Fig. 1B) the strands are antiparallel, with the TTA loops (crossed or uncrossed) at the top and bottom of the stack. However, a crystal structure of the potassium complex has revealed that this sequence can also adopt a fully parallel structure in which the loops run between the bottom of one stack and the top of the other (13). The structure may also be affected by the nature of the monovalent cation.
Intramolecular quadruplexes form stable structures, especially in the presence of potassium, though small changes in the sequence can have large effects on the structure and stability (36). The human (GGGTTA)4 and Oxytricha (GGGGTTTT)4 sequences fold to form stacks of three and four G-quartets respectively (12–14). However, the Tetrahymena sequence (GGGGTT)4 only forms three stacked quartets, which are linked by variable length loops containing TGGT, TTG and TT (37). In contrast the HIV integrase inhibitor (GGGT)4 adopts an extremely stable structure that contains only two stacked quartets, linked by GT in the loops (30–32). The bases in the loops can also stack against the terminal quartets. Intermolecular quadruplexes are known to form extremely slowly (38) and are therefore not suitable for investigation by melting experiments. In contrast, intramolecular complexes appear to fold much faster and generally do not show hysteresis between their melting and annealing curves (29). However, folding of the HIV integrase inhibitor in the presence of potassium includes a slow step, which is thought to involve a structural rearrangement (30). Other intramolecular quadruplexes have also been shown to display different melting and annealing profiles, especially in the presence of low concentrations of potassium (39).
There have been few studies that examine the effect of loops on quadruplex stability and most of these have changed the base composition and length (29,39). In the present study we have examined the properties of oligonucleotides in which the four G3-tracts are separated by non-nucleosidic linkers of different lengths. For this we have used the linkers shown in Figure 1C, containing propanediol, octanediol and hexaethylene glycol. The stability of these intramolecular complexes was determined using oligonucleotides that contain a fluorescent group (fluorescein) at one end and a quencher (methyl red) at the other end (39,40). When the oligonucleotide folds into an intramolecular complex these groups are in close proximity and the fluorescence is quenched. When the structure melts these groups become separated and there is a large increase in the fluorescence signal. This technique depends on collisional (proximal) quenching between the fluorophore and quencher and is similar to other studies which have measured the fluorescence energy transfer between fluorescein and tetramethylrhodamine (41,42). The structures adopted by these sequences have been probed by circular dichroism (CD), since parallel and antiparallel arrangements of the strands usually generate characteristic CD spectra (43–46).
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides containing fluorophores and quenchers were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer on either the 0.2 or 1 µmol scale and were purchased from Oswel DNA service (Southampton). These were purified by HPLC or on denaturing polyacrylamide gels. Methyl red and fluorescein were incorporated into the oligonucleotides using MeRed-dR or Fam-dR (40). The sequences of the oligonucleotides used in these studies are shown in Figure 1C.
Fluorescence melting
Fluorescence melting curves were determined in a Roche LightCycler, as previously described (40) in a total reaction volume of 20 µl. For each reaction the final oligonucleotide concentration was 0.25 µM, diluted in 10 mM lithium phosphate pH 7.4, supplemented with various concentrations of NaCl or KCl. The LightCycler has one excitation source (488 nm) and the changes in fluorescence were measured at 520 nm. In a typical experiment the oligonucleotides were first denatured by heating to 95°C for 5 min. They were then annealed by cooling to 30°C at a rate of 0.1°C s–1 (the slowest heating and cooling rate for the LightCycler). The samples were held at 30°C before heating to 95°C at 0.1°C s–1. Recordings were taken during both the annealing and melting steps. It was noted that for several oligonucleotides, as described in the Results, there was significant hysteresis between the annealing and melting curves, indicating the melting profile was not at thermodynamic equilibrium. Melting experiments were therefore performed at a slower rate of heating and cooling, by increasing the temperature in 1°C steps, leaving the samples to equilibrate for 5 min between each reading. No hysteresis was observed at this much slower rate of temperature change (0.2°C min–1).
Data analysis
Tm values were obtained from the minima in the first derivatives of the melting profiles using the LightCycler software or, together with ΔH, from van’t Hoff analysis of the melting profiles using FigP for Windows. This analysis assumes a simple two-state equilibrium between the folded and unfolded forms. In some instances the melting curves showed a linear change in fluorescence with temperature in regions outside the melting transition. This was accounted for by fitting a straight line to the first and last portions of the fluorescence curve. All reactions were performed at least twice and the calculated Tm values usually differed by < 0.5 K with a 5% variation in ΔH. Since ΔG = 0 at the Tm, ΔS was estimated as ΔH/Tm. Values for ΔG at 310 K were then estimated by assuming that ΔCp = 0. In some instances n, the number of specifically bound monovalent cations, was calculated from the slopes of plots of ΔG against log[M+] as previously described (30,47).
This analysis assumes that the oligonucleotides form folded, intramolecular, complexes and that the strands do not associate to generate intermolecular structures. This was tested for each oligonucleotide by examining the concentration dependence of the melting profiles, comparing the Tm values between 0.15 and 1.5 µM. As expected this revealed no significant changes in Tm, consistent with the formation of intramolecular complexes.
