Abstract
Early mitotic inhibitor 1 (Emi1) inhibits the activity of the anaphase promoting complex/cyclosome (APC/C), which is a multisubunit ubiquitin ligase that targets mitotic regulators for degradation in exit from mitosis. Levels of Emi1 oscillate in the cell cycle: it accumulates in the S phase and is rapidly degraded in prometaphase. The degradation of Emi1 in early mitosis is necessary for the activation of APC/C in late mitosis. Previous studies have shown that Emi1 is targeted for degradation in mitosis by a Skp1–Cullin1 F-box protein (SCF) ubiquitin ligase complex that contains the F-box protein β-TrCP. As with other substrates of SCFβ-TrCP, the phosphorylation of Emi1 on a DSGxxS sequence is required for this process. However, the protein kinase(s) involved has not been identified. We find that Polo-like kinase 1 (Plk1), a protein kinase that accumulates in mitosis, markedly stimulates the ligation of Emi1 to ubiquitin by purified SCFβ-TrCP. Cdk1-cyclin B, another major mitotic protein kinase, has no influence on this process by itself but stimulates the action of Plk1 at low, physiological concentrations. Plk1 phosphorylates serine residues in the DSGxxS sequence of Emi1, as suggested by the reduced phosphorylation of a derivative in which the two serines were mutated to nonphosphorylatable amino acids. Transfection with an small interfering RNA duplex directed against Plk1 caused the accumulation of Emi1 in mitotically arrested HeLa cells. It is suggested that phosphorylation of Emi1 by Plk1 is involved in its degradation in mitosis.
Ubiquitin-mediated protein degradation and protein phosphorylation are two basic, often intertwined, regulatory mechanisms in the control of the cell cycle and in other types of cellular regulation. A good example for the interrelationship between these two processes is the intricate regulation of the anaphase promoting complex/cyclosome (APC/C). APC/Cisa large, multisubunit ubiquitin ligase complex (1, 2) that targets mitotic regulators for degradation in exit from mitosis (reviewed in refs. 3 and 4). The activity of APC/C is tightly controlled: It is active from late mitosis until the end of G1 of the next cell cycle. APC/C is activated in mitosis in a process initiated by the phosphorylation of several of its subunits (5, 6), which is necessary for its interaction with the activatory protein Cdc20 (7, 8). Mitotic phosphorylation of APC/C is carried out by the protein kinases Cdk1-cyclin B and Polo-like kinase (Plk1) (8–10). Both cyclin B and Plk1 are targeted for degradation by APC/C, thus terminating the action of these mitotic kinases. Inhibitory phosphorylations are also involved in the regulation of APC/C activity. Another ancillary protein, Cdh1, which keeps the APC/C active in G1, is converted by phosphorylation to an inactive form in the G1 to S-phase transition (3, 4). From early S phase until prometaphase, the APC/C is also kept inactive by the F-box protein early mitotic inhibitor 1 (Emi1) (11, 12).
Emi1 was first discovered in Drosophila as a gene product, regulator of cyclin A, which causes the accumulation of cyclin A (13). More recently, Jackson and coworkers (11, 12, 14) have shown that in Xenopus eggs and in mammalian cells, Emi1 blocks the degradation of both cyclin A and cyclin B by inhibiting the activity of both APC/CCdc20 and APC/CCdh1. Levels of Emi1 oscillate in the cell cycle: It accumulates in the S phase and is rapidly destroyed in prometaphase (11, 12). Thus, the degradation of Emi1 in early mitosis is necessary for the activation of APC/C in late mitosis.
Two recent reports have shown that Emi1 is targeted for degradation in mitosis by a Skp1–Cullin F-box protein (SCF) ubiquitin ligase complex, which contains the F-box protein β-TrCP (15, 16). The action of SCF ubiquitin ligases is also tightly coupled to protein phosphorylation, but in this case the phosphorylation of the protein substrate is required (17). The action of SCFβ-TrCP on its specific protein substrates usually requires their prior phosphorylation on a specific DSGxxS motif (18, 19). Indeed, Emi1 has a DSGxxS sequence, and mutation of serines 145 and 149 in this sequence blocks the degradation of Emi1 in mitotic Xenopus extracts (15) and in cultured mammalian cells (16). However, the identity of the protein kinases involved remained unknown. It has been shown that mutation of all five Cdk phosphorylation sites of Emi1 slows down (although does not block completely) the degradation of Emi1, and it was proposed that initial phosphorylation of Emi1 by Cdk1 may trigger its further phosphorylation by an unknown protein kinase (15). Such a two-step phosphorylation mechanism is involved in the SCFβ-TrCP-mediated ubiquitylation of β-catenin (20). The present investigation was undertaken to identify and characterize the mode of action of the protein kinases involved in the degradation of Emi1 in mitosis.
