Abstract
Translational based medicine often utilizes animal models to study new and innovative therapeutics. In asthma and cystic fibrosis (CF), the animal models focus on airway inflammation and remodeling. The asthma model is based upon hypersensitivity induced airway disease whereas the CF model focuses on the inflammatory response to infection with Pseudomonas aeruginosa. Qualitative measures of inflammation and lung pathophysiology introduce significant variability and difficulty in interpreting interventional outcomes. This manuscript focuses on the development of a quantitative means to measure lung inflammation using the murine models of chronic asthma and CF. The highly sensitive and reproducible quantitative computational program interfaced with Image Pro Microscopy to monitor changes in lung inflammation and lung patho-physiology. The software-interfaces with image microscopy and automates the lung section review process. Results from this program recapitulated data obtained by manual point counting of inflammation, bronchoalveolar lavage differential and histology. The data shows a low coefficient of variation and high reproducibility between slides and sections. The utilization of this new microscopy program will enhance the quantitative means of establishing changes in lung structure and inflammation as a measure of therapeutic intervention with the ability of refining interpretation of in vivo models potentially short-circuiting translation into the clinical setting.
Keywords: Lung Disease, Microscopic Quantification, Inflammation, Therapeutic Response
Introduction
Lung disease has a major debilitating impact on society. Whether the decrease in pulmonary function is due to chronic reactive airway disease as in asthma; fibrosis as in interstitial pulmonary fibrosis or inflammation or infection as in CF, all processes result in a decreased quality of life and life expectancy (1,2). Research continues to focus on discovering new therapies and mechanisms of patho-physiology. The development of unique and innovate approaches to mechanisms and/or pharmacological interventions start at the bench and transitions into animal models then finally into patients if the data supports efficacy with minimal deleterious effects. Parameters of in vitro, in vivo and clinical trials involve assessment of lung structure and inflammation. Traditional measures are done by visual evaluation of bronchoalveolar lavage (BAL) cells and differential or by lung histology from tissue sections (3). Qualitative numerical values are given by technician scoring and visual point counting(4). These methods have significant inter- and intra- sample coefficients of variation (CV) as well as the potential of introducing significant subjective interpretation. The impact of these elevated CVs on determining model validity and therapeutic impact cannot be under-estimated. New methods for quantitative assessment of inflammation are essential for improving model efficacy and validation of new therapeutics (5-7).
Several companies have generated programs which allow for the development of computer generated MACROs for quantitative assessment of inflammation (5). One popular piece of software is Image Pro 7.0 Plus©. Image Pro software, combined with the correct hardware, allows one to view images from a microscope on a monitor as a live video-stream. Other capabilities of the software include, but are not limited to, the ability to take pictures of images, the ability to make on-screen measurements, and the ability to perform various data-analysis functions. We have utilized this software to write a MACRO program which autonomously operates the microscope to scan images and assign a value of inflammation based upon inflammatory cell recruitment into the lung. To validate this program we have used two standard murine models of lung disease to correlate traditional measures of lung inflammation with the MACRO generated algorithm. The first model is the pulmonary infection and inflammation model of cystic fibrosis (CF). In this model, cystic fibrosis transmembrane receptor (CFTR) deficient mice are manipulated to mimic chronic infection and inflammation with agar beads impregnated with Pseudomonas aeruginosa (8,9). The second model of in vivo lung inflammation is the ovalbumin murine model of chronic asthma. Mice are sensitized with ovalbumin and rested for a couple of weeks(10). This is followed by chronic airway challenging with ovalbumin to generate the airway remodeling and inflammation which mimics chronic asthma (4,11). Both the CF and chronic asthma models are validated in vivo models established to mimic different patho-physiologic components of these chronic airway diseases. Output parameters of both models include BAL total cell counts, differentials, and lung pathology with hematoxylin and eosin (H&E) stains (10,12).
In these studies we have combined the power of computer analysis with traditional measures of inflammation to validate a very powerful quantitative measure of airway lung patho-physiology associated with inflammatory cell recruitment. The results of this study suggest that digital microscopy is an efficient, effective, and powerful research technique with vast potential to enhance our interpretation of airway inflammation in the context of either murine models of CF lung infection and inflammation or chronic asthma airway inflammation.