Circular dichroism
CD spectra of the oligonucleotides were measured on a Jasco J-720 spectropolarimeter. The fluorescently labelled oligonucleotides were used at a concentration of 5 µM and were dissolved in 10 mM lithium phosphate pH 7.4 containing 50 mM sodium or potassium chloride. These samples were boiled for 3 min before slowly cooling to room temperature and incubating overnight. Spectra were collected between 320–200 nm, using 16 scans at 100 nm min–1, 1 s response time, 1 nm bandwidth. A buffer baseline was subtracted from each sample spectrum and the final spectra were normalized to have zero ellipticity at 320 nm.
RESULTS
Fluorescence melting profiles
The sequences of the oligonucleotides used in this work are shown in Figure 1. In these sequences several different non-nucleosidic linkers replace the TTA linker in the human telomeric repeat. In the telomeric sequence, C3′ of the terminal G in each tract is separated from C4′ of the first G in the next tract by 23 bonds and four phosphate groups. Replacing this with propanediol, octanediol or hexaethylene glycol changes the distance to 11, 16 and 25 bonds respectively and utilizes only two phosphates. The use of two hexaethylene glycol linkers increases the length to 45 bonds with three phosphates. These oligonucleotides enable us to explore how loop length and charge affect quadruplex stability.
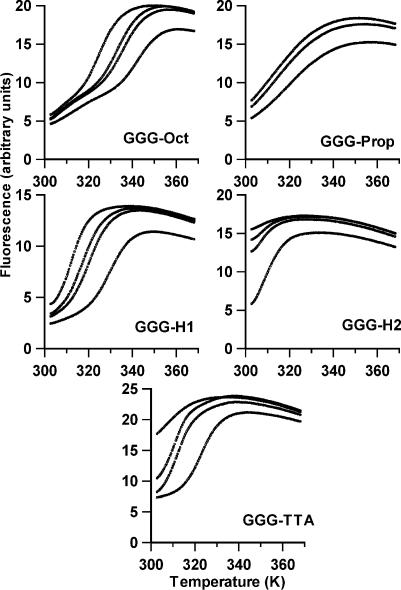
Typical fluorescence melting curves for the five oligonucleotides, in the presence and absence of sodium, are shown in Figure 2. In these experiments the buffer used was 10 mM lithium phosphate, as this cation is known to destabilize quadruplexes, and various concentrations of sodium chloride were added to this. The temperature was changed at a rate of 0.1°C s–1. All the oligonucleotides show temperature-dependent changes in fluorescence, which vary with sodium concentration. It can be seen that the least stable complexes are generated with GGG-H2 and the human telomeric repeat GGG-TTA. As expected these oligonucleotides have a high fluorescence in the presence of lithium alone, suggesting that they do not adopt a folded structure under these conditions. On addition of sodium chloride the melting curves typical of quadruplex formation are produced, from which the Tm values presented in Table 1 were estimated. The stabilization by sodium is not simply due to the increase in ionic strength, as addition of an equivalent concentration of lithium chloride has no effect. The curves in the presence of 40 mM NaCl show no hysteresis between the melting and annealing profiles, suggesting that the reactions are at thermodynamic equilibrium and that there are no slow steps in either the association or dissociation reactions. GGG-TTA shows some hysteresis at lower sodium concentrations, in which the Tm for melting is higher than for annealing. Under these conditions the quadruplex formed with GGG-H2 is too unstable to measure.
Figure 2.
Representative fluorescence melting curves for the oligonucleotides in the presence of 10 mM lithium phosphate pH 7.4 containing different concentrations of NaCl. The samples were heated at 0.1°C s–1. The y-axis shows the fluorescence (arbitrary units). In each case the sodium concentration increases from left to right in order 0, 5, 10, 40 mM (the curve for 5 mM is not shown for GGG-Prop).
Table 1. Tm values for melting and annealing of the five oligonucleotides, determined in 10 mM lithium phosphate pH 7.4, containing different concentrations of sodium chloride.
| |
10 mM Li |
+ 5 mM NaCl |
+ 10 mM NaCl |
+ 40 mM NaCl |
|
|---|---|---|---|---|---|
| Tm (K) | Tm (K) | Tm (K) | Tm (K) | ΔH (KJ mol–1)b | |
| GGG-Prop |
|
|
|
|
|
| Melt |
311.4a |
312.7a |
314.7a |
321.7 |
70 |
| Anneal |
309.8a |
311.9a |
313.6a |
319.9 |
|
| GGG-Oct |
|
|
|
|
|
| Melt |
324.8 |
333.4 |
335.9 |
342.6 |
– |
| Anneal |
324.2 |
333.4 |
334.7 |
342.7 |
|
| GGG-H1 |
|
|
|
|
|
| Melt |
312.1 |
316.9 |
320.8 |
329.2 |
149 |
| Anneal |
309.5 |
315.7 |
320.2 |
329.4 |
|
| GGG-H2 |
|
|
|
|
|
| Melt |
Too unstable |
|
|
307.6 |
129 |
| Anneal |
|
|
|
308.7 |
|
| GGG-HT |
|
|
|
|
|
| Melt |
Too unstable |
311.0 |
313.0 |
322.8 |
175 |
| Anneal | Too unstable | 308.9 | 322.0 |
aIndicates a very broad transition.
bΔH values (kJ mol–1) determined from van’t Hoff analysis of the melting profiles in the presence of 40 mM sodium chloride. The samples were heated and cooled at a rate of 0.1°C s–1.