Methods
Reagents. Extracts from HeLa S3 cells arrested in mitosis (nocodazole treatment for 18 h) or S phase (2 h after release from double thymidine block) were prepared as described (21). His-6-Cul1-Roc1 and His-6-Skp1-β-TrCP were produced by coinfection of 5B insect cells with baculoviruses encoding the corresponding proteins and were purified by nickel agarose chromatography as described (22). The approximate concentrations of these proteins (representing the proteins present at lower concentration) were His-6-Cul1-Roc1 (1 μM) and His-6-Skp1-β-TrCP (0.1 μM). The baculovirus expression vectors of His-6-Plx1 [the Xenopus homologue of Plk1 (23), referred to as “Plk1”] and of its catalytically inactive N172A mutant were generously provided by W. Dunphy (California Institute of Technology, Pasadena); these proteins were expressed and purified as described (10). Protein kinase Cdk1-GST-Δ88-cyclin B (referred to as “Cdk1-cyclin B”) was prepared and purified as described (10). E1, His-6-UbcH5c, methylated ubiquitin, and ubiquitin aldehyde were prepared as described (10, 22). 35S-labeled myc6-Emi1 and its S145N/S149N mutant were produced by in vitro transcription–translation of pCS2 plasmids expressing these proteins by using SP6 TnT Quick Kit (Promega) and [35S]methionine (Amersham Pharmacia). Constructs for bacterial expression of the GST-Emi1-131-160 fragment and of its S145/S149N mutant were prepared by PCR by using the corresponding pCS2-Emi1 plasmids (15). Inserts were cloned between BamH1 and EcoR1 sites of pGEX-2T (Amersham Pharmacia), and constructs were verified by sequencing. The plasmids were transfected into BL21 bacteria and expressed and purified by affinity chromatography on glutathione-agarose. Both proteins were >95% homogeneous, as judged by SDS/PAGE and Coomassie staining.
Assay of the Ligation of Emi1 to Ubiquitin by SCFβ-TrCP. Reaction mixtures in a volume of 10 μl contained 40 mM Tris·HCl, pH 7.6, 2mg/ml ovalbumin (carrier protein), 5 mM MgCl2, 1 mM DTT, 10 mM phosphocreatine, 0.1 mg/ml creatine phosphokinase, 0.5 mM ATP, 0.1 μM E1, 10 μM UbcH5c, 100 μM methylated ubiquitin, 0.5 μM ubiquitin aldehyde, 0.3 μl of 35S-Emi1, 0.2 μl of Cul1-Roc1, 0.5 μl of β-TrCP-Skp1, and protein kinases as indicated in the figures. After incubation at 30°C for 30 min, samples were subjected to electrophoresis on 8% SDS/polyacrylamide gels followed by radioautography or PhosphorImager analysis.
Phosphorylation of GST-Emi1-131-160 Fragments by Plk1. The reaction mixture in a volume of 10 μl contained 40 mM Tris·HCl, pH 7.6, 5 mM MgCl2, 0.5 mM DTT, 2 mg/ml ovalbumin (carrier protein), 0.5 mM [γ-32P]ATP (≈100 mCi/mmol; 1 Ci = 37 GBq), 160 nM Plk1, and GST-Emi1-131-160 fragments as specified. After incubation at 30°C for 30 min, samples were resolved on a 12.5% SDS/polyacrylamide gel, fixed, dried, and subjected to PhosphorImager analysis. Radioactivity in bands corresponding to the ≈30-kDa GST-Emi1-131-160 proteins was estimated. Control incubations without added protein substrates (but in the presence of Plk1) were carried out in parallel, and the small amount of radioactivity in the ≈30-kDa region was subtracted from results. Further controls indicated that free GST and ovalbumin carrier protein were not phosphorylated by Plk1 (data not shown).
Cell Culture and Transfection. HeLa S3 cells were cultured at 37°C in a 5% CO2 humidified atmosphere in DMEM supplemented with 10% (vol/vol) FBS, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Twenty to 24 h before transfection, 6 × 105 cells were seeded in 60-mm plates in a similar medium lacking antibiotics. For small interfering RNA (siRNA) transfection, we have used a commercially available RNA duplex directed against nucleotides 1416–1438 of human Plk1 [Dharmacon, Layfette, CO (catalog no. P-002133-04-20); ref. 24]. Before transfection, siRNA was diluted in OptiMEM 1 (GIBCO) to a concentration of 250 nM and was mixed with Oligofectamine reagent (Invitrogen) to a final reagent concentration of 1.5% (vol/vol). The mixture was incubated for 20 min at room temperature and then diluted 5-fold in OptiMEM and added to cells at 2 ml per plate. The final concentration of siRNA Plk1 was 50 nM. Control (“mock”) treatment was similar, except that treatment was in the absence of siRNA. After incubation at 37°C for 4 h in serum-free OptiMEM, 1 ml of DMEM containing 30% FCS was added, and the cells were incubated for a further 20 h. Where indicated, nocodazole (0.5 μg/ml) was added 8 h after transfection (16 h before the end of treatments). The cells were harvested, and the lysates were prepared as described (22). Samples containing 25 μg of protein were resolved on a 10% SDS/polyacrylamide gel, transferred to nitrocellulose, and blotted with the following antibodies: monoclonal anti-Plk1 (Zymed, catalog no. 33-700, 1:500), anti-cyclin B (Transduction Laboratories, Lexington, KY; catalog no. C23420, 1:1,000), and a polyclonal anti-Emi1 antibody directed against a 10-aa synthetic peptide derived from the extreme C terminus of the human Emi1 protein, which was developed in collaboration with Zymed. The anti-Emi1 antibody was affinity purified and used at a concentration of 1 μg/ml.
