Abstract
IκB kinase (IKK), discovered as the major activator of NF-κB, plays additional roles in signaling. By using mouse embryo fibroblasts (MEFs) lacking both the α and β subunits of IKK, we find that these proteins are required for induction of a major subset of IFNγ-stimulated genes and that this requirement is independent of NF-κB activation. Furthermore, there is no defect in IFNγ-stimulated signal transducer and activator of transcription 1 (Stat1) activation or function in the IKKα/β-null MEFs. Therefore, although activated Stat1 dimers are necessary for the activation of these genes in response to IFNγ, they are not sufficient. These results reveal an important additional pathway for IFNγ-stimulated gene expression in which an NF-κB-independent function of IKK is required.
Much progress has been made toward understanding how genes are activated in response to IFNγ. The major pathway involves receptor-dependent activation of the Janus protein tyrosine kinases, which in turn phosphorylate the latent transcription factor signal transducer and activator of transcription 1 (Stat1) to activate expression of IFN-stimulated genes (1). Although Stat1 is essential for cells to respond fully to IFNγ, several ancillary pathways are also activated by IFNγ (2–4), and gene expression in Stat1-null cells and mice can still mediate partial antiviral responses to IFNγ (2, 5). Additionally, in cells from mice lacking Stat1, IFNγ enhances proliferation and suppresses apoptosis, compared with cells from WT mice (3). Ultimately, the diversity of gene-expression patterns mediated by both the Stat1-dependent and -independent pathways, as well as the balance between these pathways, will determine the biological responses to IFNγ.
The inhibitor of κB (IκB) kinase (IKK) complex, responsible for the cytokine-induced activation of the latent NF-κB (6–9), consists of the two highly related kinase subunits IKKα and IKKβ, as well as a third structural subunit, IKKγ (10). All three subunits are necessary for NF-κB-dependent gene expression (11, 12). It was first thought that the only substrates of IKK were the cytoplasmic IκBs. However, recent work has revealed that the IKK complex has additional targets whose phosphorylation is necessary for NF-κB-stimulated gene expression. IKKβ and IKKγ are required for cytokine-induced degradation of IκB (13–19). Whereas IKKα and IKKβ are required to phosphorylate the p65/RelA subunit of NF-κB (11, 20–24) and for processing the p105 precursor to the p50 subunit of NF-κB (25), IKKα alone is required for processing the p100 precursor to the p52 subunit of NF-κB (26–28). Furthermore, IKK-dependent phosphorylation of the p65 subunit of NF-κB stimulates its transactivation function and IKKα is involved in the phosphorylation of histone H3 on NF-κB-dependent promoters (11, 24, 29, 30). Therefore, the IKK complex phosphorylates several target proteins in the process of activating NF-κB-dependent genes.
There are prior suggestions that IKK might contribute to IFNγ-dependent gene expression. It has been proposed that IFNs can activate NF-κB directly as a component of IFN-stimulated gene expression (31, 32). IFNβ appears to be able to activate NF-κB by phosphorylating a portion of p65 NF-κB that is constitutively free in the nucleus in some cell lines (33). However, studies in several cell types fail to reveal direct and complete activation of NF-κB by IFNγ (33, 34). There is ample evidence of important cross talk between the activation of the IKK pathway by the proinflamatory cytokines tumor necrosis factor (TNF) or interleukin 1 (IL-1) and IFN-dependent signaling, and the combination of TNF and IFNγ leads to synergistic induction of inflammatory genes (35–37). Also, IL-1 and TNF enhance the phosphorylation of serine-727 of Stat1, which occurs incompletely in response to IFNγ alone, and thus can potentiate IFNγ-mediated, Stat1-driven gene expression (38). Additionally, IFNβ and γ increase the transactivation function of constitutive basal NF-κB already free in the nucleus in several cell types by stimulating phosphorylation of the p65 subunit of NF-κB (33, 34). The results presented here define a previously unrecognized role of the IKKs in IFNγ-dependent signaling that is critical for many IFNγ-stimulated genes.
Materials and Methods
Biological Reagents and Cell Culture. Recombinant human IL-1β was from the National Cancer Institute. Recombinant murine TNF, IFNβ, and IFNγ were from PeproTech (Rocky Hill, NJ). Polyclonal anti-hemagglutinin, anti-green fluorescent protein (GFP), anti-IKKα, and anti-IKKβ were from Santa Cruz Biotechnology. Polyclonal anti-phosphoserine-727 Stat1, anti-phosphotyrosine-701 Stat1, and anti-Stat1 antibodies were from Upstate Biotechnology (Lake Placid, NY). WT and IKKα/β-null mouse embryo fibroblasts (MEFs) were provided by Inder Verma (Salk Institute for Biological Studies, La Jolla, CA). WT MEFs stably expressing IκBα super repressor (IκBαSR) were generated by transfection and selection with G418 for the pLXIN retroviral vector plasmid (Clontech) encoding a mutant IκBα that contains serine-to-alanine substitutions at positions 32 and 36. The p65NFκB-null and corresponding WT MEFs were provided by David Baltimore (California Institute of Technology, Pasadena). All cells were maintained in the Dulbecco–Vogt-modified Eagle's medium, supplemented with 10% FCS, 100 μg/ml penicillin G, and 100 μg/ml streptomycin. For all experiments, unless otherwise indicated, cells at 80% confluence on 100-mm dishes were stimulated with IL-1 (2 ng/ml), TNF (25 ng/ml), IFNβ (1,000 units/ml), or IFNγ (1,000 units/ml) at 37°C. All results are typical of at least three independent experiments.