It can be seen that GGG-H1 adopts a much more stable structure than GGG-H2, and a clear melting profile is obtained with this oligonucleotide, even in the absence of added sodium. Once again there is no hysteresis, except at the lowest concentrations of sodium. GGG-Oct and GGG-Prop also produce melting profiles in the absence of sodium, though these have different shapes. The profiles with GGG-Prop, which has the shortest linker, are much shallower, as evidenced by the low value for ΔH, and show a smaller dependence on sodium ion concentration. The melting profiles with GGG-Oct have an unusual shape. Although this oligonucleotide shows a clear melting transition in the absence of sodium, suggesting that it has adopted a folded structure, the initial part shows a linear increase in fluorescence with temperature. The effect is even more pronounced on addition of sodium. This clearly cannot represent a simple two-state equilibrium, suggesting that this sequence adopts multiple structural forms. We were therefore unable to obtain ΔH values for this transition from a simple van’t Hoff analysis.
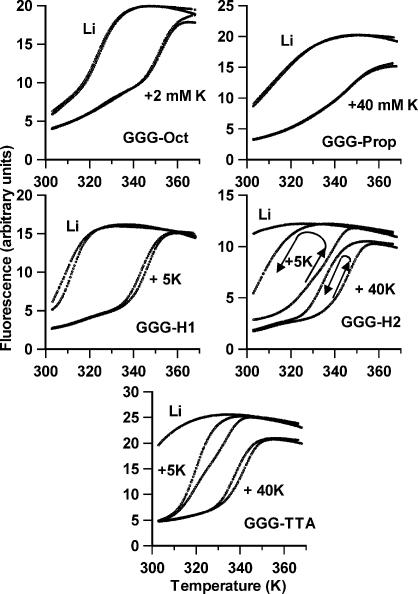
Similar melting experiments were performed with these oligonucleotides in the presence of potassium chloride. Representative melting and annealing curves for these are shown in Figure 3 and the Tm values are summarized in Table 2. It can be seen that, under these conditions, several of the oligonucleotides show hysteresis between the heating and cooling curves. The curves for GGG-Oct and GGG-Prop are fully reversible, while the melting Tms for GGG-H1 are ∼3– 4 K higher than the annealing curves. In contrast GGG-H2 shows considerable hysteresis, and the difference between the heating and annealing curves is greatest at low potassium concentration. In the presence of 5 mM KCl the apparent melting Tm is 339 K while the Tm for the annealing reaction is too low to measure (i.e. <310 K), implying a difference of at least 30 K. The annealing reactions can be measured at higher potassium concentrations, and the ΔTm between melting and annealing is 27 and 11 K at 10 and 40 mM KCl respectively. It is clear that, at this rate of heating (0.1°C s–1), the melting curves are not at thermodynamic equilibrium and the folding reaction must include at least one slow step. A similar, though less pronounced, hysteresis is evident with the human telomere sequence GGG-TTA, with a ΔTm of 4 K in the presence of 40 mM KCl. In this case the melting (but not annealing) curves are an unusual biphasic shape at intermediate potassium concentrations (this is most clearly evident in Fig. 3 for the melting curve in the presence of 5 mM KCl) and two melting transitions are reported in Table 2. The relative amplitudes of the two components vary with the potassium concentration; the lower Tm is more abundant at low concentrations, while the higher Tm dominates at high potassium. The lowest of these Tm values is similar to the annealing Tm.
Figure 3.
Representative fluorescence melting and annealing curves for the oligonucleotides in the presence of 10 mM lithium phosphate pH 7.4 containing different concentrations of KCl. The samples were heated and cooled at 0.1°C s–1. In each case, where there are differences, the melting curve is to the right of the annealing curve. The two curves are superimposable for GGG-Oct and GGG-Prop. The arrows show the direction of the temperature change for GGG-H2.
Table 2. Tm values for melting and annealing of the five oligonucleotides, determined in 10 mM lithium phosphate pH 7.4, containing different concentrations of potassium chloride.