Results
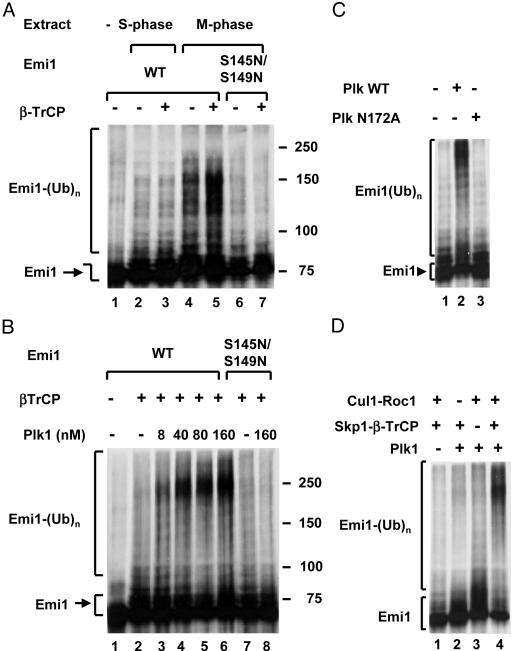
Plk1 Stimulates the Ubiquitylation of Emi1 by the SCFβ-TrCP Complex. It has been shown previously that Emi1 is targeted for degradation by the SCFβ-TrCP ubiquitin ligase complex and that its phosphorylation on a DSGxxS consensus site is required for this process (15, 16). However, the protein kinase(s) involved in this phosphorylation has not been identified. In the experiment shown in Fig. 1A, the ubiquitylation of Emi1 in extracts derived from HeLa cells arrested in S phase or in mitosis have been examined. In agreement with previous results (16), we found that M-phase extracts promote the ligation of Emi1 to ubiquitin and that this process is markedly stimulated by the supplementation of β-TrCP (Fig. 1 A, lanes 4 and 5). By contrast with extracts from S-phase cells, only slight ubiquitylation of Emi1 could be detected, even in the presence of β-TrCP (lanes 2 and 3). As observed previously in vivo (15, 16), we found that ubiquitylation of Emi1 by mitotic extracts in the presence or absence of β-TrCP was abolished by S145N/S149N mutation in the DSGxxS motif (lanes 6 and 7). These results are consistent with the notion that some protein kinases, which are more active in mitosis than in S phase, are required for the ligation of Emi1 to ubiquitin.
Fig. 1.
Plk1 is required for the ligation of Emi1 to ubiquitin (Ub) by SCFβ-TrCP. (A) The ubiquitylation of Emi1 is stimulated by β-TrCP in extracts from M-phase cells but not from S-phase cells. The ligation of ubiquitin to wild-type (lanes 1–5) or S145N/S149N mutant (lanes 6 and 7) is shown. Emi1 was determined as described in Methods in the presence of 30 μg of protein extract from HeLa cells arrested in the S phase (lanes 2 and 3) or M phase (lanes 4–7). Where indicated, 0.5 μlof β-TrCP-Skp1 was added. (B) Plk1 stimulates the ubiquitylation of Emi1 by purified SCFβ-TrCP. Ligation of wild-type or S145/S149N mutant Emi1 to ubiquitin was determined as described in Methods in the presence of the indicated concentrations of Plk1. (C) Requirement for enzymatic activity of Plk1. The ubiquitylation of Emi1 by SCFβ-TrCP was determined in the presence of a 160 nM wild-type or catalytically inactive (N172A) mutant of Plk1, as indicated. (D) Requirement for different components of SCFβ-TrCP. Where indicated, Cul1-Roc1 or β-TrCP-Skp1 was added at the amounts specified in Methods. Plk1 was added at 160 nM, and the ligation of Emi1 to ubiquitin was assayed.