Methods
Animal Models
All procedures involving mice were reviewed and approved by Case Western Reserve University, Institutional Animal Care and Use Committee. Case Western Reserve University Animal Assurance #: A3145-01, with IACUC # 2009-0128. Ovalbumin Murine Model of Chronic Asthma: Balb/c mice were purchased from Jackson Laboratories (Bar Harbor, Maine) and sensitized by intra-peritoneal injections (100 μL) of 10 μg of ovalbumin emulsified in 1.5 mg of Al (OH)3. On day 14, mice were exposed to 1% wt/vol ovalbumin (4,12) in PBS by aerosolization every other day for 4 weeks. Sham sensitization and challenges were carried out with sterile Al(OH)3 in PBS. Murine Model of CF Infection and Inflammation: To generate a transient chronic infection, CFTR KO mice (Cftrtm1UNC –TgN(FABPCFTR)#Jaw: gut corrected Cftr KO) were infected with 5 x103 to 9 x104 colony-forming units (CFU) Pseudomonas aeruginosa, strain M5715 (a clinical isolate) embedded on agarose beads and suspended in 20 μL PBS (9,13). Cftr KO and controls were anesthetized then inoculated with bacteria into the trachea with a plastic catheter angled toward the right mainstream bronchus(13). Cultures were verified by bacteriology before inoculation and BAL was cultured to determine level of infection at the study's termination.
Murine Cell Source: (a) Lung Inflammation
Mice were injected inter-peritoneally with ketamine (80 mg/kg) and xylazine (10 mg/kg) as previously described (14). The thoracic cavity was opened and lungs exposed followed by inserting a cannula through the trachea into the bronchi and infusing 1 x 1ml aliquot of warm PBS containing 0.2% lidocaine to do the BAL. The BAL fluid sample was recovered by aspirating the liquid with a syringe and evaluated for total cell count, cellular differential. (b) Lung Histology. Lungs were perfused with paraformaldehyde, sectioned while controlling for proximal airways for comparison purposes(14). Differential cell counts were obtained from cytospins stained with hematoxylin and eosin (H&E). Animals were assessed for inflammation by BAL and a separate set of animals were evaluated for lung histology.
Hardware, Software and Scripting
We used an Olympus BX50 microscope, which was connected to a Prior OptiScan II machine. The Prior OptiScan II machine served to motorize features of the microscope, such as the stage. The script for the software developed was written under the “MACRO” feature within the Image Pro 7.0 Plus© software. The Image Pro 7.0 Plus© software enabled communication between the computer, the microscope, and the Prior OptiScan II machine. The MACRO was designed specifically to test for inflammation within a lung section by scanning a lung section for the number of inflammatory nuclei. Inflammatory nuclei were defined to the computer based on size (in microns) and on color (purple-based due to the use of H&E stain). H&E defines neutrophils, macrophages, lymphocytes, eosinophils and other inflammatory nuclei. Although interstitial cellular nuclei are also stained they are considered background. Overall, the value of nuclei in the control (not treated tissue sections) was not significantly different than the saline treated inflammation control so this manuscript focused on the treatment control group for ease of interpretation and presentation. All samples were evaluated as a comparison to the saline control. The essential function of the software was to measure and indicate inflammation levels in varying samples by realizing that non-inflamed lung sections would have less inflammatory nuclei than inflamed lung sections. Baseline inflammatory nuclei were established on all study non-treated controls before initiation of studies.
Software Operation and Procedure
The software definition is based upon the following: The process is utilizes the Opti-Scann II machine of the Image –Pro MC software. Using the MACRO function, the Stage-Pro will initiate. This is where the physical limits of the automated state are established. At this point the slide image is place on the microscope stage and set for 60X magnification. The program operates on two levels: collecting data and then analyzing data. The data collection process is initiated by setting scan parameters along a given slide. The program then divides the area encompassed by these parameters in to 196 subsections and autonomously takes pictures of these 196 subsections. The program then analyzes each of these 196 images individually. The program scans each image for “inflammatory nuclei.” The parameters for what defines inflammatory nuclei are pixel-shade and object size. Currently the program scans for objects that are the approximate size range of white blood cells and are a shade of purple (due to H&E). Using the pixel definition of choice, the color most associated with the recruitment of inflammatory cells: neutrophils, lymphocytes, macrophages, eosinophils are counted based upon recognition. The microscope will proceed to take 144 pictures covering 195 areas of the several (3-sections) on the given slide. After the pictures have been minimized, the microscope will scan through each image to assess the amount of inflammation as a comparison to the control. The nuclei counted for each section exported to Excel® for evaluation. The MACRO operated based on the following scripted (C++) procedure. (a) Stage Alignment. The first process is aligning and formatting the mechanical microscope stage. This step serves to ensure functionality of the mechanical stage, as well as to return the stage to the origin position. (b) Scan Area Parameters. The next step is to set the desired scan parameters. The user defines the parameters by defining the top-left and bottom-right corners of the desired scan area. This step serves to establish what section of the slide to be examined. (c) Automated Image Capture. Now that the scan parameters have been defined the MACRO divides the desired area into 196 sections and has the microscope proceed to take a picture of each of the 196 sections. The ensuing microscope movement and image-capture process are controlled entirely by the computer. (d) Image Analysis. The MACRO scans each of the 196 images for objects that match the definition of inflammatory nuclei and export the data into the default data-analysis program, Microsoft Office Excel 2007©.