Northern Analysis. Cells were stimulated with IL-1, TNF, IFNβ, or IFNγ for 6 h, and total RNA was isolated by using the TRIzol reagent (Invitrogen). RNA was fractionated by electrophoresis in a formaldehyde gel and transferred to Hybond-N membranes according to the procedures provided by Amersham Pharmacia Biosciences. cDNA probes for mRNAs encoding murine IFN-inducible protein 10 (IP10), guanylate binding protein 2, IFN regulatory factor 1 (provided by Thomas Hamilton and Bryan Williams, Cleveland Clinic Foundation), and human GAPDH were made by using the random priming kit from Amersham Pharmacia Biosciences. Probe hybridization and washing were performed according to procedures provided by Amersham Pharmacia Biosciences and the signals were visualized by autoradiography.
Western Analysis. Cells, untreated or treated with IFNγ, were washed once with PBS and lysed for 30 min at 4°C in 1 ml of 0.5% Nonidet P-40 lysis buffer, as described in ref. 39. Cellular debris was removed by centrifugation at 16,000 × g for 15 min. Cell extracts were fractionated by SDS/PAGE and transferred to nitrocellulose membranes. Analysis was performed with the indicated primary antibodies, which were visualized with horse-radish peroxidase-coupled goat anti-rabbit or anti-mouse immunoglobulins, by using the ECL Western-blotting detection system (Perkin–Elmer).
Transfection and Reporter Assays. The Stat1-dependent reporter plasmid p7XmycGas contains seven tandem copies of the γ-activated sequence (GAS) site from the c-Myc gene (3). The NF-κB-dependent reporter plasmid p5XIP10κB (provided by Bryan Williams, Cleveland Clinic Foundation) contains five tandem copies of the NF-κB site from the IP10 gene. WT and IKKα/β-null MEFs were transfected transiently by using Fugene (Roche Diagnostics) with 2 μg of p7XmycGas or p5XIP10κB with 0.5 μg of pSV2-βgal. Cells were divided into separate plates for treatment 8 h after transfection and allowed to readhere for 16 h. The cells were then stimulated with IL-1, TNF, or IFNγ for 6 h and harvested. Luciferase or galactosidase activity was determined with the luciferase-assay system or chemiluminescent reagents (Promega). Luciferase activity was normalized to β-galactosidase activity to control for transfection efficiency.
Adenovirus Infections. MEFs were infected with adenoviruses, provided by Frank Mercurio (Celgene, San Diego), expressing GFP, hemagglutinin-tagged IκBαSR, IKKα, IKKβ, a combination of IKKα and IKKβ, or kinase-inactive (KI) IKKβ. The titer of each adenovirus stock used for infection was as follows: GFP = 3 × 108, IκBαSR = 5 × 108, IKKα = 5 × 108, IKKβ = 5 × 108, and KI IKKβ = 1 × 109 virus particles per ml. After 18 h, the infected cells were stimulated with IFNγ for 6 h or were unstimulated.
Affymetrix GeneChip Analysis. WT and IKKα/β-null cells were untreated or treated with IFNγ for 2 or 6 h. Total RNA was isolated by using the TRIzol reagent (Invitrogen). Preparation of complementary RNA and hybridization to MG-U74A murine genome arrays was performed as described by Affymetrix (Santa Clara, CA). Stained chips were read and analyzed by using an Affymetrix GeneChip scanner and the accompanying software.
Real-Time Semiquantitative PCR Confirmation of Selected Genes. To validate a subset of genes with apparent significant changes, real-time RT-PCR was performed. WT and IKKα/β-null cells were either untreated or treated with IFNγ for 2 or 6 h. Total RNA was isolated by using the TRIzol reagent. Total RNA (≈1 μg) from each sample was reverse transcribed as described by the manufacturer by using the iScript cDNA Synthesis Kit (Bio-Rad). Gene sequences available from the National Center for Biotechnology Information GenBank and Unigene databases were selected to design primers. Optimum primer sequences were selected after verification for gene-specific complementation using the National Center for Biotechnology Information blast program. Semiquantitative analysis of gene expression was performed by using the Bio-Rad iCycler IQ system and the manufacturer's protocol for the iQ SYBR Green Supermix kit supplied by Bio-Rad. cDNA concentrations for each sample were normalized by using two control genes that showed no change in expression on the arrays: β-actin and GAPDH. Standard curves were generated by preparing serial dilutions, and the relative level of expression of each of the verified genes was determined.
