Abstract
Aging is thought to be caused by the accumulation of damage, primarily from oxidative modifications of cellular components by reactive oxygen species (ROS). Here we used yeast methionine sulfoxide reductases MsrA and MsrB to address this hypothesis. In the presence of oxygen, these antioxidants could increase yeast lifespan and did so independent of the lifespan extension offered by caloric restriction. However, under ROS-deficient, strictly anaerobic conditions, yeast lifespan was shorter, not affected by MsrA or MsrB, and further reduced by caloric restriction. In addition, we identified changes in the global gene expression associated with aging in yeast, and they did not include oxidative stress genes. Our findings suggest how the interplay between ROS, antioxidants, and efficiency of energy production regulates the lifespan. The data also suggest a model wherein factors implicated in aging (for example, ROS) may influence the lifespan yet not be the cause of aging.
Aging is typically viewed as the accumulation of damage that results in death. This damage is thought to be caused by reactive oxygen species (ROS). Indeed, ROS can damage biomolecules, accelerate aging, and shorten lifespan (1–3), whereas antioxidant enzymes alleviate these effects, providing support for a free radical theory of aging (4). The contrasting effects of ROS and antioxidants, as well as correlation between accumulation of oxidative damage in cellular components and the lifespan of an organism, have been the main arguments in favor of this theory.
Proteins become oxidatively damaged by oxidation of their amino acid residues and cofactors. Methionine residues in proteins are particularly susceptible to oxidation by ROS, resulting in methionine-S-sulfoxides (Met-S-SO) and methionine-R-sulfoxides (Met-R-SO) (5). Oxidized methionines can be repaired by antioxidant enzymes Met-S-SO reductase (MsrA) and Met-R-SO reductase (MsrB) (6). Recent studies revealed that methionine sulfoxide reduction provides lifespan extension in animals: deletion of the MsrA gene in mice reduced lifespan by ≈40% (7), whereas MsrA overexpression, predominantly in the nervous system, extended fruit fly lifespan by ≈70% (8). The correlation between methionine sulfoxide reductase activity and lifespan has been attributed to antioxidant function of MsrA. The role of MsrB in lifespan regulation has not been previously addressed.
Most known modulators of the rate of aging are conserved across species, suggesting that common, conserved processes regulate and cause aging in diverse organisms. For example, caloric restriction (CR) is a dietary regimen that is known to extend lifespan in organisms from yeast to mammals (3, 9, 10). Saccharomyces cerevisiae has been extensively used as a model organism in studies on the mechanisms of aging (9, 10). Yeast express methionine sulfoxide reductases and can grow both aerobically and anaerobically, and in the present study, we used these features to examine the casual role of ROS-dependent processes in aging.
Materials and Methods
Yeast, Growth, and Media. S. cerevisiae parental strain (referred to as WT) BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and its isogenic MsrA and MsrB mutants lacking methionine sulfoxide reductases (msraΔ, msrbΔ, and msraΔmsrbΔ) were described in ref. 11. The pnc1Δ mutant was kindly provided by D. Sinclair (Harvard Medical School, Boston). Cells were grown in either YPD media (1% yeast extract/2% peptone/2% dextrose) or supplemented YNB (yeast nitrogen base) media containing 0.05 mg/ml required amino acids and methionine sulfoxides. Samples that grew in the presence of methionine sulfoxides did not receive methionine.
Preparation of Recombinant MsrA and MsrB. MsrA (YER042W; MXR1) gene was amplified from yeast genomic DNA by PCR with primers MsrA_F1 and MsrA_B1, which contained NheI and XhoI restriction sites, respectively. The amplified fragment was first cloned into the SmaI site of pBS II KS (Stratagene) and then subcloned into NheI/XhoI sites of bacterial expression vector pet28(a). MsrA protein carrying an N-terminal 6×His tag was purified from BL-21 Escherichia coli cells by using Talon metal affinity resin (Clontech). MsrB (YCL033C) gene was amplified by using primers MsrB_F1 and MsrB_B1 containing NdeI and XhoI restriction sites and cloned, expressed, and purified as described for MsrA. Primer sequences were as follows: MsrA_F1, CTCGCTAGCGTAAATCAAGTTGCAATGTCG; MsrA_B1, CTCCTCGAGCTACATTTCTCTCAGATAATGAG; MsrB_F1, CATATGGAATAAGTGGAGCAGGCTGTAC; and MsrB_B1, CTCGAGCATTTAGTCCTTCTTGAGGTTTAAAG.
Overexpression of MsrA and MsrB in Yeast. Both MsrA and MsrB genes were subcloned from corresponding pBS constructs to BamHI/XhoI sites of p423-GPD high copy yeast expression vector. Yeast cells were transformed with the plasmids by a standard lithium acetate method, and the transformants were selected for histidine prototrophy on supplemented YNB media.