Data Analysis
The “average-if” function calculated the mean number of inflammatory nuclei per image, thus giving an overall inflammatory nuclei count for the entire lung section. The “average-if” function excluded any inflammatory nuclei count which was less than five inflammatory nuclei in calculating the mean. This served to eliminate any blank images (images with no lung tissue on them) as well as any images with debris on them from the final mean. This number was then used to compare levels of inflammations. Data analysis was validated using Prism 3.0 (San Jose, CA); with 95% confidence intervals with p<0.05.
Results
Murine Model of Chronic Asthma and The ImagePro MACRO
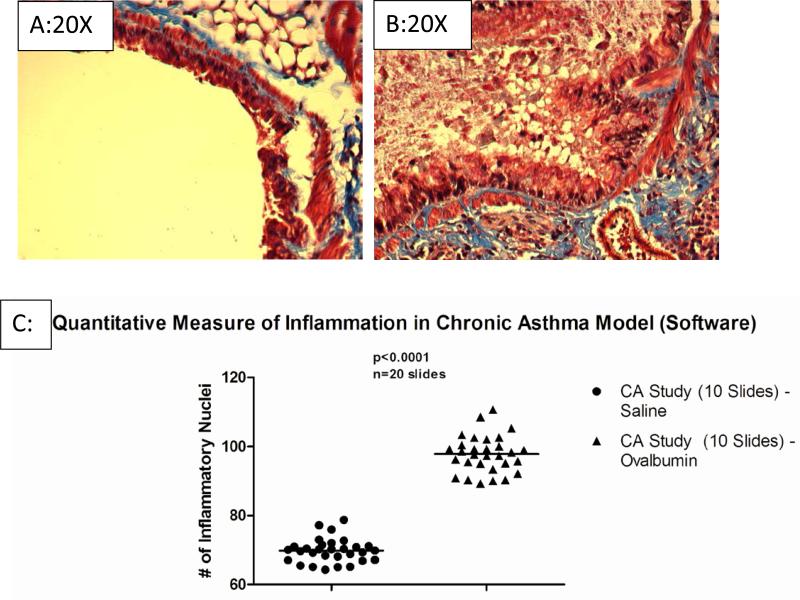
To validate our software we used two different murine models of airway disease. The ovalbumin murine model of chronic asthma was used to measure changes in airway cell recruitment (10). In this model, Balb/c mice were sensitized with ovalbumin, rested for 2 weeks followed by every-other day challenges for 4 weeks. The control group consists of ovalbumin sensitized animals challenged with saline (Figure 1A). At 4 weeks the lung histology showed extensive inflammatory infiltrate (Figure 1B), compared to the saline challenge control. This was consistent with the total cell recruitment in the BAL fluid (Figure 1C). The software examined 30 lung sections that were treated with saline and 30 lung sections that were treated with ovalbumin. The 30 lung sections that were treated with saline were not expected to show signs of significant inflammation due to the fact that saline has been proven to not precipitate a large inflammatory reaction. The 30 lung sections that were treated with ovalbumin were expected to show signs of significant inflammation due to the fact that ovalbumin has been proven to precipitate a large inflammatory reaction. The mean number of inflammatory nuclei for the ovalbumin-treated lung sections was 98, while the mean number of inflammatory nuclei for the saline-treated lung sections was only 69. This indicates the viability of the software in direct applications, specifically the chronic asthma lung model. The histological evaluation of the model was consistent with the observations obtained by using the quantification of the inflammatory nuclei 97.8±5.3 which was 30±7% more than the inflammatory nuclei obtained with the controls sections (Figure 1C, 69.8±3.4 inflammatory nuclei, p<0.002).
Figure 1. Visual Representation of Automated-Imaging Software.