| KCl (mM) | GGG-TTA | GGG-H2 | GGG-H1 | GGG-Prop | GGG-Oct | |
|---|---|---|---|---|---|---|
| 40 |
Melt |
342.2 |
346.7 |
Too high |
347.0 |
Too high |
| |
Anneal |
338.0 |
335.9 |
Too high |
346.0 |
Too high |
| 10 |
Melt |
324.0/335.1 |
342.0 |
Too high |
336.5 |
Too high |
| |
Anneal |
324.0 |
314.9 |
Too high |
338.7 |
Too high |
| 5 |
Melt |
319.4/333.9 |
339.4 |
347.4 |
335.1 |
358.8 |
| |
Anneal |
320.4 |
Too low |
343.1 |
334.9 |
358.7 |
| 2 |
Melt |
314.0/329.4 |
334.5 |
341.2 |
328.8 |
352.0 |
| |
Anneal |
314.3 |
Too low |
336.7 |
328.0 |
352.1 |
| 1 |
Melt |
309.3/323.4 |
329.4 |
335.6 |
324.2 |
347.3 |
| |
Anneal |
309.0 |
Too low |
332.3 |
323.5 |
345.9 |
| 10 mM Li |
Melt |
Too low |
Too low |
312.1 |
311.4a |
324.7 |
| Anneal | Too low | Too low | 309.5 | 309.8a | 324.2 |
aIndicates a very broad transition.
The samples were heated and cooled at a rate of 0.1°C s–1.
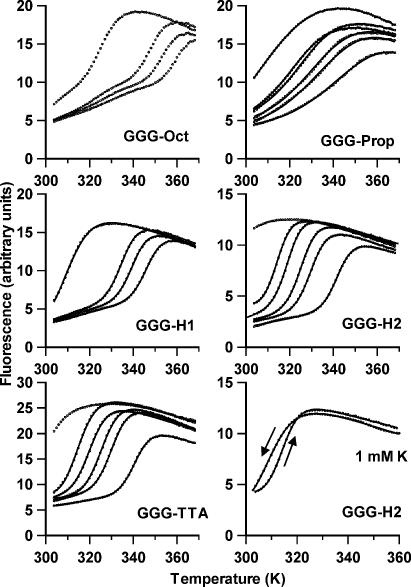
It is clear from these results that this rate of temperature change is too fast to permit a proper analysis of the melting profiles. The experiments were therefore repeated at a much slower rate of heating and cooling (0.2°C min–1). Since the slowest rate of heating for the LightCycler is 0.1°C s–1 we achieved this by increasing the temperature in 1°C steps, leaving the reaction to equilibrate for 5 min before measuring the fluorescence. This produces melting and annealing profiles with fewer data points and which are therefore only accurate to within 1°C. The results of these experiments are shown in Figure 4 and the Tm values are summarized in Table 3. Examination of this table reveals that there is almost no hysteresis between the melting and annealing profiles for all the oligonucleotides, except GGG-H2 in the presence of low potassium concentrations, which shows ΔTm values of 6, 5 and 2 K in the presence of 1, 2 and 5 mM KCl respectively. This residual hysteresis is illustrated by comparing the melting and annealing profiles in the bottom right panel of Figure 4. It can be seen that these two curves also have different shapes, and this is emphasized by the very different ΔH and ΔG values estimated by van’t Hoff analysis of the melting and annealing profiles. It is clear that the kinetics of folding and unfolding of this intramolecular quadruplex are extremely slow in the presence of low potassium concentrations. All the other oligonucleotides show no hysteresis and there are no significant differences between the parameters derived from the heating or annealing curves.
Figure 4.
Fluorescence melting curves for the oligonucleotides in the presence of 10 mM lithium phosphate pH 7.4 containing different concentrations of KCl. The samples were heated and cooled at 0.2°C min–1. In each case the potassium concentration increases from left to right; 0, 1, 2 and 5 mM KCl for GGG-Oct and GGG-H1 and 0, 1, 2, 5, 10 and 40 mM KCl for GGG-Prop, GGG-H2 and GGG-TTA. The bottom right hand panel compares the melting and annealing curves for GGG-H2 in the presence of 10 mM lithium phosphate, containing 1 mM KCl. In each case the solid lines show van’t Hoff fits to the data points as described in the Materials and Methods section.
Table 3. Tm (K), ΔH (kJ mol–1) and ΔG (kJ mol–1 at 300 K) values for melting and annealing of the five oligonucleotides, determined in 10 mM lithium phosphate pH 7.4, containing different concentrations of potassium chloride (mM).