Because the major mitotic protein kinase Cdk1-cyclin B phosphorylates S/TP sites and thus cannot phosphorylate the DSGxxS sequence, we searched for other mitotic protein kinases that may be involved in this process. A possible candidate was Polo-like kinase 1 (Plk1). Levels of Plk1 oscillate markedly in the cell cycle, with a peak in mitosis (25). We have therefore tested whether recombinant, purified Plk stimulates the ligation of Emi1 to ubiquitin by purified recombinant SCFβ-TrCP complex. For this purpose, we have used the recombinant Xenopus homologue Plx1, which is functionally similar to mammalian Plk1 (23). As shown in Fig. 1B, the addition of increasing amounts of Plk1 markedly stimulated the formation of polyubiquitylated derivatives of Emi1 in the purified system. In accordance with the results obtained in crude mitotic extracts (Fig. 1 A), we found that Plk1-stimulated ubiquitylation of Emi1 by purified SCFβ-TrCP complex was also prevented by mutation in S145 and S149 residues (Fig. 1B, lanes 7 and 8). Control experiments showed that the action of Plk1 to stimulate the ubiquitylation of Emi1 required its protein kinase activity, given that no significant ubiquitylation was observed with its catalytically inactive N172A mutant (Fig. 1C) and that all components of the SCFβ-TrCP complex had to be present for Plk1-stimulated ubiquitylation of Emi1 (Fig. 1D). We concluded that phosphorylation by Plk1 stimulates the ligation of Emi1 to ubiquitin by SCFβ-TrCP complex in vitro in a process that requires phosphorylatable S145 and S149 amino acid residues.
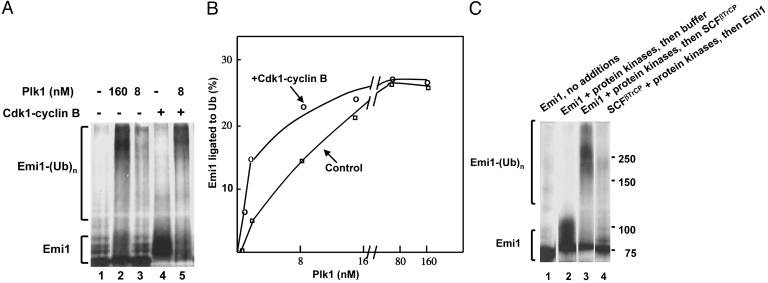
Protein Kinases Plk1 and Cdk1-Cyclin B Synergistically Promote the Ubiquitylation of Emi1. The results described above suggested that Plk1 might have a role in the ubiquitylation of Emi1. We noted, however, that the concentrations of recombinant Plk1 required for robust ubiquitylation of Emi1 (160 nM) were about 20-fold higher than those present in mitotic extracts of similar ubiquitylation activity (data not shown). A possible explanation was that another factor, present in mitotic extracts, potentiates the action of Plk1 at low concentrations. A likely candidate was Cdk1-cyclin B. It has been reported previously that mutation of all five Cdk phosphorylation sites of Emi1 slows down its degradation in mitotic extracts from Xenopus eggs, and it was suggested that primary phosphorylation of Emi1 by Cdk1 may allow its further phosphorylation by another protein kinase (15). Other reports suggested that Plk1 binds tightly to proteins previously phosphorylated by Cdks, resulting in increased catalytic efficiency of Plk1 in further phosphorylations (26, 27). In fact, synergistic action of Plk1 and Cdk1 in the mitotic phosphorylation of several proteins have been reported (see ref. 27 and references therein). We have therefore tested whether Cdk1-cyclin B promotes the ubiquitylation of Emi1 at low, physiological concentrations of Plk1. The addition of Cdk1-cyclin B by itself strongly phosphorylated Emi1, as indicated by marked retardation of electrophoretic migration, but did not promote the formation of high molecular weight ubiquitylated derivatives (Fig. 2A, lane 4). By contrast, when Cdk1-cyclin B was supplemented together with Plk1 at low concentration, there was marked synergistic stimulation of the formation of Emi1–ubiquitin conjugates (Fig. 2 A, compare lanes 3–5). This concentration of Plk1 was 20-fold lower than that required for maximal effect (compare lanes 2 and 3) and, therefore, was in its physiological range. Quantitation of the effect of Cdk1-cyclin B on Emi1–ubiquitin ligation at different concentrations of Plk1 (Fig. 2B) showed a marked (2- to 3-fold) stimulation by Cdk1-cyclin B at low concentrations of Plk1. With increasing concentrations of Plk1, there was a gradual decrease in additional stimulation by Cdk1-cyclin B, and there was no additional stimulation at high concentrations of Plk1.
Fig. 2.
Plk1 and Cdk1-cyclin B synergistically stimulate the ubiquitylation of Emi1 by SCFβ-TrCP. (A) Synergistic effect of the two protein kinases on the ligation of Emi1 to ubiquitin (Ub). The ubiquitylation of Emi1 by SCFβ-TrCP was determined as described in Methods in the presence of the indicated concentrations of Plk1. Where indicated, 500 units of Cdk1-cyclin B were added. (B) Influence of concentrations of Plk1 on the synergistic action of Cdk1-cyclin B. Plk1 was supplemented at the indicated concentrations in the absence or presence of 500 units of Cdk1-cyclin B. The ligation of Emi1 to ubiquitin was assayed as described in Methods in the presence of 1 mM okadaic acid and was quantified by PhosphorImager analysis. Background values of Emi1 ubiquitylation in the absence of Plk1 (≈10% of Emi1 ubiquitylated) were subtracted. (C) Phosphorylation of Emi1 and not of some other component of the ubiquitylation system is required for Emi1–ubiquitin ligation. In a two-stage experiment, the first incubation contained Tris buffer, MgCl2, DTT, ovalbumin, an ATP-regenerating system in concentrations similar to those described in Methods, and okadaic acid at 1 μM. Where indicated, 0.1 μlof 35S-Emi1, 500 units of Cdk1-cyclin B, and 20 nM Plk1 were added. Where indicated, SCFβ-TrCP was added in a mixture that also included 0.1 μM E1, 10 μM UbcH5, 100 μM methylated ubiquitin, and 0.5 μM ubiquitin aldehyde. After incubation at 30°C for 30 min, 10 μM staurosporine was added to stop kinase action. Subsequently, the indicated components were added, and Emi1–ubiquitin ligation was determined after a second incubation (30°C, 30 min).