A representative control (Figure 1A) and chronic asthma (Figure 1B), show a significant increase in inflammation induced by ovalbumin challenge. The histology shows enhanced inflammatory cell recruitment and extracellular matrix deposition (arrows of designation). The software then analyzed each image and determined the number of inflammatory nuclei. The scatter plot shows the relative number of inflammatory nuclei for sections from saline challenge (Figure 1C, n=10 slides) and the ovalbumin challenge samples positive control (n=10 slides). The ImagePro program defined the inflammatory difference to be significant at p<0.002.
Murine Model of Lung Infection and Inflammation in Cystic Fibrosis
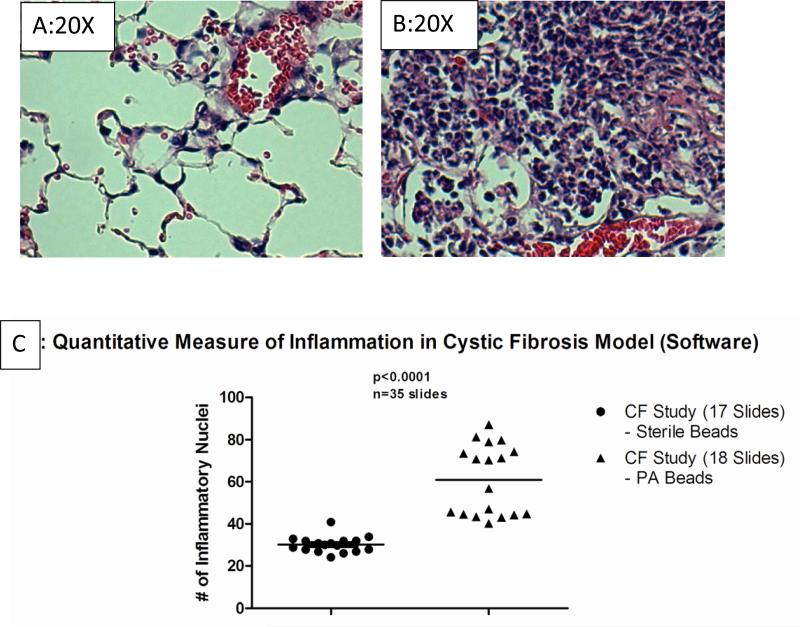
For the second model of inflammation we used murine the Pseudomonas aeruginosa lung infection in Cftr knockout mice (3,13). In order to sustain the infection, Pseudomonas aeruginosa was impregnated onto agar beads and intra-tracheally administered to mice. Once infected the animals were followed for up to 10 days post-infection before euthanasia for lung histology or BAL. Cftr deficient mice without infection have healthy lung histology (Figure 2A). However, post-infection the lungs become infiltrated with neutrophils and lymphocytes (Figure 2B). The software examined 17 sets of lungs from mice that were treated with sterile beads and 18 sets of lungs from mice that were treated with Pseudomonas aeruginosa agarose beads. The 17 lungs that were treated with sterile beads were not expected to show any inflammation, while the lungs treated with Pseudomonas aeruginosa agarose beads were expected to have excessive numbers of white blood cells. The software validated this hypothesis. Over several sections, the Cftr KO lungs consistently had greater numbers of inflammatory nuclei (Figure 2C, 60±20, n=35, p<0.001) over the non-treated control (32±4, n=35). Interestingly the controls in the Cftr KO infection study (C57BL/6) background consistently had less inflammatory infiltrate than the saline challenged Balb/c mice in the asthma studies emphasizing the requirement for species specific controls in evaluating inflammation. The models were then used to test the precision, accuracy and reproducibility of the microscope quantified inflammation.
Figure 2. A Representative Section from the Cftr KO Murine Model.
Cftr knockout mice without infection are shown for comparison (Figure 2A) and are similar to wild type mice (data not shown). These studies show enhanced inflammatory cell recruitment (Figure 2B) in response to agar beads impregnated with Pseudmonas aeruginosa. The software then analyzed each image and determined the number of inflammatory nuclei. The mean value is designated to each slide after viewing 196 images. The scatter plot shows the relative number of inflammatory nuclei for sections from Cftr KO mice given sterile agarose beads (Figure 2C, n=17 slides) versus Pseudomonas aeruginosa impregnated agarose beads (Figure 2C, n=18 slides). The ImagePro program defined the inflammatory difference to be significant at p<0.001.