| |
|
GGG-TTA |
|
GGG-H2 |
|
GGG-H1 |
|
GGG-Prop |
|
GGG-Oct |
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔH | Tm | ΔG | ΔH | Tm | ΔG | ΔH | Tm | ΔG | ΔH | Tm | ΔG | ΔH | Tm | ΔG | ||
| 40 |
Melt |
262 |
342/341 |
31.8 |
276 |
343/342 |
34.2 |
|
|
|
|
–/347 |
|
|
|
|
| |
Anneal |
273 |
341/340 |
32.5 |
295 |
343/342 |
36.6 |
|
|
|
|
–/344 |
|
|
|
|
| 10 |
Melt |
234 |
331/330 |
22.0 |
240 |
330/330 |
22.1 |
|
|
|
|
–/339 |
|
|
|
|
| |
Anneal |
248 |
331/330 |
23.1 |
236 |
330/328 |
21.2 |
|
|
|
|
–/337 |
|
|
|
|
| 5 |
Melt |
219 |
327/326 |
17.8 |
238 |
326/325 |
18.9 |
279 |
347/346 |
37.9 |
76.9 |
337/334 |
8.3 |
|
–/359 |
|
| |
Anneal |
227 |
327/326 |
18.5 |
210 |
324/323 |
15.6 |
287 |
346/345 |
38.1 |
81.1 |
337/336 |
8.9 |
|
–/361 |
|
| 2 |
Melt |
200 |
321/320 |
12.9 |
243 |
320/319 |
14.9 |
264 |
341/340 |
31.5 |
87.9 |
328/330 |
7.6 |
354 |
355/353 |
54.7 |
| |
Anneal |
193 |
320/319 |
12.1 |
168 |
315/314 |
7.7 |
285 |
340/339 |
33.4 |
88.6 |
329/328 |
7.9 |
|
–/355 |
|
| 1 |
Melt |
178 |
314/315 |
7.9 |
233 |
314/315 |
10.5 |
250 |
336/334 |
31.9 |
91.0 |
324/323 |
6.7 |
308 |
349/348 |
43.2 |
| |
Anneal |
167 |
314/314 |
7.2 |
146 |
308/309 |
3.7 |
271 |
335/335 |
28.5 |
92.4 |
325/326 |
7.1 |
306 |
348/348 |
42.4 |
| 10 mM Li |
Melt |
|
|
|
|
|
|
135 |
310/311 |
4.4 |
52.0 |
314/– |
2.4 |
167 |
328/325 |
14.1 |
| Anneal | 151 | 311/311 | 5.5 | 59.0 | 316/– | 3.0 | 197 | 328/326 | 17.0 | |||||||
The samples were heated and cooled at a rate of 0.2 K min–1. The first value for Tm was determined from van’t Hoff analysis of the melting profiles, while the second (in italics) was estimated from the maximum in the first derivative of the melting profile, using the LightCycler programme.
Comparison of the Tm values presented in Tables 1 and 3 shows that all these oligonucleotides adopt more stable structures in the presence of potassium than sodium, as expected for DNA quadruplexes. GGG-TTA and GGG-H2 have very similar stabilities (though clearly different kinetics), while GGG-H1 is more stable. GGG-Prop produces shallow melting profiles, as noted in the presence of sodium. The most stable complex appears to be GGG-Oct, though the melting profiles again show a steady increase in fluorescence at lower temperatures.
The ΔH values for these transitions show a strong dependence on the ionic strength. This is an unusual observation since simple polyelectrolyte theory suggests that there should be no change in enthalpy with salt concentration. However, this has been observed in other studies with quadruplexes and is consistent with the presence of specific ion binding sites (30). The slopes of plots of ΔG against log[M+] can be used to determine the stoichiometry of ion binding as previously described (30,47) these yielded values of n (the number of specifically bound K+) of 2.6 ± 0.1 for GGG-TTA, 2.5 ± 0.4 for GGG-H1 and 3.6 ± 0.3 and 2.5 ± 0.2 for GGG-H2, derived from the annealing and melting profiles respectively. These compare favourably to values ∼3, which were determined for T30695 and related intramolecular quadruplexes (30). Similar plots for the data with GGG-Prop give a value of 0.42 ± 0.04, suggesting that this oligonucleotide folds into a structure that contains fewer potassium ions, and presumably fewer G-quartets. A similar analysis is not possible for GGG-Oct as it produces melting profiles with an unusual shape.
CD spectroscopy
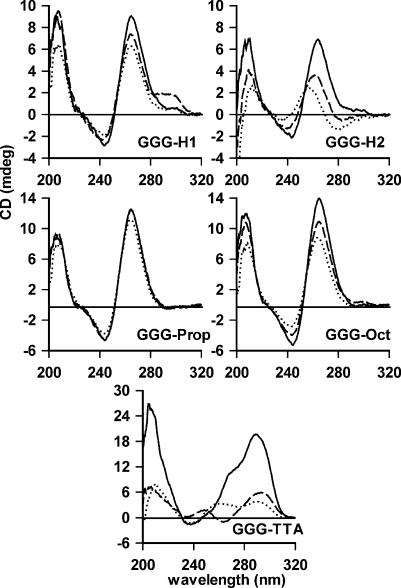
The melting profiles demonstrate that these oligonucleotides adopt stable complexes that are characteristic of quadruplex formation. These appear to be intramolecular and not intermolecular complexes, as the melting profiles are independent of oligonucleotide concentration (between 0.15 and 1.5 µM) and they run as a single band on polyacrylamide gels. However these do not provide information on the structure of the complexes, in particular whether the strands are arranged in a parallel or antiparallel orientation. We attempted to address this question by measuring their CD spectra. Parallel quadruplexes, generated by the association of four G-rich strands, are characterized by a positive ellipticity maximum at 264 nm and a negative minimum at 240 nm, while antiparallel (intramolecular) complexes usually possess a positive maximum at 295 nm and a negative minimum at 265 nm (43–46). CD spectra for these oligonucleotides in the presence of different cations are shown in Figure 5. It can be seen that all the oligonucleotides with non-nucleosidic linkers generate CD signatures with a positive maximum ∼264 nm and a minimum ∼244 nm, which is characteristic of parallel quadruplex formation. In contrast the human telomeric sequence (GGG-TTA) has maxima at ∼290 nm. The ellipticity of GGG-TTA at 290 nm is much greater in the presence of potassium than sodium and this is accompanied by a shoulder ∼270 nm and a small negative minimum ∼240 nm. The spectrum in sodium has a peak at 293 nm and a negative minimum at 263 nm, while the spectrum in lithium alone is much weaker. These results are similar to those previously reported (45). GGG-H1, GGG-Prop and GGG-Oct show similar spectra in the presence of all three cations, though the ellipticities increase in the order potassium > sodium > lithium. These differences are more pronounced for GGG-H2 and the spectrum in the presence of lithium alone is rather different. These results suggest that G-rich sequences containing non-nucleosidic linkers adopt a parallel structure. These results will be considered further in the Discussion.