Possible Additional Component in the Yeast Methionine Sulfoxide Reduction Pathway. The ability of msraΔmsrbΔ double mutant cells to grow on Met-R-SO (Figs. 7 and 8, which are published as supporting information on the PNAS web site) suggested the presence of an additional (unknown) MsrB activity in yeast cells.
Methionine Sulfoxide Reductase Activity. The ability of yeast MsrA to reduce protein-bound Met-S-SO and the ability of MsrB to reduce Met-R-SO was determined by the reduction of dabsylated methionine sulfoxides to dabsylated methionine with an HPLC assay as described in ref. 11. Reaction mixtures had 5 μg of purified enzymes and 25–400 μM methionine sulfoxide substrates, and were incubated for 30 min before HPLC analysis. Met-S-SO and Met-R-SO were separated from l-methionine-R,S-sulfoxide according to the method of Lavine (12), and its purity (>95% for each sulfoxide) was assessed by an HPLC procedure as described in ref. 11.
To assay for Met-S-SO and Met-R-SO activities in parental cells and in cells overexpressing MsrA or MsrB, yeast cells harboring p423 (empty vector), pMsrA, or pMsrB were cultured in YNB containing either 2% or 0.5% glucose at 30°C for 22 h. Crude extracts were prepared with glass beads, followed by sonication. Enzyme activities were assayed by using dabsylated L-Met-S-SO or L-Met-R-SO as substrates. The reaction mixture (100 μl) contained 50 mM sodium phosphate (pH 7.5), 20 mM DTT, 200 μM substrate, and 200 μg of crude extracts. The reaction was carried out at 37°C for 60 min and stopped by adding 200 μl of acetonitrile, and products were assayed by the HPLC procedure (11).
Yeast Aging Assay. Cells were kept growing on fresh media for 2 days before analysis. For each strain, 35 daughter cells (starter mothers) were collected and lined up by a micromanipulator on agar plates. New buds (daughters) from these virgin cells were removed and discarded as they formed. This process continued until cells ceased dividing. Lifespan was determined as the total number of daughter cells that each mother cell generated. In experiments involving parental cells and deletion mutants, aging was assayed by using cells grown on YPD, whereas cells overexpressing MsrA or MsrB were assayed by using YNB media. This media factor contributed to differences in lifespan. Therefore, lifespan comparisons could only be made for cells grown on the same medium.
Anaerobic aging assays were performed as described above except that they were performed in an anaerobic glove chamber (Coy Laboratory Products, Ann Arbor, MI) operating at less than 1 ppm of oxygen. Supplies and cells were incubated for 24 h under anaerobic conditions before aging experiments.
RNA Isolation from Old Cells. Large-scale isolation of old yeast cells was described in detail by Park et al. (13). This method is based on the observation that the cell surface of new daughter cells is made de novo at the budding site. Briefly, logarithmically growing cells (20-ml culture) were harvested and washed with 1× PBS and incubated with 15 mg of sulfo-NHS-LC-biotin (Pierce) in 2 ml of PBS buffer for 10 min at room temperature to label the cell surface. After washing with PBS, cells were grown in YPD at 30°C until 1.5 OD600 was reached. Labeled old cells were sorted by 1 ml of streptavidin-coated magnetic beads (PerSeptive Biosystems, Framingham, MA). To determine the age of sorted cells, bud scars of samples were stained with Calcoflour white M2R (fluorescent brightener 28, Sigma) and counted under UV fluorescence. By using this method, ≈10 million cells were collected that were 8–12 generations old (designated as 10-generation-old cells). The sorting procedure was repeated twice to obtain the same amount of 18- to 24-generation-old cells (designated as 20-generation-old cells). To extract RNA, cells were incubated with PBS containing 5% β-glucoronidase-HP-2S (Sigma) for 5 min at room temperature to digest the cell wall. For cell lysis and total RNA isolation, the PicoPure RNA isolation system (Arcturus, Mountain View, CA) was used to purify ≈5 μg of total RNA from 10 million cells. mRNAs were selected and amplified by RiboAmp mRNA amplification system (Arcturus) for DNA microarray analysis. Logarithmically growing cells that were either biotinylated or unlabeled were also prepared and used as controls.