Histological Precision Testing in the Murine Models of Chronic Asthma and Cystic Fibrosis
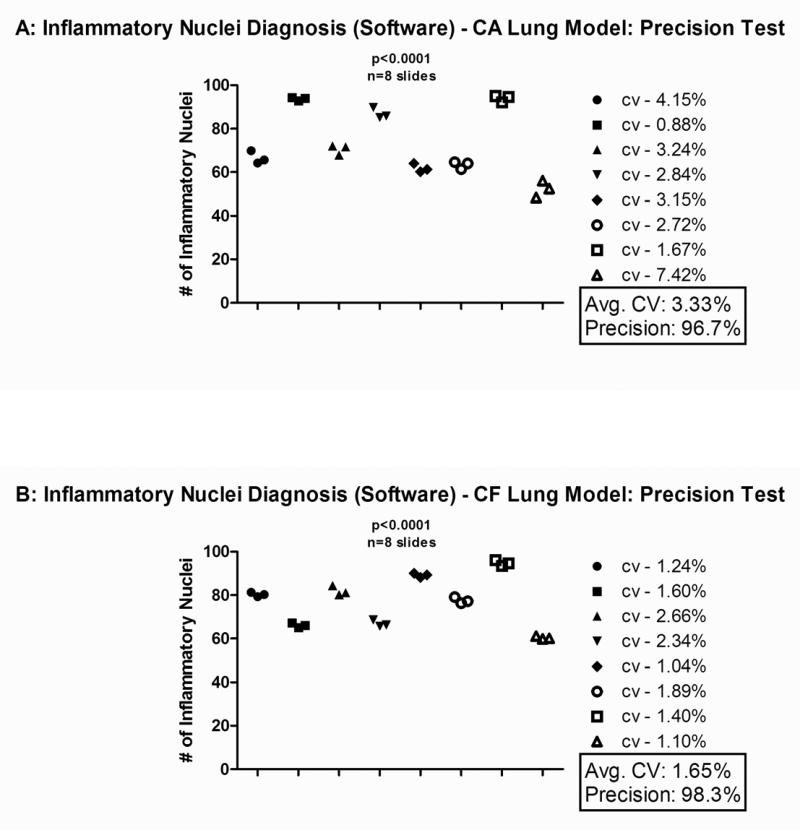
The overall level of precision for the software was determined by examining eight sets of three different lung sections from different depths within a given lung using the process demonstrated in Figure 3. This was done for both the chronic asthma and Cftr KO lung infection/inflammation models. Each lung section was examined by the software and diagnosed with a number of inflammatory nuclei. The number of inflammatory nuclei for each of the three lung sections from a given lung was then analyzed by calculating the coefficient of variation for those values. The level of precision was then calculated from the coefficient of variation. The mean coefficient of variation for the chronic asthma lung model (Figure 4A) and the Cftr KO lung model (Figure 4B) was 3.33% and 1.65%, respectively. The level of precision for the chronic asthma lung model (Figure 4A) and the Cftr KO lung model (Figure 4B) was 96.7% and 98.3%, respectively.
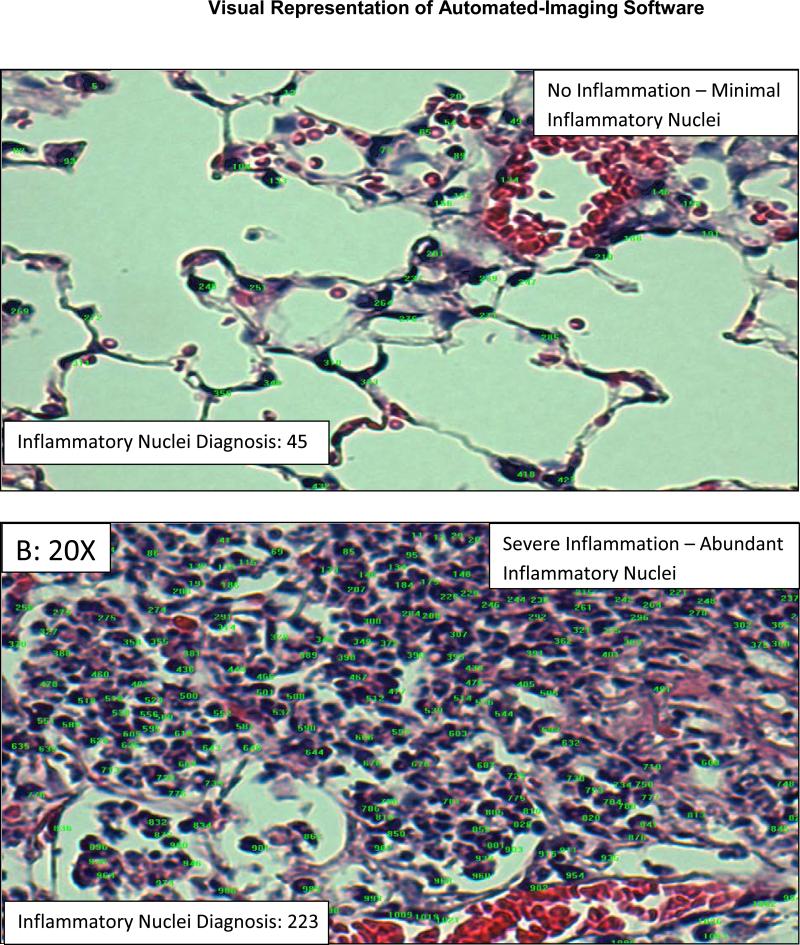
Figure 3. Inflammatory Nuclei Diagnosis (Software) – chronic asthma/Cftr KO Lung Model: Precision Test.