Results
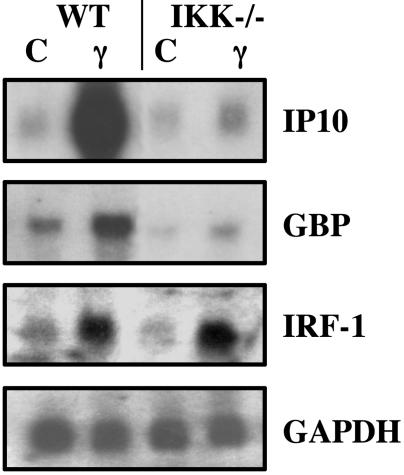
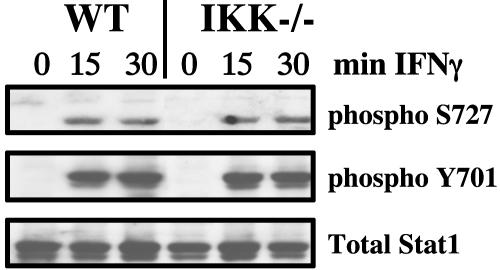
IKKα and IKKβ Are Required to Activate a Subset of IFNγ-Stimulated Genes. IFNγ fails to induce the expression of IP10 and guanylate binding protein 2 mRNAs but not IFN regulatory factor 1 mRNA in IKKα/β-null MEFs, compared with the normal induction of all three genes in matched WT MEFs (Fig. 1). However, Stat1 is activated normally in the IKKα/β-null MEFs, as shown by the normal phosphorylation of tyrosine-701 and serine-727 (Fig. 2) and activation of a Stat1-dependent GAS reporter plasmid in response to IFNγ (Fig. 3a).
Fig. 1.
IKK-dependent responses to IFNγ. WT or IKKα/β-null cells were treated with IFNγ (γ) for 6 h or were untreated (C), and total RNA was analyzed by the Northern procedure.
Fig. 2.
Normal activation of Stat1 by IFNγ in IKKα/β-null cells. WT or IKKα/β-null cells were treated for 15 or 30 min with IFNγ or were untreated. Western analysis was performed with antibodies against phosphoserine-727 Stat1, phosphotyrosine-701 Stat1, and total Stat1.
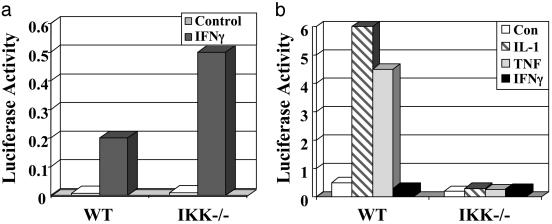
Fig. 3.
Responses of GAS and NF-κB elements to IFNγ.(a) Normal activation of a GAS-dependent reporter by IFNγ in IKKα/β-null cells. WT or IKKα/β-null cells were transfected transiently with p7XmycGAS-luciferase and pSV2βgal and treated with IFNγ as shown. (b) IFNγ fails to activate an NF-κB-dependent promoter. WT or IKKα/β-null cells were transfected transiently with p5XIP10κB luciferase and pSV2βgal. After 8 h, the cells were divided into different plates and incubated for 16 h more. The cells were treated as indicated with IFNγ, IL-1, or TNF for 6 h or were untreated, and the luciferase activity was compared with β-galactosidase activity to normalize for transfection efficiency.
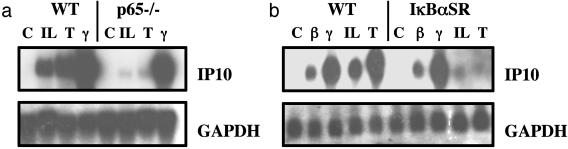
The Function of IKK in IFNγ-Dependent Gene Expression Does Not Involve NF-κB. In comparison with the positive controls of IL-1 and TNF, IFNγ failed to induce any detectable NF-κB-dependent promoter activity in WT MEFs (Fig. 3b). As expected, all three cytokines failed to induce NF-κB-dependent promoter activity in IKKα/β-null MEFs (Fig. 3b). We also investigated the potential role of NF-κB in IFNγ-dependent signaling by stimulating WT or p65NFκB-null MEFs with IL-1, TNF, or IFNγ. As expected, the p65NFκB-null MEFs were deficient in the IL-1- and TNF-stimulated induction of IP10 mRNA, but the IFNγ-stimulated induction of IP10 mRNA was normal in these cells (Fig. 4a). Additionally, we generated WT MEFs stably expressing the IκBαSR, which contains serine-to-alanine mutations at positions 32 and 36. The IκBαSR cannot be phosphorylated on these residues or degraded and thus is stably bound to NF-κB, keeping it trapped in an inactive state in the cytoplasm. We stimulated WT or IκBαSR MEFs with IFNβ, IFNγ, IL-1, or TNF. Whereas the IκBαSR MEFs were deficient in IL-1- and TNF-stimulated induction of IP10 mRNA, both IFNβ- and IFNγ-stimulated induction of this mRNA was normal (Fig. 4b). All of these data indicate that the role of IKK in IFNγ-dependent gene expression does not involve NF-κB.