DNA Microarray Analysis. Cells were grown to 0.8 OD600 in 200 ml of medium under indicated conditions, harvested by centrifugation, and kept at –80°C. Total cellular RNA (≈2 mg) was isolated by the hot phenol method, and mRNA was isolated from total RNA by using the PolyATtract mRNA Isolation System IV kit (Promega) according to the manufacturer's instructions. mRNA (1–2 μg) was used to generate cDNA probes by reverse transcription (Superscript II, GIBCO) with incorporation of Cy3-dCTP or Cy5-dCTP (Amersham Biosciences). The arrayed slides were produced as follows. The PCR amplification products of all S. cerevisiae ORFs were purchased from Research Genetics (Huntsville, AL). Each PCR product was reamplified and purified on glass fiber filter plates (Millipore). The reamplified products representing ≈6,000 ORFs (4% not represented) were diluted to 50% DMSO and spotted in duplicate on 3-aminopropyl-methyldiethoxy silane-coated slides by using a Generation III Microarray Spotter or a Lucidea Array Spotter (Amersham Biosciences). The cDNA-labeled probes were then hybridized onto an arrayed slide, and fluorescence was captured with a GEN III dual-laser confocal scanner (Molecular Dynamics) or a GenePix4000B Scanner (Axon Instruments, Foster City, CA). Fluorescent intensities were quantified by using arrayvision 6.0 (Imaging Research, St. Catherine's, ON, Canada) or imagene 5.5 (BioDiscovery, El Segundo, CA).
Clustering Methods. DNA microarray data were mean centered and hierarchically clustered by using cluster (14), using array weights calculated with a cutoff of 0.8 and an exponent 1, as described in the cluster manual (http://rana.lbl.gov/manuals/ClusterTreeView.pdf).
Identification of Genes Affected During Aging. To identify genes whose expression was changed during aging, the duplicate data for each experiment were averaged, the 10- and 20-generation-old cells were averaged, and then genes for which the averaged old data were 1.8-fold different (either higher or lower) in expression compared to the control cells were selected. This identified 150 genes that were more strongly affected in aged cells or that were more strongly affected in the control.
Results and Discussion
Homology analyses revealed that S. cerevisiae has single MsrA (MXR1, YER042W) and MsrB (YCL033C) genes (11). We cloned and prepared recombinant forms of these proteins and found that, as expected, MsrA stereospecifically reduced dabsylated Met-S-SO and MsrB dabsylated Met-R-SO in in vitro assays (Fig. 1). Dabsylated methionine sulfoxides mimic methionine sulfoxide residues in proteins, in contrast to free methionine sulfoxides, which are also reduced by methionine sulfoxide reductases, but at much slower rates. Overexpression of MsrA in a parental strain grown in YNB resulted in the specific increase in dabsyl-MsrA activity, whereas overexpression of MsrB specifically increased dabsyl-MsrB activity (Table 1).
Fig. 1.
Stereospecific activities of yeast MsrA and MsrB. (Upper) MsrA activity of recombinant yeast MsrA assayed as reduction of dabsylated l-methionine-S-sulfoxide (Met-S-SO) to dabsylated methionine. (Lower) MsrB activity of recombinant yeast MsrB assayed as reduction of dabsylated l-methionine-R-sulfoxide (Met-R-SO) to dabsylated methionine. DTT-dependent activities were detected by absorbance at 436 nm in an HPLC procedure. Locations of dabsylated methionine sulfoxide substrates (Met-S-SO and Met-R-SO) and dabsylated methionine product (Met) are indicated.
Table 1. MsrB and MsrA activities in yeast cells overexpressing MsrA or MsrB.
| MsrA activity, pmol/min per mg of crude protein
|
MsrB activity, pmol/min per mg of crude protein
|
|||
|---|---|---|---|---|
| Strain | 2% Glucose | 0.5% Glucose | 2% Glucose | 0.5% Glucose |
| Vector only | 7.5 | 6.6 | ND | ND |
| pMsrA | 34.2 | 28.6 | ND | ND |
| pMsrB | 9.4 | 5.0 | 44.4 | 44.0 |
Parental cells expressing either vector (Vector only), MsrA (pMsrA), or MsrB (pMsrB) were grown on either 2% glucose or under CR conditions (0.5% glucose), and MsrA and MsrB activities were determined by using dabsylated methionine sulfoxides as substrates. ND, not detectable.
Contributions of MsrA and MsrB to the reduction of dabsylated methionine sulfoxides were markedly different. Recombinant MsrA was 11-fold more active with dabsyl-Met-S-SO (specific activity 54.7 nmol/min/mg of protein) than recombinant MsrB with dabsyl-Met-R-SO (specific activity 4.9 nmol/min/mg protein). In addition, MsrA was required for efficient in vivo reduction of free methionine sulfoxides by yeast cells, whereas MsrB did not contribute to this process, unless yeast lacked the MsrA gene (Figs. 7 and 8). Taken together, these data suggested that MsrA was a major contributor to the availability of methionine from free methionine sulfoxides and to the reduction of protein methionine sulfoxides in yeast cells, whereas MsrB played a minor role in these processes. The fact that the MsrA/MsrB double mutant grew on Met-R-SO (Fig. 8C) suggested the presence of a minor MsrB activity in yeast. This finding was also consistent with the observation of additional methionine sulfoxide reductase activities in E. coli (15, 16).