Figure 5.
CD spectra of the fluorescently labelled oligonucleotides. The oligonucleotides (5 µM) were dissolved in 10 mM lithium phosphate pH 7.4 (dotted lines) containing 50 mM potassium chloride (solid lines) or 50 mM sodium chloride (dashed lines). The spectra for GGG-Prop in the presence of sodium and potassium are superimposed.
DISCUSSION
The results presented in this paper demonstrate that oligonucleotides with non-nucleosidic linkers can adopt secondary structures, which are consistent with quadruplex formation. In each case the Tm values are higher in potassium than sodium containing buffers and both these cations are much more effective than lithium. However, the melting temperature, the shape of the melting profile and the kinetics of quadruplex formation are affected by the nature of the linker.
The CD spectra of the oligonucleotides with non-nucleosidic linkers are typical of those with parallel structures, displaying positive maxima at ∼264 nm, in contrast to maxima at 295 nm seen with antiparallel structures, such as the human telomeric repeat (GGG-TTA). The only exception is seen in the spectrum of GGG-H1, which shows a positive shoulder at ∼295 nm in the presence of sodium, suggesting that this may contain a mixture of parallel and antiparallel forms. It therefore appears that the composition of the loops affects the overall structure, and that the presence of bases favours the formation of antiparallel structures. Even though these linkers are short, they are sufficient to bridge between one end and the other of a G3-stack; indeed this distance is shorter than that diagonally across a G-quartet. Although it seems likely that these sequences adopt a parallel structure, this conclusion should be treated with some caution. Firstly, crystallographic studies on the human telomeric repeat have indicated that this adopts a parallel structure in the presence of potassium (13), yet its CD spectrum still displays the typical antiparallel signature (positive peak ∼95 nm). Secondly there are several examples of intramolecular quadruplexes that are thought to adopt an antiparallel structure, yet which display CD spectra that are characteristic of parallel complexes (30,32,48,49), including the HIV integrase aptamer (G3T)4. The CD spectra do not only represent the parallel or antiparallel arrangement per se, but also reflect the glycosidic torsion angles, which have been shown to be syn or trans in different structures (3). In general, the guanines adopt an anti configuration in parallel structures, and an alternating anti/syn arrangement in with antiparallel strands, though several different combinations of glycosidic torsion angles have been described.
The stability of the sequences that contain a single non-nucleosidic linker in each loop decreases in order GGG-Oct > GGG-H1 > GGG-Prop, in both sodium and potassium containing buffers, and these are all more stable than the human telomeric repeat sequence GGG-TTA. All three of these sequences produce stable complexes in the presence of lithium alone and there is little or no hysteresis in their melting and annealing profiles under any of the conditions examined. The shallower melting profile of GGG-Prop and its smaller dependence on potassium concentration suggest that this folded sequence contains fewer G-quartets. The linker in this sequence is the same length as that in the HIV integrase aptamer (G3T)4, which contains a single T between each of the G3 tracts (30–32). Despite the similarity between these sequences GGG-Prop is less stable, suggesting that stacking of bases in the loops plays a significant part in quadruplex stability. The loops may also affect the coordination of metal ions, since GGG-Prop is less affected by the potassium ion concentration. The loop in GGG-H1 is only slightly longer than that in the human telomere sequence, yet it forms a more stable quadruplex and displays much faster kinetics of folding and unfolding (no hysteresis). This could be because of the reduced charge (it contains only two phosphates in each linker, instead of four), but also may reflect the presence or absence of base stacking.
Although there is no hysteresis with GGG-Prop, GGG-Oct and GGG-H1, the faster kinetics is not merely a property of oligonucleotides that contain non-nucleosidic linkers as the most pronounced hysteresis is observed with GGG-H2. Clearly the formation of the G-quartets alone cannot account for this slow step, which presumably results from a metal ion-dependent rearrangement of the loops. The interaction with the metal ion does not seem to be responsible for the slow kinetics since the potassium dependency of the Tms for GGG-H1, GGG-TTA and GGG-H2 is very similar, and each complex appears to contain three specially bound potassium ions. It is possible that these kinetic differences are related to the different structures that have been proposed in the presence of sodium and potassium (13).