The above results were consistent with the interpretation that the two protein kinases phosphorylate Emi1 in a manner that allows its efficient ubiquitylation by Emi1. However, it was also possible that one or both protein kinases do not act on Emi1, but rather phosphorylate SCFβ-TrCP, or some other component of the ubiquitylation system and increase their activity in Emi1–ubiquitin ligation. This problem was examined by a two-stage experiment shown in Fig. 2C. In the first stage, the two protein kinases were incubated with Emi1, or with SCFβ-TrCP together with other components of the ubiquitin ligation system. Subsequently, staurosporine was added to stop the action of both protein kinases, the missing components were supplemented and Emi1–ubiquitin ligation was allowed to proceed in a second incubation. When Emi1 was first incubated with the two protein kinases and SCFβ-TrCP was added in the second incubation, significant accumulation of Emi1–ubiquitin conjugates could be observed (lane 3). This finding was compatible with the interpretation that phosphorylation of Emi1 by the two protein kinases in the first stage converted it to an efficient substrate for SCFβ-TrCP in the second stage. By contrast, when SCFβ-TrCP was first incubated with the two protein kinases (in the presence of E1 and E2) and Emi1 was added in the second incubation, there was no significant accumulation of Emi1–ubiquitin conjugates beyond background levels (lane 4). Control experiments indicated that the addition of staurosporine before the first incubation effectively prevented the phosphorylation and ubiquitylation of Emi1 in all cases (data not shown). We concluded that the phosphorylation of Emi1, and not of some component of the ubiquitin ligase system, is required for its ligation to ubiquitin.
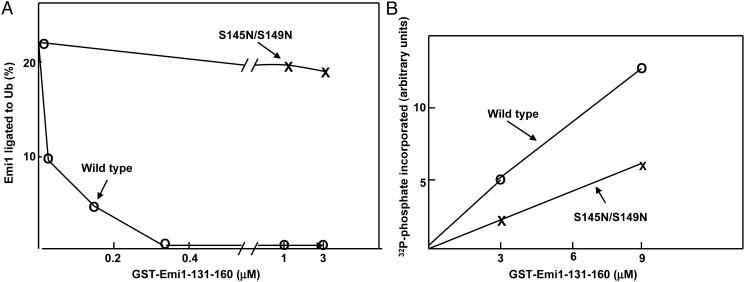
Phosphorylation of Emi1 by Plk1 in the DSGxxS Sequence. The data described above indicate that the phosphorylation of Emi1 and the presence of phosphorylatable serine residues in its DSGxxS sequence are required for Plk1-stimulated ubiquitylation. However, those data do not directly show that Plk1 phosphorylates the DSGxxS motif. A survey of presently available information on sites in different proteins phosphorylated by Emi1 does not reveal a strictly conserved consensus motif, except for the presence of acidic (D/E) amino acid residues in the vicinity of the phosphorylated T/E in most, but not in all, cases (see Discussion). Because full-length Emi1 may contain several sites that are phosphorylated by Plk1, we have examined this problem with a 30-aa fragment similar to amino acid residues 131–160 of Emi1, which contains the DSGxxS sequence. A mutant fragment in which serines 145 and 149 in this sequence were changed to asparagines was also prepared. Both fragments were expressed in bacteria as GST fusion proteins and were purified to >95% homogeneity. Control experiments indicated that GST is not phosphorylated by Plk1 (data not shown). We have first examined whether the 131–160 fragment of Emi1 is recognized as a specific substrate, by competition on the ligation of Emi1 to ubiquitin by the purified system. As shown in Fig. 3A, the wild-type 131–160 fragment effectively inhibited the ligation of full-length Em1 to ubiquitin, whereas similar concentrations of the S145N/149N mutant fragment had no significant influence. These data indicate that the 131–160 fragment interacts specifically with the system that ligates Emi1 to ubiquitin. We next tested the phosphorylation of these fragments of Emi1 by Plk1 by the incorporation of 32P-phosphate from [γ-32P]ATP (see Methods). As shown in Fig. 3B, Plk1 phosphorylated both wild-type and S145N/149N mutant fragments of Emi1, but the phosphorylation of the mutant fragment was significantly reduced to ≈45% of the value obtained with the wild-type fragment. The reduction in the phosphorylation of the mutant suggests that Plk1 phosphorylates the DSGxxS sequence, whereas the residual phosphorylation in the mutant shows that it contains an additional site(s) that is phosphorylated by Plk1. Because of the high background caused by the additional phosphorylation sites, we did not investigate further whether S145, S149, or both are phosphorylated by Plk1.