Fig. 4.
The requirement of IKKα/β for IFNγ-stimulated IP10 gene expression is independent of NF-κB activation. (a) The response of IP10 to IFNγ does not require p65. WT or p65NFκB-null cells were treated with IL-1, TNF, or IFNγ for 6 h or were untreated, and total RNA was analyzed by the Northern procedure. (b) The response of IP10 to IFNγ is not inhibited by the IκBα super repressor. WT cells or cells expressing the IκBαSR were treated with IFNβ, IFNγ, IL-1, or TNF for 6 h or were untreated, and total RNA was analyzed by the Northern procedure.
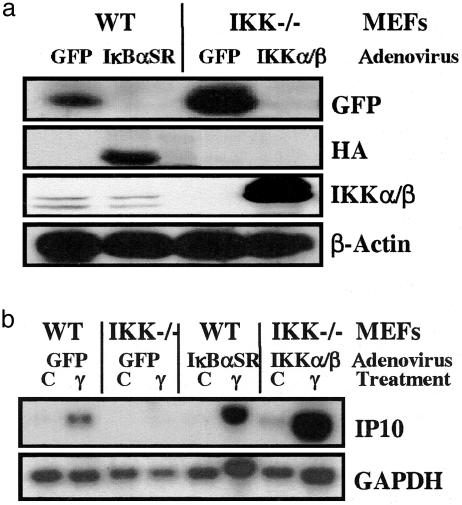
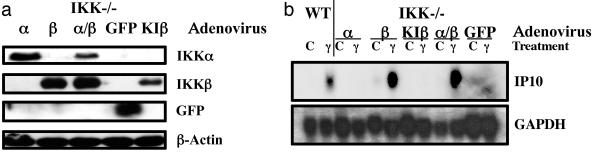
Reexpression of IKKα and β in IKKα/β-null MEFs Restores IFNγ-Dependent Transcription. IKKα/β-null MEFs were infected with recombinant adenoviruses, encoding GFP (as a control) or with equal amounts of viruses encoding IKKα and IKKβ (separately or together), or KI IKKβ. Additionally, WT MEFs were infected with either control GFP adenovirus or IκBSR adenovirus, to inhibit NF-κB activation. Eighteen hours after infection, the cells were untreated or treated with IFNγ for 6 h. Western analysis demonstrated that the appropriate proteins (IKKα, IKKβ, KIIKKβ, GFP, or IκBSR) were expressed efficiently in the adenovirus-infected cells (Figs. 5a and 6a). Again, inhibition of NF-κB activity by IκBSR had no effect on the ability of IFNγ to activate gene expression in the WT MEFs (Fig. 5b) but completely inhibited IL-1- or TNF-induced IP10 mRNA expression (data not shown). Reexpression of IKKα and IKKβ together in IKKα/β-null MEFs completely restored, and even enhanced, IFNγ-stimulated IP10 mRNA expression (Figs. 5b and 6b), as well as IL-1- or TNF-induced IP10 mRNA expression (data not shown). These data indicate that the requirement of IKK for IFNγ-dependent gene expression in the IKKα/β-null MEFs is specific. We additionally investigated the roles of IKKα and IKKβ separately by reexpressing these proteins individually in IKKα/β-null MEFs. IKKβ, but not IKKα, restored IFNγ-mediated induction of IP10 mRNA (Fig. 6b). Additionally, reexpression of KI IKKβ failed to restore IFNγ-stimulated induction of IP10 mRNA (Fig. 6b). However, neither IKKα nor IKKβ alone was able to restore IL-1- or TNF-induced expression of IP10 (data not shown). These data distinguish the role of the IKKs in IFNγ-induced gene expression from their role in activating NF-κB and indicate that, for IP10 mRNA induction, kinase-active IKKβ alone is able to restore responsiveness to IFNγ.
Fig. 5.