Following this initial characterization of the yeast methionine sulfoxide reduction pathway, we tested the effects of deletion and overexpression of MsrA and MsrB genes on the replicative lifespan of yeast cells (Fig. 2 and Tables 2 and 3), determined as the number of daughter cells produced by each mother cell (9, 10). Deletion of the MsrA gene shortened lifespan by 26% (Fig. 2A), and overexpression increased it by 25% (Fig. 2B). Deletion or overexpression of MsrB had little effect on the lifespan; however, the lifespan of the msraΔmsrbΔ double mutant was further reduced compared to that of msraΔ cells. Thus, under standard growth conditions (aerobic growth in the presence of 2% glucose), both proteins contributed to longevity, with MsrA being the major contributor to regulation of yeast lifespan by methionine sulfoxide reduction. The fact that the increased capacity for methionine sulfoxide reduction can increase lifespan in such diverse organisms as fruit flies (8) and yeast suggests evolutionary conservation of this function in aging.
Fig. 2.
Effects of methionine sulfoxide reductases and CR on yeast lifespan under aerobic and anaerobic conditions. (A) Lifespan analyses of parental (shown as WT) cells and isogenic cells lacking MsrA (msraΔ) or MsrB (msrbΔ) genes grown on YPD (contains 2% glucose) or on YPD under CR conditions (0.5% glucose). (B) Lifespan analyses of parental (WT) cells overexpressing p423GPD vector only (v only), MsrA (pMsrA), or MsrB (pMsrB) grown on supplemented YNB (2% glucose) or corresponding CR media. (C) Lifespan analyses of parental (WT) msraΔ and msrbΔ cells grown on YPD and parental cells grown on the CR medium under anaerobic conditions. (D) Lifespan analyses of parental (WT) cells with vector only, pMsrA, and pMsrB grown on YNB and parental cells with vector only and pMsrB grown on the CR medium under anaerobic conditions. Experiments shown in A and C were performed with cells grown on YPD (see Table 2), whereas those shown in B and D were performed with cells grown on YNB media (see Table 3). This media factor contributes to differences in lifespans of parental strains (compare data shown A and C to data shown in B and D).
Table 2. Lifespan analyses of parental (WT), msraΔ, msrbΔ, and msraΔmsrbΔ cells grown on 2% glucose (YPD) or 0.5% glucose (caloric restriction medium, CR) under aerobic and anaerobic conditions.
| Aerobic conditions
|
Anaerobic conditions
|
|||
|---|---|---|---|---|
| Strain | 2% glucose | 0.5% glucose (CR) | 2% glucose | 0.5% glucose (CR) |
| WT | 23.5 | 25.9 | 15.3 | 12 |
| msraΔ | 17.4 | 22.8 | 14.6 | ND |
| msrbΔ | 22.7 | 27.4 | 15.7 | ND |
| msraΔmsrbΔ | 8.7 | 17.7 | ND | ND |
ND, not determined. Lifespans (average number of daughter cells produced by mother cells) were calculated from the data shown in Fig. 2 A and C.
Table 3. Lifespan analyses of parental (WT) cells expressing an empty vector, MsrA (pMsrA), or MsrB (pMsrB) grown on 2% glucose (supplemented YNB) or 0.5% glucose (caloric restriction medium, CR) under aerobic and anaerobic conditions.
| Aerobic conditions
|
Anaerobic conditions
|
|||
|---|---|---|---|---|
| Strain | 2% glucose | 0.5% glucose (CR) | 2% glucose | 0.5% glucose (CR) |
| Vector only | 10.9 | 14.8 | 12 | 7.4 |
| pMsrA | 13.6 | 16 | 11.6 | ND |
| pMsrB | 12.1 | 23.9 | 10.3 | 6.8 |
ND, not determined. Lifespans (average number of daughter cells produced by mother cells) were calculated from the data shown in Fig. 2 B and D.
We next addressed the role of MsrA and MsrB in yeast longevity under CR conditions (growth in the presence of 0.5% glucose) (17) (Fig. 2 A and B and Tables 2 and 3). We found that CR did not abrogate the effects of MsrA deletion or overexpression: although CR extended the lifespan of all strains tested, the lifespan of the msraΔ strain was shorter than the lifespan of the parental strain in both standard and CR growth conditions. Similarly, the lifespan of cells was extended by MsrA overexpression (compared to the parental cells expressing an empty vector) when cells were grown in either standard or CR media. Thus, the influence of MsrA on yeast lifespan was independent of that of CR.