Fig. 3.
Phosphorylation of DSGxxS sequence of Emi1 by Plk1. (A) Emi1–ubiquitin (Ub) ligation is competed by the wild-type 131–160 fragment but not by the mutant fragment. Ubiquitylation of 35S-Emi1 by SCFβ-TrCP was determined in the presence of the indicated concentrations of wild-type or S145/S149N mutant GST-131-160 fragment of Emi1. (B) Phosphorylation of wild-type and mutant GST-Emi1 131–160 fragments by Plk1. The Plk1-dependent incorporation of 32P-phosphate from [γ-32P]ATP into wild-type or S145/S149N mutant GST-Emi1 131–160 fragments was determined as described in Methods. Results were expressed in arbitrary units.
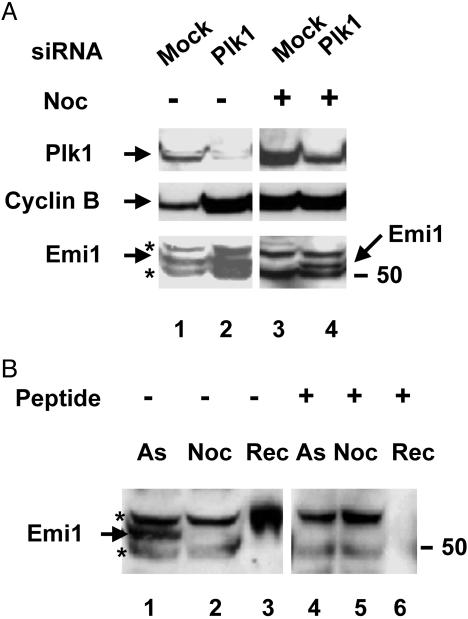
Influence of Plk1 on Levels of Emi1 in Vivo. We next sought to examine the question of whether Plk1 indeed has a role in the degradation of Emi1 in intact cells. For this purpose, we used siRNA to deplete Plk1 in HeLa cells. In accordance with previous results (24), we observed that transfection of asynchronous cells with low concentrations of an RNA duplex corresponding to nucleotides 1416–1438 of the coding region of Plk1 effectively lowered, although did not abolish completely, levels of Plk1 protein (Fig. 4A Top, compare lanes 1 and 2). Also as reported (24, 28), we observed that depletion of Plk1 arrests cells in mitosis, as indicated both by cell morphology (data not shown) and by marked increase in levels of cyclin B (Fig. 4A Middle, compare lanes 1 and 2). When cells were arrested in mitosis by treatment with nocodazole, levels of Plk1 were greatly increased, as expected from the known oscillations in levels of Plk1 in the cell cycle (25). In nocodazole-treated cells, too, siRNA directed against Plk1 caused a marked decrease in levels of Plk1 protein (Fig. 4A Top, compare lanes 3 and 4).
Fig. 4.
Influence of depletion of Plk1 on levels of Emi1 in mitotically arrested HeLa cells. (A) Effects of transfection with siRNA duplex directed against Plk1 on levels of Plk1, cyclin B, and Emi1 proteins. HeLa cells were either mock-treated or transfected with siRNA directed against human Plk1, as described in Methods. Where indicated, nocodazole (Noc, 0.5 μg/ml) was added 16 h before collection of cells. Cells were collected 24 h after transfection, and samples were immunoblotted with antibodies directed against Plk1 (Top), cyclin B (Middle), and C-terminal peptide of Emi1 (Bottom). (B) Specificity of anti-Emi1 antibody. Extracts from asynchronous (As) or nocodazole-arrested cells and 0.1 ng of recombinant Emi1 (Rec) were separated on 10% SDS/polyacrylamide gels, transferred to nitrocellulose, and blotted with an antibody directed against a C-terminal peptide of Emi1 (1 μg/ml) in the absence (lanes 1–3) or presence (lanes 4–6) of C-terminal peptide (0.1 μg/ml). Asterisks denote crossreacting proteins; the number on the right indicates the position of molecular mass marker protein (kDa).
Levels of Emi1 were determined in these samples with an antibody directed against a C-terminal peptide of this protein. This antibody reacts well with recombinant Emi (Fig. 4B, lane 3). In extracts from HeLa cells, the antibody reacts with three protein bands in the molecular mass region of 50–60 kDa (Fig. 4B, lane 1). Of these, the middle band (Mr ≈ 55,000) was identified as Emi1, by the following observations: (i) This protein, but not the two others, disappeared in cells treated with nocodazole (Fig. 4B, lane 2), as would be expected for Emi1 (11, 12). (ii) Treatment of antibody with the C-terminal peptide abolished the reaction with recombinant Emi1 (Fig. 4B, lane 6) and with the ≈55-kDa endogenous protein (lane 4) but not with the two other crossreacting proteins (lanes 4 and 5). It should be noted that our preparation of recombinant Emi1 has somewhat slower electrophoretic migration than endogenous Emi1 because of the presence of an additional 20-aa fragment in the construct.