The software then analyzed each image and determined the number of inflammatory nuclei; this number is shown in the bottom left-hand corner of each image. The green numbers represent nuclei that are of the right color, while the purple highlights represent nuclei that are of the right size. Inflammatory nuclei are only counted if they are of the right color and size.
Figure 4. Data Analysis of the Chronic Asthma and Cftr KO Murine Model Inflammation.
The following data was obtained from eight different slides, each containing three lung sections from the same mouse at varying depths. The software assigned each lung section with a number of inflammatory nuclei. These numbers were plotted based on the slide/mouse from which they came. The coefficient of variation (the standard deviation divided by the mean) was calculated for each mouse based on the number of inflammatory nuclei that each of the three lung sections possessed. This value is represented next to each symbol denoting a set of lung sections as the “CV” value. From this the level of precision (in %) was determined by multiplying the CV value by 100 and then subtracting that number from 100. Next to the graph is the mean CV and the mean precision for the 8 slides shown. This test was applied to the murine ovalbumin chronic asthma lung model in Figure 4A and to the Cftr KO Pseudomonas aeruginosa infection model in Figure 4B. The mean CV was 3.33% and the mean precision was 96.7% for the chronic asthma lung model. The mean CV was 1.65% and the mean precision was 98.3% for the Cftr KO Pseudomonas aeruginosa infection model.
Histological Accuracy Testing in the Murine Models of Chronic Asthma and Cystic Fibrosis
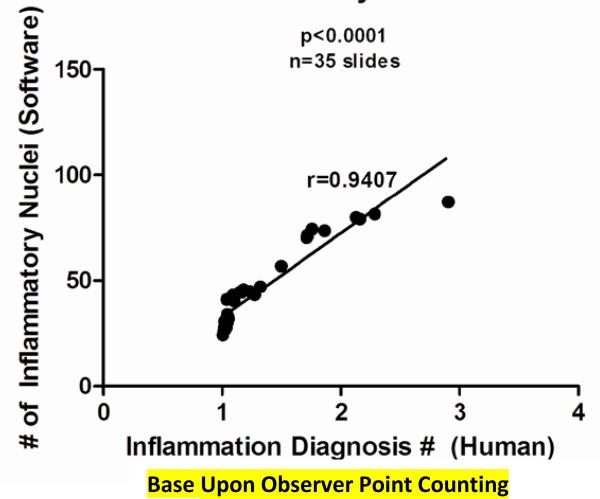
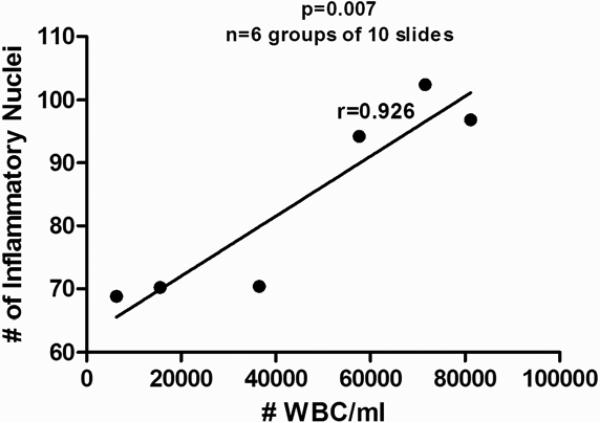
Accuracy was measured by comparing manual point counting and the BAL white blood cell infiltrate relative to the image obtained using our MACRO. The accuracy of the software, relative to the point counting and BAL infiltrate was compared using correlation measurements. This was done by comparing 35 sets of independent slides using manual point counting and automated software. The coefficient of correlation (the r-value) for the data was 0.94, p<0.0001. The proximity of this value to 1.0 indicates that there is a very strong positive correlation, thus establishing the accuracy of the software relative to manual point counting. This is demonstrated in Figure 5. The accuracy of the software compared to traditional measures of inflammation (number of white blood cells (#WBC)/ml of BALF) was also evaluated. This was done by comparing the mean #WBC/ml for 6 groups of mice to the mean number of inflammatory nuclei for those same 6 groups of mice. The coefficient of correlation (the r-value) for the data was 0.926, p=0.007, Figure 6.