Although MsrB had no effect on the lifespan under standard growth conditions, overexpression of MsrB extended lifespan by 62% under CR conditions. In addition, CR prolonged the lifespan of both msrbΔ mutant (21%) and MsrB overexpressing (97%) cells. Interestingly, the combination of MsrB overexpression and CR extended the lifespan by 119%. Thus, CR and MsrB overexpression could synergistically extend the yeast lifespan. Cells missing both MsrA and MsrB genes showed a 104% increase in their life span by CR.
To better understand the relationship between lifespan extensions by CR and MsrB overexpression, we analyzed cells lacking pyrazinamidase/nicotinamidase 1 (PNC1) gene. PNC1 was recently shown to govern lifespan extension by CR (18). However, the extension of the lifespan by MsrB overexpression under CR conditions was still evident in pnc1Δ mutant cells (data not shown). Overall, these data further suggested that methionine sulfoxide reductases and CR could independently extend yeast lifespan.
The lifespan extensions offered by MsrA and MsrB were not due to increased supply of methionine. Indeed, the lifespan of yeast cells grown on low methionine, or even on methionine sulfoxide, was slightly higher compared to cells grown on standard concentrations of methionine. Furthermore, an increase in methionine concentration in the media above normal decreased the lifespan (Fig. 3).
Fig. 3.
Lifespan of yeast cells grown in the presence of different concentrations of methionine in the medium. Replicative yeast aging assay was performed to determine whether availability of methionine could alter lifespan. Parental cells were grown and analyzed on YNB media, including all supplements. The concentration of methionine was changed from 0.005 mg/ml to 0.5 mg/ml (1×= 0.05 mg/ml), but all of the other supplements were kept at the same concentration. Average life spans were 23.6 generations for 0.1×, 21 generations for 0.5×, 18.3 generations for 1×, 18 generations for 2×, 17 generations for 10×, and 22.5 generations for 1× methionine-R,S-sulfoxide.
ROS are viewed as by-products of aerobic metabolism, and the free radical theory of aging proposed that ROS are the cause of aging (2, 4). The effects of MsrA and MsrB on the lifespan of yeast cells during aerobic growth (Fig. 2 A and B) were consistent with the idea that ROS decrease the lifespan, whereas antioxidants increase it. If so, cells characterized by the lack of ROS would be expected to live longer. To examine this issue, we studied the lifespan of parental BY4741 cells, as well as cells deficient in and overexpressing MsrA or MsrB, under strictly anaerobic conditions, where oxygen concentration was controlled at below detection limit (<1 ppm). Surprisingly, we found that under these ROS-deficient conditions parental cells exhibited a 35% decline in the lifespan (Fig. 2C) compared to cells grown in air (Fig. 2A).
Under anaerobic conditions, oxidative damage to cellular components is lower, and expression of major antioxidant genes, such as catalases CTA1 and CTT1, is decreased (19). Consistent with the lack of ROS and diminished role of methionine sulfoxide reductases in protection against oxidative stress under anaerobic conditions, neither deletion nor overexpression of MsrA or MsrB had any effect on the lifespan (Fig. 2 C and D).
We further examined how CR affects the lifespan under anaerobic conditions. Unexpectedly, we observed that under anaerobic conditions, CR decreased lifespans of all strains tested by 22–38% (Fig. 2 C and D). The decreased lifespan observed under anaerobic CR conditions highlights the interplay between efficiency of energy production, oxidative stress, and antioxidant defense in regulating the aging process. CR has been thought to extend yeast lifespan by decreasing metabolic rate, but a recent study revealed that it extends the lifespan by increasing respiration (17). Because respiration depends on oxygen, anaerobic yeast cells lose the choice of shifting metabolic fluxes between respiration and glycolysis to produce energy.
Thus, a lower metabolic rate due to reduced glucose in a growth medium appears to decrease lifespan, whereas a more efficient energy production through respiration appears to increase lifespan. Furthermore, aerobic metabolism is accompanied by the production of ROS, which decrease lifespan via damage caused by various oxidative modifications, such as methionine sulfoxidation. However, antioxidant enzymes, such as MsrA and MsrB, can counteract the toxic levels of ROS and therefore extend lifespan by repairing ROS targets (or eliminating ROS). Taken together, the data suggest a model wherein a ROS-independent pathway(s) causes aging, the rate of which can be modulated by efficiency of energy production, ROS, and antioxidants. For example, CR might increase lifespan because the longevity offered by respiration outweighs the detrimental effect of ROS damage. Under anaerobic conditions, ROS and antioxidants do not play major roles in lifespan regulation, and CR decreases lifespan because of reduction in energy source under conditions where the compensatory increase in the efficiency of energy production is not possible.