Examination of the levels of Emi1 in cells depleted of Plk1 by siRNA treatment showed a moderate increase compared with asynchronous, mock-treated cells (Fig. 4A Bottom, lanes 1 and 2). However, in view of the fact that Plk1 depletion arrests cells in mitosis, it is notable that Emi1 levels were not decreased, as is usually the case in mitotically arrested cells. Indeed, more pronounced differences were observed in nocodazole-treated cells: Levels of Emi1 were practically undetectable in this situation in mock-transfected cells and were significantly increased by Plk1 depletion (Fig. 4A Bottom, lanes 3 and 4). It was concluded that depletion of Plk1 increases the steady-state levels of Emi1 in mitosis. This conclusion is most likely due to inhibition of the rapid degradation of Emi1, which takes place in this stage of the cell cycle. It was technically impossible to compare rates of degradation of Emi1 under these conditions, because of the practically undetectable levels of Emi1 in nocodazole-arrested cells.
Discussion
In this investigation, we have examined the possible involvement of Plk1 in SCFβ-TrCP-mediated degradation of Emi1 in mitosis. We suspected that Plk1 may be involved, because of the following considerations: (i) The timing of the rise in levels of Plk1 in early mitosis (25) coincides with the degradation of Emi1 in prometaphase (11, 12); and (ii) the phosphorylation of proteins by Plk is potentiated by their prior phosphorylation by Cdks, because of the interaction of the Polo-box domain with phosphorylated S/TP sites (26, 27). This finding was consistent with the observed requirement for both Cdk and DSGxxS phosphorylation sites for the degradation of Emi1 (15). We indeed found that Plk1 stimulated the ubiquitylation of Emi1 by purified SCFβ-TrCP, a process that required serines 145 and 149 in the DSGxxS sequence (Fig. 1) and that Cdk1-cyclin B potentiated this action at low, physiological levels of Plk1 (Fig. 2). That Plk1 may phosphorylate serine residues in the DSGxxS sequence is suggested by the reduction of the phosphorylation of a fragment in which these two serines were mutated (Fig. 3). Based on mutagenesis studies of peptides derived from Cdc25C, it has been reported recently that the consensus motif for Plk phosphorylation is D/ExS/TxxD (29). However, examination of the literature on sites in different proteins phosphorylated by Plk1 does not reveal such a strict consensus phosphorylation motif. In most cases, Plk1 prefers acidic regions in proteins, but the position of D/E residues relative to the phosphorylated S/T residue is variable (see ref. 30 and references therein). Thus, the acidic amino acid residue may be adjacent or may be at several residues amino-terminal or carboxy-terminal to the phosphorylated S/T residue. In still other, although more rare, cases there is no acidic amino acid residue among the six that surround the S/T phosphorylated by Plk1 (see Table 1 in ref. 6 for an extensive list of Plk phosphorylation sites in different subunits of APC/C). Thus, the DSGxxS sequence may be a Plk1 phosphorylation site because of its D residue or because of some other, as yet unrecognized features in the structure of Emi1.
We examined the question of whether Plk1 is indeed involved in the regulation of levels of Emi1 in cells by the use of RNA interference. Although some siRNAs produce nonspecific effects on the expression of unrelated proteins (31), in other cases they appear to act quite specifically (32). In accordance with previous reports using different siRNAs against Plk1 (24, 25), we observed that an RNA duplex that effectively lowers levels of Plk1 arrests cells in mitosis (Fig. 4). Levels of Emi1 were significantly increased in Plk1-depleted, mitotically arrested cells (Fig. 4), consistent with the notion that Plk1 is required for the degradation of Emi1 in mitosis.
Acknowledgments
We thank Gil Bornstein and Ramla Benmaamar for help and reagents. This work was supported by grants from the Israel Science Foundation and the Israel Cancer Research Fund (to A.H.), National Institutes of Health Grants R01-CA76584 and R01-GM57587, and an Irma T. Hirschl Scholarship (to M.P.).
Abbreviations: Emi1, early mitotic inhibitor 1; APC/C, anaphase promoting complex/cyclosome; Plk, Polo-like kinase; SCF, Skp1–Cullin1 F-box protein; siRNA, small interfering RNA.