Reexpression of IKKα and IKKβ in the IKKα/β-null cells restores IFNγ-stimulated IP10 expression. Three plates each of WT or IKKα/β-null cells were infected with adenoviruses capable of expressing GFP, IκBSR, or IKKα and IKKβ together. After 18 h, the cells were treated with IFNγ for 6 h or were untreated. (a) Western analysis, performed on equal amounts of total protein from each infection, for GFP, the hemagglutinin (HA) tag of IκBSR, IKKα/IKKβ, and β-actin (loading control). (b) Northern analysis, performed with an equal amount of total RNA from each sample.
Fig. 6.
Reexpression of kinase-active IKKβ, but not IKKα or KI IKKβ, in IKKα/β-null cells restores IFNγ-stimulated IP10 expression. Three plates each of the cells were infected with adenoviruses expressing IKKα (α), IKKβ (β), GFP, or kinase-inactive IKKβ (KIβ). After 18 h, the cells were treated with IFNγ for 6 h or were untreated. (a) Western analysis, performed on an equal amount of total protein from each infection for GFP, IKKα, IKKβ, and β-actin (loading control). (b) Northern analysis, performed with an equal amount of total RNA from each sample.
Many IFNγ-Stimulated Genes Require IKK Function. To assess the extent of involvement of IKKα/β in IFNγ-stimulated gene expression more comprehensively, we conducted an Affymetrix-based expression analysis of WT and IKKα/β-null MEFs after stimulation with IFNγ for 0, 2, or 6 h. Labeled cRNA probes were hybridized to MG-U74A arrays, which contain sequences corresponding to ≈6,000 genes and 6,000 ESTs. Many IFNγ-stimulated genes require the presence of IKK. The number of genes induced 2-fold or more by IFNγ in the WT MEFs was 82 at 2 h and 200 at 6 h. The number of genes whose expression was either present but not induced by IFNγ or absent in the IKKα/β-null MEFs as compared with the WT controls was 45 at 2 h and 128 at 6 h. The expression of a subset of these genes was then confirmed by Northern analysis and semiquantitative real-time PCR (data not shown). The fold induction (from the array experiment) of each confirmed gene is shown in Table 1 as the ratio of intensities in the IFNγ-treated WT and IKKα/β-null MEFs compared with their untreated samples. Genes that were not induced in the IKKα/β-null MEFs as compared with the WT controls are included in Table 1, along with examples of genes that do not depend on IKKα/β for induction by IFNγ.
Table 1. IKK-dependent and -independent IFNγ-stimulated genes.
| Fold induction
|
|||||
|---|---|---|---|---|---|
| WT
|
IKK-null
|
||||
| Gene | Accession no. | 2 h | 6 h | 2 h | 6 h |
| IP10 | M33266 | 19.7 | 10.5 | NI | NI |
| LMP-7 | U22033 | 6.9 | 42.0 | NI | NI |
| SOCS-2 | U88327 | 3.2 | 4.0 | 1.2 | NI |
| Mx1 | M21038 | NI | 3.7 | NI | NI |
| Ndr1 | U52073 | NI | 4.5 | NI | NI |
| Spp1 | X13986 | NI | 2.8 | NI | NI |
| Dub1 | U41636 | NI | 4.1 | NI | NI |
| LY-6 | X04653 | 2.3 | 2.6 | 1.5 | NI |
| GBP2 | AJ007970 | 238 | 55.7 | 24.2 | 5.6 |
| Fas | M83649 | 4.3 | 4.3 | 1.3 | 2.4 |
| Mgl1 | U15635 | 3.2 | 18.4 | NI | 5.6 |
| IRF-1 | M21065 | 12.9 | 45.0 | 18.4 | 37.2 |
| Tapasin | AF110520 | NI | 2.6 | 1.2 | 2.8 |
| IFN202 | M31418 | 3.0 | 2.3 | 1.9 | 7.5 |
| IFN204 | M31419 | 2.3 | 4.0 | 2.6 | 6.9 |
| GARG-49 | U43086 | 3.0 | 4.3 | 1.2 | 13.9 |
mRNAs induced in WT or IKKα/β-null cells were identified on Affymetrix MG-U74A arrays after stimulation with IFNγ for 0, 2, or 6 h and confirmed independently. The fold inductions shown are from the arrays. NI, not induced.
Discussion
It is well known that Stat1 plays a major and essential role in IFNγ-dependent signaling and in the consequent biological responses. However, recent evidence indicates that several ancillary pathways are also important. Stat1-independent gene expression plays a role in antiviral responses to IFNγ, and IFNγ enhances proliferation and suppresses apoptosis of cells lacking Stat1 (3). Here, we demonstrate that in addition to activating Stat1, IFNγ engages an additional signal-transduction pathway that involves IKK and is essential for the induction of a subset of IFNγ-stimulated genes. Despite the failure of IFNγ to induce this important subset of IFNγ-stimulated genes in IKKα/β-null MEFs, our data indicate normal, and even enhanced, transcriptional activation of Stat1 in these cells. Therefore, a critical signaling component necessary in addition to Stat1 to activate this subset of IFNγ-stimulated genes is missing in the absence of IKK. We also demonstrate that only kinase-active IKKβ, and not IKKα, is required to restore the IFNγ-stimulated expression of IP10. Future studies are needed to evaluate the separate dependence of additional genes on IKKβ and IKKα.