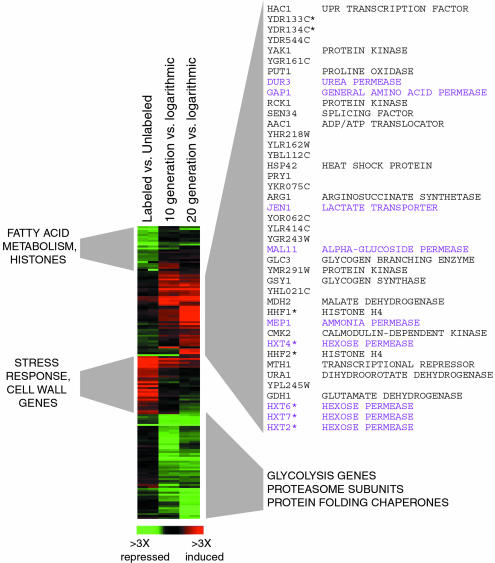
To understand the changes that occur during aging in yeast, we prepared, by a protocol involving biotinylation of cell membranes (13), young, medium old (10 generation), and old (20 generation) yeast cells and compared their gene expression patterns. Surprisingly, gene expression was little affected by the age of cells: only 150 genes (≈2.5% of all genes) showed ≥2 fold difference in expression between young and old cells (the full set can be found in Data Set 1, which is published as supporting information on the PNAS web site). We found that several genes involved in glycolysis and protein synthesis, folding, and degradation were repressed in old cells, whereas several transporter genes were induced (Fig. 4). In addition, we found that biotinylation resulted in decreased expression of several fatty acid metabolism and histone genes and increased expression of stress response and cell wall biogenesis genes, and that the expression of these genes normalized in old cells. The altered gene expression in aged cells is consistent with decreased flux through glycolysis, reduced protein turnover, more active utilization of transport systems, and, possibly, reduced stress response. We suggest that this gene expression pattern could serve as a signature of aging in yeast cells. It should be noted that this signature did not include oxidative stress response genes, or MsrA and MsrB genes, because expression of these genes was not changed during aging (Figs. 5 and 9, which is published as supporting information on the PNAS web site). Previously, gene expression changes and major metabolic pathways were assessed in yeast cells at generation 7–8 (compared to generation 0–1), revealing increased gluconeogenesis and energy storage (20).
Fig. 4.
Genes whose expression is affected by aging in yeast. Genes whose average expression in duplicate experiments was ≥2-fold different in old cells (20 generations) or in both 10- and 20-generation-old cells relative to the logarithmically growing cells are displayed. Ten- and 20-generation-old cells were isolated by a protocol involving biotinylation of cell membranes (see Materials and Methods). The labeled versus unlabeled data show a control experiment, in which biotinylated young cells were compared to young unlabeled cells. Annotations for the differentially expressed genes are shown next to the gene expression diagram. Genes that are highly similar and therefore could cross-hybridize on the microarrays are annotated with an asterisk. Clusters of genes whose expression was elevated (shown in red) or decreased (shown in green) during aging are shown on the left and right. Genes with elevated expression in aging cells are listed. Of these, genes involved in transport are shown in purple and other genes are shown in black.
Fig. 5.
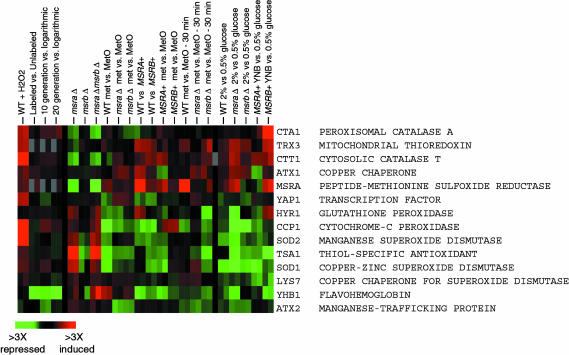
Expression of oxidative stress response genes is not affected during aging in yeast. Genes annotated by the Saccharomyces Genome Database as involved in oxidative stress are shown. Their expression was compared in parental (WT) cells versus same cells treated with 100 μM H2O2 for 30 min (WT + H2O2), parental versus msraΔ cells (msra), parental versus msrbΔ cells (msrb), parental versus msraΔmsrbΔ cells (msra msrb), parental cells grown in the presence of Met versus same cells grown in the presence of Met-R,S-SO (WT met vs. MetO), msraΔ cells grown in the presence of methionine versus same cells grown in the presence of Met-R,S-SO (msra met vs. MetO), msrbΔ cells grown in the presence of methionine versus same cells grown in the presence of Met-R,S-SO (msrb met vs. MetO), parental cells expressing vector versus parental cells expressing MsrA (WT vs. MSRA+), parental cells expressing vector versus parental cells expressing MsrB (WT vs. MSRB+), parental cells expressing MsrA grown on methionine versus same cells grown on Met-R,S-SO (MSRA+ met vs. MetO), parental cells expressing MsrB grown on methionine versus same cells grown on Met-R,S-SO (MSRB+ met vs. MetO), parental cells grown on methionine and then treated with methionine for 30 min versus same cells treated with Met-R,S-SO (WT met vs. MetO - 30 min), msraΔ cells grown on methionine and then treated with methionine for 30 min versus same cells treated with Met-R,S-SO (msra met vs. MetO - 30 