References
- 1.Sudakin, V., Ganoth, D., Dahan, A., Heller, H., Hershko, J., Luca, F. C., Ruderman, J. V. & Hershko, A. (1995) Mol. Biol. Cell 6, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King, R., Peters, J. M., Tugendreich, S. M., Rolfe, M., Hieter, P. & Kirschner, M. W. (1995) Cell 81, 279–288. [DOI] [PubMed] [Google Scholar]
- 3.Harper, J. W., Burton, J. L. & Solomon, M. J. (2002) Genes Dev. 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- 4.Peters, J. M. (2002) Mol. Cell 5, 931–943. [DOI] [PubMed] [Google Scholar]
- 5.Lahav-Baratz, S., Sudakin, V., Ruderman, J. V. & Hershko, A. (1995) Proc. Natl. Acad. Sci. USA 92, 9303–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraft, C., Herzog, F., Gieffers, C., Mechtler, K., Hagting, A., Pines, J. & Peters, J. M. (2003) EMBO J. 22, 6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shteinberg, M., Protopopov, Y., Listovsky, T., Brandeis, M. & Hershko, A. (1999) Biochem. Biophys. Res. Commun. 260, 193–198. [DOI] [PubMed] [Google Scholar]
- 8.Rudner, A. D. & Murray, A. W. (2000) J. Cell. Biol. 149, 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descombes, P. & Nigg, E. A. (1998) EMBO J. 17, 1328–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golan, A., Yudkovsky, Y. & Hershko, A. (2002) J. Biol. Chem. 277, 15552–15557. [DOI] [PubMed] [Google Scholar]
- 11.Reimann, J. D. R., Freed, E., Hsu, J. Y., Kramer, E. R., Peters, J. M. & Jackson, P. K. (2001) Cell 105, 645–655. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, J. Y., Reimann, J. D. R., Sorensen, C. S., Lukas, J. & Jackson, P. K. (2002) Nat. Cell Biol. 4, 358–366. [DOI] [PubMed] [Google Scholar]
- 13.Dong, X., Zavitz, K. H., Thomas, B. J., Lin, U., Campbell, S. & Zipursky, S. L. (1997) Genes Dev. 11, 94–105. [DOI] [PubMed] [Google Scholar]
- 14.Reimann, J. D. R., Gardner, B. E., Margottin-Goguet, F. & Jackson, P. K. (2001) Genes Dev. 15, 3278–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margottin-Goguet, F., Hsu, J. Y., Loktev, A., Hsieh, H. M., Reimann, J. D. R. & Jackson, P. K. (2003) Dev. Cell 4, 813–826. [DOI] [PubMed] [Google Scholar]
- 16.Guardavaccaro, D., Kudo, Y., Boulaire, J., Barchi, M., Busino, L., Donzelli, M., Margottin-Goguet, F., Jackson, P. K., Yamasaki, K. & Pagano, M. (2003) Dev. Cell 4, 799–812. [DOI] [PubMed] [Google Scholar]
- 17.Guardavaccaro, D. & Pagano, M. (2004) Oncogene 23, 2037–2049. [DOI] [PubMed] [Google Scholar]
- 18.Laney, J. D. & Hochstrasser, M. (1999) Cell 97, 427–430. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis, T. (1999) Genes Dev. 1 3, 505–510. [DOI] [PubMed] [Google Scholar]
- 20.Amit, S., Hatzubai, A., Birman, Y., Andersen, J. S., Ben-Shushan, E., Mann, M., Ben-Neriah, Y. & Alkalay, I. (2002) Genes Dev. 16, 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yudkovsky, Y., Shteinberg, M., Listovsky, T., Brandeis, M. & Hershko, A. (2000) Biochem. Biophys. Res. Commun. 271, 299–304. [DOI] [PubMed] [Google Scholar]
- 22.Bornstein, G., Bloom, J., Sitry-Shevah, D., Nakayama, K., Pagano, M. & Hershko, A. (2003) J. Biol. Chem. 278, 25752–25757. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai, A. & Dunphy, W. G. (1996) Science 273, 1377–1380. [DOI] [PubMed] [Google Scholar]
- 24.Spankuch-Schmitt, B., Breiter-Hahn, J., Kaufman, M. & Strebhart, K. (2002) J. Natl. Cancer Inst. 94, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 25.Nigg, E. (1998) Curr. Opin. Cell Biol. 10, 776–783. [DOI] [PubMed] [Google Scholar]
- 26.Elia, A. E., Cantley, L. C. & Yaffe, M. B. (2003) Science 299, 1228–1231. [DOI] [PubMed] [Google Scholar]
- 27.Elia, A. E., Rellos, P., Haire, L. F., Chao, J. W., Ivins, F. J., Hoepker, K., Mohammad, D., Cantley, L. C., Smerdon, S. J. & Yaffe, M. B. (2003) Cell 115, 83–95. [DOI] [PubMed] [Google Scholar]
- 28.Liu, X. & Erikson, R. L. (2002) Proc. Natl. Acad. Sci. USA 99, 8672–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajima, H., Tokoshima-Morimoto, F., Taniguchi, E. & Nishida, E. (2003) J. Biol. Chem. 278, 25277–256280. [DOI] [PubMed] [Google Scholar]
- 30.Yarm, F. R. (2002) Mol. Cell. Biol. 22, 6209–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scacheri, P. C., Rozenblatt-Rosen, O., Caplen, N. J., Wolfsberg, T. G., Umayam, L., Lee, J. C., Hughes, C. M., Sharimugam, K. S., Bhattacharjee, A., Meyerson, M. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D. N. & Fesik, S. W. (2003) Proc. Natl. Acad. Sci. USA 100, 6347–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]