The proteins encoded by this subset of IFNγ-stimulated genes play important roles in biological responses to IFNγ, including the inflammatory and antiviral responses, the expression of MHC class I, and growth control. IP10, also known as CXCL10, is a CXC chemokine that attracts T lymphocytes and natural killer cells through activation of CXCR3, the only chemokine receptor identified to date that binds to this ligand (40). Low molecular mass protein (LMP) 7 is required for expression of the cell-surface MHC class I–peptide complex, which requires the coordinated expression of MHC class I heavy chain, β2-microglobulin, transporters associated with antigen-processing (TAP)-1, and the proteosome components LMP-2 and LMP-7 (41). Suppressor of cytokine signaling (SOCS) 2 is a member of a family of intracellular proteins involved in the negative regulation of cytokine signaling. The phenotype of SOCS-2-deficient mice, which grow to 1½ times the size of their WT littermates, suggests that SOCS-2 may attenuate growth hormone signaling (42). The Mx proteins are IFN-induced GTPases and have activity against a variety of viruses. The murine Mx1 protein accumulates in the nucleus of IFN-treated cells and is active against members of the Orthomyxoviridae family, such as the influenza viruses (43). The mechanism through which Mx1 exerts its antiviral action is still unclear, but undefined nuclear factors have been postulated to be involved. Nuclear Dbf2-related 1 is a serine/threonine protein kinase belonging to a subfamily of kinases that are involved in regulating cell division and morphology (44). Secreted phosphoprotein 1, or osteopontin, is a ligand with pleomorphic immunologic activities, including the activation of macrophage chemotaxis, promotion of T helper 1 responses, and activation of the B1 subset of B cells (45). It has been implicated in the development of murine lupus and is overexpressed in humans with systemic lupus erythematosus (45). The cytokine-inducible immediate early gene deubiquitinating enzyme 1 is related to members of the UBP superfamily of deubiquitinating enzymes and seems to have growth-suppressing activity (46). The GBPs are a family of 65- to 67-kDa proteins induced by both type I and II IFNs, GBP-2 is believed to contribute to IFNγ-stimulated antiviral activity and regulation of cell proliferation (47, 48).
Phosphorylation of serine-727 of Stat1 in response to IL-1 or TNF, as well as the ability of IFNs to increase the transactivation function of p65NFκB, indicate that the signaling components of these two pathways interact in complex and intricate ways (33, 34, 38). We demonstrated previously that the phosphatidylinositol 3-kinase (PI3K)/AKT pathway plays an important role in activating the IKK complex to phosphorylate p65NFκB, helping to activate NF-κB-dependent gene transcription in response to IL-1 or TNF (49). We also established that the PI3K/AKT pathway is required for the phosphorylation of Stat1 on serine-727 in response to IFNγ and thus for a full transcriptional response (50). Are PI3K and AKT involved in IKK-dependent IFNγ-stimulated gene expression as well? We have not been able to answer this question yet because PI3K/AKT inhibitors and dominant negative mutants block Stat1 serine-727 phosphorylation, which is required in addition to the IKK-dependent signal for the IFNγ-mediated stimulation of these genes. This question can be addressed by using alternative assays to elucidate the mechanisms through which IKK participates in IFNγ-dependent signaling. In general, IFNγ-dependent signaling does not seem to activate NF-κB directly. This conclusion is consistent with our data indicating that the requirement of IKK for IFNγ-dependent signaling is independent of classical NF-κB activation. However, we cannot yet rule out the possibility that an NF-κB subunit other than p65, not inhibited by the overexpression of the IκBαSR, may be involved in this pathway in a way that does not result in the activation of the NF-κB-dependent promoter.
Acknowledgments
This work was supported by National Institutes of Health Grants P01 CA 62220 (to G.R.S.) and R01 CA 100748 and grants from the American Cancer Society and the Ohio Cancer Research Associates (to N.S.).
Abbreviations: IκB, inhibitor of κB; IKK, IκB kinase; MEFs, mouse embryo fibroblasts; Stat1, signal transducer and activator of transcription 1; TNF, tumor necrosis factor; KI, kinase-inactive; GAS, γ-activated sequence; IκBαSR, IκBα super repressor; IP10, IFN-inducible protein 10.