min), msrbΔ cells grown on methionine and then treated with methionine for 30 min versus same cells treated with Met-R,S-SO (msrb met vs. MetO - 30 min), parental cells grown on standard 2% glucose versus same cells grown under 0.5% glucose CR conditions (WT 2% vs. 0.5% glucose), msraΔ cells grown on 2% glucose versus same cells grown under 0.5% glucose CR conditions (msraΔ 2% vs. 0.5% glucose), msrbΔ cells grown on 2% glucose versus same cells grown under 0.5% glucose CR conditions (msrbΔ 2% vs. 0.5% glucose), parental cells expressing MsrA and grown on 2% glucose versus same cells grown on 0.5% glucose (MSRA+ YNB vs. 0.5% glucose), and parental cells expressing MsrB and grown on 2% glucose versus same cells grown on 0.5% glucose (MSRB+ YNB vs. 0.5% glucose). In addition, aging microarray data (as described in Fig. 4) are shown for the oxidative stress response genes.
To obtain better insights into gene expression patterns characteristic of aging and methionine sulfoxide reduction, we also analyzed gene expression under 22 conditions relevant to these processes (Data Set 1). Deletion of MsrA or MsrB genes (or both), overexpression of MsrA or MsrB, CR, hydrogen peroxide treatment, and short- or long-term treatment of parental or mutant cells with methionine sulfoxide resulted in significant changes in gene expression, including oxidative stress response genes (Figs. 5 and 9). However, these expression changes did not correlate with the effects each treatment had on lifespan, underscoring that the expression of oxidative stress defense genes is not a signature of aging.
We also analyzed previous microarray data of yeast cells responding to various stresses (21, 22) and found that the MsrA gene clustered with methionine biosynthesis genes (Fig. 6A). In our data set, MsrA did not cluster with these genes (Fig. 6B), because of the expression differences specific to this gene due to its deletion or overexpression. However, methionine biosynthesis genes had an elevated expression in msraΔ mutant cells grown in the presence of methionine sulfoxide, consistent with MsrA providing methionine to yeast cells. Taken together, our data suggest that MsrA functions in both antioxidant defense and methionine biosynthesis. In contrast, MsrB clustered neither with known antioxidant genes nor with methionine biosynthesis genes, and was not required for these processes unless the MsrA gene was absent, suggesting that MsrB plays a minor role in antioxidant defense and methionine biosynthesis under standard growth conditions.
Fig. 6.
MsrA is a member of methionine biosynthesis cluster and contributes to providing cells with methionine. (A) The data set of parental cells responding to diverse stressful conditions (indicated on top of the figure) was analyzed, and the top 23 genes that are most closely correlated to MsrA gene expression are shown. The genes are organized by descending Pearson correlation with MsrA first. The genes identified are either known to be involved in Met biosynthesis or sulfur fixation or thought to be induced by the methionine transcription factors. Some genes are also involved in the cellular response to nitrogen limitation, and the function of several additional genes is not known. (B) Expression of genes in yeast methionine biosynthesis cluster is shown at various conditions and mutations relevant to methionine sulfoxide reduction pathway. Gene expression was examined in samples that are described in the legend to Fig. 5.
In conclusion, our data revealed that the rate of aging in S. cerevisiae can be modulated by oxidants and antioxidants of the methionine sulfoxide reduction pathway. This finding is consistent with the involvement of ROS and antioxidants in regulation of aging under aerobic conditions. However, this effect is not equivalent to the cause of aging, if defined as factors (or processes) that are both necessary and sufficient for aging. For example, if (hypothetically) aging in yeast is caused by ROS-independent senescence factors (23), and protein(s) that function by repressing their synthesis have essential methionine residues, ROS could influence aging by generating methionine sulfoxides in these proteins, thus inhibiting their function. Consequently, methionine sulfoxide reductases would increase lifespan by repairing methionine residues in these proteins.