Figure 5. Correlation Scatter Diagram of Manual Point Counting VS Quantified Inflammation Software: Accuracy Test.
The following data was based on two values. A value based upon manual point counting was given to each of the 35 sets of lungs using the Cftr KO Pseudmonas aeruginosa infection model. The point counting value was based on looking at 500 frames from a single lung and diagnosing each frame as either a 1, 2, 3, or 4. 1 represented no inflammation, 2 represented mild inflammation, 3 represented moderate inflammation, and 4 represented severe inflammation. These values were then averaged to determine an inflammation diagnosis value for each of the 35 sets of lungs. This value served as the x-coordinate for each lung. Second, a software-based diagnosis of inflammation was given to each of the 35 sets by determining the average number of inflammatory nuclei per lung. This value served as the y-coordinate for each lung. These points were then plotted and a trend-line (best-fit-line) was drawn. The coefficient of correlation (r-value) was then calculated. The coefficient of correlation was 0.94, indicating a very strong positive correlation between manual point counting and the software with p<0.0001.
Figure 6. Correlation Scatter Diagram of BAL White Blood Cells VS Quantified Inflammation Software: Accuracy Test.
The average number of white blood cells per milliliter of BAL fluid (#WBC/ml) was determined for six groups of mice, each group containing 10 mice using the chronic asthma murine model. This value served as the x-coordinate for each group. The average number of inflammatory nuclei was then determined by the software for each group. This value served as the y-coordinate for each group. These points were then plotted and a trend-line (best-fit-line) was drawn. The coefficient of correlation (r-value) was then calculated. The coefficient of correlation was 0.926, indicating a very strong positive correlation between the number of white blood cells in a given sample and the software with p<0.0001.
Discussion
In vivo models of lung inflammation and infection are essential in the development of new directions for molecular approaches to understanding pathophysiology. Inherent in the establishment of in vivo models is to understand and quantify the inflammation within histological sections. In these models, often an objective reviewer is used to define inflammation by “point-counting” or generalized nuclei assessment. Although, manual evaluation is also key to visual interpretation, applying addition non-subjective models is also essential. This becomes especially true when new therapeutics are introduced and measurement are key in the definition of efficacy and impact on the disease pathology. We have developed a sensitive, quantitative and reproducible microscope directed computer MACRO, which systematically counts inflammatory nuclei over several sections of lung sections stained with H&E. This new computer macro with the imaging system will enhance in vivo model accuracy and response to therapeutics.
Digital microscopy is a unique technology in that it is actually a combination of two very different technologies (5). Digital microscopy takes the power of the computer and combines it with the power of the microscope to form a dual-technology that functions on not one, but two unique paradigms. The microscope allows one to examine specimens in high-detail at very small scales, whereas the computer allows one to automate processes and quantify/analyze data. The many faults that are associated with the microscope, such as experimenter bias, are solved when combined with the computer. Our study has shown that digital microscopy has the ability to produce highly precise and accurate quantifiable data. The software that we have created has the unique ability to assess levels of inflammation. In addition, the software that we have created is precise to 96.7% and has demonstrated high levels of correlation to human and experimental data.
In this study, we have developed the unique ability to assess inflammation, but that is only one of the many possible abilities of the MACRO. The program can be easily tweaked so that it would be able quantify the presence of any cell desired. This can be simply done by redefining the attributes of the cells that the software scans for. Thus, the MACRO can serve not only as a tool to assess inflammation, but as a tool to assess the presence of specific types of cells based upon their specific staining pattern. Digital microscopy also has a wide scope of possible applications. The program that we developed can not only function as a tool to assess inflammation, but as a tool to validate animal models. We can test for this by measuring if one animal has the same average number of inflammatory nuclei as another whether or not there is involvement of anti-inflammatory drug testing. By applying the software to a control group, an infected group, and an infected group treated with an anti-inflammatory drug, one can assess levels of inflammation in each group. The number of inflammatory nuclei present in each group can be used to determine and quantify the success of an anti-inflammatory drug. The program can also be applied to other species simply by editing the program to scan for cells of different sizes and colors. Further, increased precision and accuracy can be instituted although these parameters cost time and hardware storage.