The observation that GGG-Oct has the highest stability, under all conditions, suggests that this linker is the optimal length for quadruplex formation. However, this oligonucleotide produces melting profiles with an unusual biphasic shape. This effect has been noted previously for the intramolecular quadruplex formed by oligonucleotide (G3T2)4 (39), which has the same number of bonds, but one more phosphate in each linker. The biphasic profiles suggest that these oligonucleotides fold to form several stable structures, possibly with different numbers of stacked guanines. For GGG-Oct the first part of the melting curve is very shallow, suggesting that this represents dissociation of a structure that contains fewer stacked G-quartets. This structure might contain two quartets, with G-Oct (or Oct-G) in each loop, while the more stable structure could consist of three stacked G-quartets with a single octanediol residue in each loop.
Previous studies have shown that the bases between the G-tracts can significantly affect quadruplex stability (29,39), and demonstrates that these are not simply neutral linkers. For example, changing the loops from TTA to AAA abolishes quadruplex formation. The present study shows that their complete removal, replacing them with non-nucleosidic linkers, affects quadruplex stability. However, while GGG-Prop appears to be less stable than G3T, GGG-Oct has a similar stability to G3T2, and GGG-H1 is more stable than GGG-TTA, even though each pair has a similar length loop. It appears that for short loops the presence of bases enhances quadruplex stability, while longer loops are more stable with non-nucleosidic linkers. It should also be noted that the hexaethylene glycol linker might also coordinate a potassium ion (since this can adopt a similar structure to a crown ether). However, this does not explain the differences between GGG-H1 and GGG-H2.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the European Union.
REFERENCES
- 1.Williamson J.R. (1994) G-quartet structures on telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 2.Venczel E.A. and Sen,S. (1996) Synapsable DNA. J. Mol. Biol., 257, 219–224. [DOI] [PubMed] [Google Scholar]
- 3.Simonsson T. (2001) G-quadruplex DNA structures—variations on a theme. Biol. Chem., 382, 621–628. [DOI] [PubMed] [Google Scholar]
- 4.Keniry M.A. (2000) Quadruplex structures in nucleic acids. Biopolymers, 56, 123–146. [DOI] [PubMed] [Google Scholar]
- 5.Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 6.Laughlan G., Murchie,A.I.H., Norman,D.G., Moore,M.H., Moody,P.C., Lilley,D.M.J. and Luisi,B. (1994) The high-resolution crystal structure of a parallel stranded guanine tetraplex. Science, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- 7.Phillips K., Dauter,Z., Murchie,A.I., Lilley,D.M.J. and Luisi,B. (1997) The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Angstrom resolution. J. Mol. Biol., 273, 171–182. [DOI] [PubMed] [Google Scholar]
- 8.Sundquist W.I. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 9.Keniry M.A., Strahan,G.D., Owen,E.A. and Shafer,R.H. (1995) Solution structure of the Na+ form of the dimeric guanine tetraplex [d(G3T4G3)]2. Eur. J. Biochem., 233, 631–643. [DOI] [PubMed] [Google Scholar]
- 10.Henderson E., Hardin,C.C., Walk,S.K., Tinoco,I. and Blackburn,E.H. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine–guanine base pairs. Cell, 51, 899–908. [DOI] [PubMed] [Google Scholar]
- 11.Macaya R.F., Schultze,P., Smith,F.W., Roe,J.A. and Feigon,J. (1993) Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA, 90, 3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y. and Patel,D.J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 76–94. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson G.N., Lee,M.P.H. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y. and Patel,D.J. (1995) Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol., 251, 263–282. [DOI] [PubMed] [Google Scholar]
- 15.Williamson J.R., Raghuraman,M.K. and Cech,T.R. (1989) Monovalent cation induced structure of telomeric DNA—the G-quartet model. Cell, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- 16.Sen D. and Gilbert,W. (1990) A sodium–potassium switch in the formation of 4-stranded G4-DNA. Nature, 344, 410–414. [DOI] [PubMed] [Google Scholar]
- 17.Guschlabauer W., Chantot,J.F. and Thiele,D. (1990) Four stranded nucleic acid structures 25 years later: from guanosine gels to telomere DNA. J. Biomol. Struct. Dynam., 8, 491–511. [DOI] [PubMed] [Google Scholar]
- 18.Sen D. and Gilbert,W. (1992) Guanine quartet structures. Methods Enzymol., 211, 191–199. [DOI] [PubMed] [Google Scholar]
- 19.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes D. and Giraldo,R. (1995) Telomere structure and function. Curr. Opin. Struct. Biol., 5, 311–322. [DOI] [PubMed] [Google Scholar]
- 21.Wellinger R.J. and Sen,D. (1997) The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer, 33, 735–749. [DOI] [PubMed] [Google Scholar]