References
- 1.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 2.Meraz, M. A., White, J. M., Sheehan, K. C., Bach, E. A., Rodig, S. J., Dighe, A. S., Kaplan, D. H., Riley, J. K., Greenlund, A. C., Campbell, D., et al. (1996) Cell 84, 431–442. [DOI] [PubMed] [Google Scholar]
- 3.Ramana, C. V., Grammatikakis, N., Chernov, M., Nguyen, H., Goh, K. C., Williams, B. R. & Stark, G. R. (2000) EMBO J. 19, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramana, C. V., Gil, M. P., Han, Y., Ransohoff, R. M., Schreiber, R. D. & Stark, G. R. (2001) Proc. Natl. Acad. Sci. USA 98, 6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil, M. P., Bohn, E., O'Guin, A. K., Ramana, C. V., Levine, B., Stark, G. R., Virgin, H. W. & Schreiber, R. D. (2001) Proc. Natl. Acad. Sci. USA 98, 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma, I. M., Stevenson, J. K., Schwarz, E. M., Van Antwerp, D. & Miyamoto, S. (1995) Genes Dev. 9, 2723–2735. [DOI] [PubMed] [Google Scholar]
- 7.Thanos, D. & Maniatis, T. (1995) Cell 80, 529–532. [DOI] [PubMed] [Google Scholar]
- 8.Siebenlist, U., Franzoso, G. & Brown, K. (1994) Annu. Rev. Cell Biol. 10, 405–455. [DOI] [PubMed] [Google Scholar]
- 9.Baeuerle, P. A. & Henkel, T. (1994) Annu. Rev. Immunol. 12, 141–179. [DOI] [PubMed] [Google Scholar]
- 10.Rothwarf, D. M. & Karin, M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 11.Sizemore, N., Lerner, N., Dombrowski, N., Sakurai, H. & Stark, G. R. (2002) J. Biol. Chem. 277, 3863–3869. [DOI] [PubMed] [Google Scholar]
- 12.Li, X., Massa, P. E., Hanidu, A., Peet, G. W., Aro, P., Savitt, A., Mische, S., Li, J. & Marcu, K. B. (2002) J. Biol. Chem. 277, 45129–45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Q., Van Antwerp, D., Mercurio, F., Lee, K. F. & Verma, I. M. (1999) Science 284, 321–325. [DOI] [PubMed] [Google Scholar]
- 14.Li, Z. W., Chu, W., Hu, Y., Delhase, M., Deerinck, T., Ellisman, M., Johnson, R. & Karin, M. (1999) J. Exp. Med. 189, 1839–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Q., Estepa, G., Memet, S., Israel, A. & Verma, I. M. (2000) Genes Dev. 14, 1729–1733. [PMC free article] [PubMed] [Google Scholar]
- 16.Makris, C., Godfrey, V. L., Krahn-Senftleben, G., Takahashi, T., Roberts, J. L., Schwarz, T., Feng, L., Johnson, R. S. & Karin, M. (2000) Mol. Cell 5, 969–979. [DOI] [PubMed] [Google Scholar]
- 17.Rudolph, D., Yeh, W. C., Wakeham, A., Rudolph, B., Nallainathan, D., Potter, J., Elia, A. J. & Mak, T. W. (2000) Genes Dev. 14, 854–862. [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt-Supprian, M., Bloch, W., Courtois, G., Addicks, K., Israel, A., Rajewsky, K. & Pasparakis, M. (2000) Mol. Cell 5, 981–992. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, M., Fuentes, M. E., Yamaguchi, K., Durnin, M. H., Dalrymple, S. A., Hardy, K. L. & Goeddel, D. V. (1999) Immunity 10, 421–429. [DOI] [PubMed] [Google Scholar]
- 20.Yang, F., Tang, E., Guan, K. & Wang, C. Y. (2003) J. Immunol. 170, 5630–5635. [DOI] [PubMed] [Google Scholar]
- 21.Madrid, L. V., Wang, C. Y., Guttridge, D. C., Schottelius, A. J., Baldwin, A. S., Jr., & Mayo, M. W. (2000) Mol. Cell. Biol. 20, 1626–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madrid, L. V., Mayo, M. W., Reuther, J. Y. & Baldwin, A. S., Jr. (2001) J. Biol. Chem. 276, 18934–18940. [DOI] [PubMed] [Google Scholar]
- 23.Sakurai, H., Chiba, H., Miyoshi, H., Sugita, T. & Toriumi, W. (1999) J. Biol. Chem. 274, 30353–30356. [DOI] [PubMed] [Google Scholar]
- 24.Sakurai, H., Suzuki, S., Kawasaki, N., Nakano, H., Okazaki, T., Chino, A., Doi, T. & Saiki, I. (2003) J. Biol. Chem. 278, 36916–36923. [DOI] [PubMed] [Google Scholar]