Acknowledgements and Funding
This work was funded by The David and Virginia Baldwin Fund and The Cystic Fibrosis Foundation and P30 DK 027651. We would also like to thank Hawken School, and the STEMM program.
Reference List
- 1.Chmiel JF, Konstan MW. Inflammation and anti-inflammatory therapies for cystic fibrosis. Clin. Chest Med. 2007;28:331–346. doi: 10.1016/j.ccm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu. Rev. Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- 3.van Heeckeren AM, Schluchter MD. Murine models of chronic Pseudomonas aeruginosa lung infection. Lab Anim. 2002;36:291–312. doi: 10.1258/002367702320162405. [DOI] [PubMed] [Google Scholar]
- 4.Braun A, Tschernig T. Animal models of asthma: innovative methods of lung research and new pharmacological targets. Exp. Toxicol. Pathol. 57 Suppl. 2006;2:3–4. doi: 10.1016/j.etp.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 5.McCullough B, Ying X, Monticello T, Bonnefoi M. Digital microscopy imaging and new approaches in toxicologic pathology. Toxicol. Pathol. 32 Suppl. 2004;2:49–58. doi: 10.1080/01926230490451734. [DOI] [PubMed] [Google Scholar]
- 6.Brain JD. Free cells in the lungs. Some aspects of their role, quantitation, and regulation. Arch. Intern. Med. 1970;126:477–487. doi: 10.1001/archinte.126.3.477. [DOI] [PubMed] [Google Scholar]
- 7.Baak JP. Quantitative pathology today--a technical view. Pathol. Res. Pract. 1987;182:396–400. doi: 10.1016/S0344-0338(87)80076-6. [DOI] [PubMed] [Google Scholar]
- 8.Chmiel JF, Konstan MW, Knesebeck JE, Hilliard JB, Bonfield TL, Dawson DV, Berger M. IL-10 attenuates excessive inflammation in chronic pseudomonas infection in mice. Am. J. Respir. Crit. Care Med. 1999;160:2040–2047. doi: 10.1164/ajrccm.160.6.9901043. [DOI] [PubMed] [Google Scholar]
- 9.van Heeckeren AM, Tscheikuna J, Walenga RW, Konstan MW, Davis PB, Erokwu B, Haxhiu MA, Ferkol TW. Effect of Pseudomonas infection on weight loss, lung mechanics, and cytokines in mice. Am. J. Respir. Crit Care Med. 2000;161:271–279. doi: 10.1164/ajrccm.161.1.9903019. [DOI] [PubMed] [Google Scholar]
- 10.Bonfield TL, Koloze M, Lennon DP, Zuchowski B, Yang SE, Caplan AI. Human mesenchymal stem cells suppress chronic airway inflammation in the murine ovalbumin asthma model. Am. J. Physiol Lung Cell Mol. Physiol. 2010;299:L760–L770. doi: 10.1152/ajplung.00182.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar RK, Foster PS. Modeling allergic asthma in mice: pitfalls and opportunities. Am. J. Respir. Cell Mol. Biol. 2002;27:267–272. doi: 10.1165/rcmb.F248. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita N, Tashimo H, Ishida H, Kaneko F, Nakano J, Kato H, Hirai K, Horiuchi T, Ohta K. Attenuation of airway hyperresponsiveness in a murine asthma model by neutralization of granulocyte-macrophage colony-stimulating factor (GM-CSF). Cell Immunol. 2002;219:92–97. doi: 10.1016/s0008-8749(02)00565-8. [DOI] [PubMed] [Google Scholar]
- 13.van Heeckeren AM, Schluchter MD, Xue W, Davis PB. Response to acute lung infection with mucoid Pseudomonas aeruginosa in cystic fibrosis mice. Am. J. Respir. Crit Care Med. 2006;173:288–296. doi: 10.1164/rccm.200506-917OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonfield TL, Thomassen MJ, Farver CF, Abraham S, Koloze MT, Zhang X, Mosser DM, Culver DA. Peroxisome proliferator-activated receptor-gamma regulates the expression of alveolar macrophage macrophage colony-stimulating factor. J. Immunol. 2008;181:235–242. doi: 10.4049/jimmunol.181.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]