- 22.Schaffitzel C., Berger,I., Postberg,J. Hanes,J., Lipps,H.J. and Plückthun,A. (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonchia lemnae macronuclei. Proc. Natl Acad. Sci. USA, 98, 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonsson T., Pecinka,P. and Kubista,M. (1998) DNA tetraplex formation in the control regions of c-myc. Nucleic Acids Res., 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqui-Jain A., Grand,C.L., Bearss,D.J. and Hurley,L.H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catasti P., Chen,X., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Structure–function correlations of the insulin-linked polymorphic region. J. Mol. Biol., 264, 534–545. [DOI] [PubMed] [Google Scholar]
- 26.Darnell J.C., Jensen,K.B., Jin,P., Brown,V., Warren,S.T. and Darnell,R.B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 27.Sundquist W.I. and Heaphy,S. (1993) Evidence for interstrand quadruplex formation in the dimerization of immunodeficiency virus-1 genomic RNA. Proc. Natl Acad. Sci. USA, 90, 3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultze P., Macaya,R.F. and Feigon,J. (1993) Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. J. Mol. Biol., 235, 1532–1547. [Google Scholar]
- 29.Smirnov I. and Shafer,R.H. (2000) Effect of loop sequence and size on DNA aptamer stability. Biochemistry, 39, 1462–1468. [DOI] [PubMed] [Google Scholar]
- 30.Jing N., Rando,R.F., Pommier,Y. and Hogan,M.E. (1997) Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry, 36, 12498–12505. [DOI] [PubMed] [Google Scholar]
- 31.Jing N. and Hogan,M.E. (1998) Structure–activity of tetrad forming oligonucleotides as a potential anti-HIV integrase therapeutic drug. J. Biol. Chem., 273, 34992–34999. [DOI] [PubMed] [Google Scholar]
- 32.Jing N., Marchand,C., Liu,J., Mitra,R., Hogan,M.E. and Pommier,Y. (2000) Mechanism of inhibition of HIV-1 integrase by G-tetrad forming oligonucleotides in vitro. J. Biol. Chem., 275, 21460–21467. [DOI] [PubMed] [Google Scholar]
- 33.Ueyama H., Takagi,M. and Takenaka,S. (2002) A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. Fluorescence resonance energy transfer associated with guanine quartet–potassium ion complex formation. J. Am. Chem. Soc., 124, 14286–14287. [DOI] [PubMed] [Google Scholar]
- 34.Calzolari A., Di Felice,R., Molinari,E. and Garbesi,A. (2002) G-quartet biomolecular nanowires. Appl. Phys. Lett., 80, 3331–3333. [Google Scholar]
- 35.Haider S., Parkinson,G.N. and Neidle,S. (2002) Crystal structure of the potassium form of the Oxytricha nova G-quadruplex. J. Mol. Biol., 320, 189–200. [DOI] [PubMed] [Google Scholar]
- 36.Èrnugelj M., Šket,P. and Plvec,J. (2003) Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc., 125, 7866–7871. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y. and Patel,D.J. (1994) Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure, 2, 1141–1156. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt J.R., Davis,P.W. and Freier,S.M. (1996) Kinetics of G-quartet mediated tetramer formation. Biochemistry, 35, 8002–8008. [DOI] [PubMed] [Google Scholar]
- 39.Risitano A. and Fox,K.R. (2003) Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry, 42, 6507–6513. [DOI] [PubMed] [Google Scholar]
- 40.Darby R.A.J., Sollogoub,M., McKeen,C., Brown,L., Risitano,A., Brown,N., Barton,C., Brown,T. and Fox,K.R. (2002) High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res., 30, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonsson T. and Sjöback,R. (1999) DNA tetraplex formation studied with fluorescence resonance energy transfer. J. Biol. Chem., 274, 17379–17383. [DOI] [PubMed] [Google Scholar]
- 42.Mergny J.-L., Lacroix,L., Teulade-Fichou,M.-P., Hounsou,C., Guittat,L., Moarau,M., Arimondo,P.B., Vigneron,J.-P., Lehn,J.-M., Riou,J.F., Garestier,T. and Hélène,C. (2001) Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl Acad. Sci. USA, 98, 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu M., Guo,Q. and Kallenback,N.R. (1992) Structure and stability of sodium and potassium complexes of dG4T4 and dT4G4T. Biochemistry, 31, 2455–2459. [DOI] [PubMed] [Google Scholar]
- 44.Balagurumoorthy P., Brahmachari,S.K. Mohanty,D., Bansal,M. and Sasisekharan,V. (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balagurumoorthy P. and Brahmachari,S.K. (1994) Structure and stability of human telomeric sequence. J. Biol. Chem., 269, 21858–21869. [PubMed] [Google Scholar]
- 46.Lu M., Guo,Q. and Kallenbach,N.R. (1993) Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry, 32, 598–601. [DOI] [PubMed] [Google Scholar]
- 47.Cantor C.R. and Schimmel,P.R. (1980) Biophysical Chemistry. W.H.Freedman and Company, New York. [Google Scholar]
- 48.Ðapić V., Bates,P.J. Trent,J.O., Rodger,A., Thomas,S.D. and Miller,D.M. (2002) Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry, 41, 3676–3685. [DOI] [PubMed] [Google Scholar]
- 49.Ðapić V., Abdomerović,V., Marrington,R., Peberdy,J., Rodger,A., Trent,J.O. and Bates,P.J. (2003) Biophysical and biological properties of quadruplex oligdeoxyribonucleotides. Nucleic Acids Res., 31, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]