- 25.Heissmeyer, V., Krappmann, D., Wulczyn, F. G. & Scheidereit, C. (1999) EMBO J. 18, 4766–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller, J. R. & Siebenlist, U. (2003) J. Biol. Chem. 278, 12006–12012. [DOI] [PubMed] [Google Scholar]
- 27.Mordmuller, B., Krappmann, D., Esen, M., Wegener, E. & Scheidereit, C. (2003) EMBO Rep. 4, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claudio, E., Brown, K., Park, S., Wang, H. & Siebenlist, U. (2002) Nat. Immunol. 3, 958–965. [DOI] [PubMed] [Google Scholar]
- 29.Anest, V., Hanson, J. L., Cogswell, P. C., Steinbrecher, K. A., Strahl, B. D. & Baldwin, A. S. (2003) Nature 423, 659–663. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, Y., Verma, U. N., Prajapati, S., Kwak, Y. T. & Gaynor, R. B. (2003) Nature 423, 655–659. [DOI] [PubMed] [Google Scholar]
- 31.Yang, C. H., Murti, A., Pfeffer, S. R., Kim, J. G., Donner, D. B. & Pfeffer, L. M. (2001) J. Biol. Chem. 276, 13756–13761. [DOI] [PubMed] [Google Scholar]
- 32.Deb, A., Haque, S. J., Mogensen, T., Silverman, R. H. & Williams, B. R. (2001) J. Immunol. 166, 6170–6180. [DOI] [PubMed] [Google Scholar]
- 33.Rani, M. R., Asthagiri, A. R., Singh, A., Sizemore, N., Sathe, S. S., Li, X., DiDonato, J. D., Stark, G. R. & Ransohoff, R. M. (2001) J. Biol. Chem. 276, 44365–44368. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee, M., Agrawal, S. & Agarwal, S. S. (1996) Cytokine 8, 357–364. [DOI] [PubMed] [Google Scholar]
- 35.Benveniste, E. N., Sparacio, S. M. & Bethea, J. R. (1989) J. Neuroimmunol. 25, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malek, T. R., Danis, K. M. & Codias, E. K. (1989) J. Immunol. 142, 1929–1936. [PubMed] [Google Scholar]
- 37.Pujol-Borrell, R., Todd, I., Doshi, M., Bottazzo, G. F., Sutton, R., Gray, D., Adolf, G. R. & Feldmann, M. (1987) Nature 326, 304–306. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, H., Chatterjee-Kishore, M., Jiang, Z., Qing, Y., Ramana, C. V., Bayes, J., Commane, M., Li, X. & Stark, G. R. (2003) J. Interferon Cytokine Res. 23, 183–192. [DOI] [PubMed] [Google Scholar]
- 39.Welch, P. J. & Wang, J. Y. (1993) Cell 75, 779–790. [DOI] [PubMed] [Google Scholar]
- 40.Soejima, K. & Rollins, B. J. (2001) J. Immunol. 167, 6576–6582. [DOI] [PubMed] [Google Scholar]
- 41.Guo, Y., Yang, T., Liu, X., Lu, S., Wen, J., Durbin, J. E., Liu, Y. & Zheng, P. (2002) Int. Immunol. 14, 189–200. [DOI] [PubMed] [Google Scholar]
- 42.Greenhalgh, C. J., Metcalf, D., Thaus, A. L., Corbin, J. E., Uren, R., Morgan, P. O., Fabri, L. J., Zhang, J. G., Martin, H. M., Willson, T. A., et al. (2002) J. Biol. Chem. 277, 40181–40184. [DOI] [PubMed] [Google Scholar]
- 43.Engelhardt, O. G., Ullrich, E., Kochs, G. & Haller, O. (2001) Exp. Cell Res. 271, 286–295. [DOI] [PubMed] [Google Scholar]
- 44.Tamaskovic, R., Bichsel, S. J., Rogniaux, H., Stegert, M. R. & Hemmings, B. A. (2003) J. Biol. Chem. 278, 6710–6718. [DOI] [PubMed] [Google Scholar]
- 45.Kikuchi, K., Tanaka, A., Miyakawa, H., Kawashima, Y., Kawaguchi, N., Matsushita, M. & Gershwin, M. E. (2003) Hepatol. Res. 26, 87–90. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Y., Carroll, M., Papa, F. R., Hochstrasser, M. & D'Andrea, A. D. (1996) Proc. Natl. Acad. Sci. USA 93, 3275–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakash, B., Praefcke, G. J., Renault, L., Wittinghofer, A. & Herrmann, C. (2000) Nature 403, 567–571. [DOI] [PubMed] [Google Scholar]
- 48.Vestal, D. J., Gorbacheva, V. Y. & Sen, G. C. (2000) J. Interferon Cytokine Res. 20, 991–1000. [DOI] [PubMed] [Google Scholar]
- 49.Sizemore, N., Leung, S. & Stark, G. R. (1999) Mol. Cell. Biol. 19, 4798–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen, H., Ramana, C. V., Bayes, J. & Stark, G. R. (2001) J. Biol. Chem. 276, 33361–3368. [DOI] [PubMed] [Google Scholar]