However, under anaerobic conditions, methionine sulfoxide reductases would not have the modulatory effect on the lifespan because, in the absence of ROS, methionine sulfoxides would not be formed in target proteins. Instead, these proteins might be influenced by other cellular components (resulting in a shorter lifespan under anaerobic conditions than in air), and the hypothetical ROS-independent senescence factors would still cause aging. Thus, the modulatory effects of MsrA and MsrB (and by analogy certain other antioxidant enzymes) on the yeast lifespan are not equivalent to ROS being the cause of aging. Instead, our data suggest that other, unknown processes cause aging in yeast and that these processes can be modulated by ROS and antioxidants.
Supplementary Material
Acknowledgments
We thank the University of Utah Microarray Core Facility, led by Dr. Brian Dalley and supported by the Huntsman Cancer Foundation, for excellent technical assistance; Dennis Winge for helpful comments and support; and Audrey Atkin and Stephen Ragsdale for the use of equipment. This work was supported by National Institutes of Health Grant AG021518 (to V.N.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CR, caloric restriction; Met-R-SO, methionine-R-sulfoxide; Met-S-SO, methionine-S-sulfoxide; MsrA, Met-S-SO reductase; MsrB, Met-R-SO reductase; ROS, reactive oxygen species; YNB, yeast nitrogen base; YPD, 1% yeast extract/2% peptone/2% dextrose.
References
- 1.Stadtman, E. R. (1992) Science 257, 1220–1224. [DOI] [PubMed] [Google Scholar]
- 2.Sohal, R. S. & Weindruch, R. (1996) Science 273, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel, T. & Holbrook, N. J. (2000) Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- 4.Harman, D. J. J. (1956) Gerontology 11, 298–300. [DOI] [PubMed] [Google Scholar]
- 5.Levine, R. L., Moskovitz, J. & Stadtman, E. R. (2000) IUBMB Life 50, 301–307. [DOI] [PubMed] [Google Scholar]
- 6.Weissbach, H., Etienne, F., Hoshi, T., Heinemann, S. H., Lowther, W. T., Matthews, B., St. John, G., Nathan, C. & Brot, N. (2002) Arch. Biochem. Biophys. 397, 172–178. [DOI] [PubMed] [Google Scholar]
- 7.Moskovitz, J., Bar-Noy, S., Williams, W. M., Requena, J., Berlett, B. S. & Stadtman, E. R. (2001) Proc. Natl. Acad. Sci. USA 98, 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan, H., Tang, X. D., Chen, M. L., Joiner, M. L., Sun, G., Brot, N., Weissbach, H., Heinemann, S. H., Iverson, L., Wu, C. F. & Hoshi, T. (2002) Proc. Natl. Acad. Sci. USA 99, 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tissenbaum, H. A. & Guarente, L. (2002) Dev. Cell 2, 9–19. [DOI] [PubMed] [Google Scholar]
- 10.Jazwinski, S. M. (2002) Annu. Rev. Microbiol. 56, 769–792. [DOI] [PubMed] [Google Scholar]
- 11.Kryukov, G. V., Kumar, R. A., Koc, A., Sun, Z. & Gladyshev, V. N. (2002) Proc. Natl. Acad. Sci. USA 99, 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavine, T. F. (1947) J. Biol. Chem. 169, 477–491. [PubMed] [Google Scholar]
- 13.Park, P. U., McVey, M. & Guarente, L. (2002) Methods Enzymol. 351, 468–777. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne, F., Spector, D., Brot, N. & Weissbach, H. (2003) Biochem. Biophys. Res. Commun. 300, 378–382. [DOI] [PubMed] [Google Scholar]
- 16.Spector, D., Etienne, F., Brot, N. & Weissbach, H. (2003) Biochem. Biophys. Res. Commun. 302, 284–289. [DOI] [PubMed] [Google Scholar]
- 17.Lin, S. J., Kaeberlein, M., Andalis, A. A., Sturtz, L. A., Defossez, P. A., Culotta, V. C., Fink, G. R. & Guarente, L. (2002) Nature 418, 344–348. [DOI] [PubMed] [Google Scholar]
- 18.Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O. & Sinclair, D. A. (2003) Nature 423, 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper, M. D., Daran-Lapujade, P., Bro, C., Regenberg, B., Knudsen, S., Nielsen, J. & Pronk, J. T. (2002) J. Biol. Chem. 277, 37001–37008. [DOI] [PubMed] [Google Scholar]
- 20.Lin, S. S., Manchester, J. K. & Gordon, J. I. (2001) J. Biol. Chem. 276, 36000–36007. [DOI] [PubMed] [Google Scholar]
- 21.Gasch, A. P. & Eisen, M. B. (2002) Genome Biol. 3, RESEARCH0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinclair, D. A. & Guarente, L. (1997) Cell 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.