SUMMARY
One of the most powerful ways to test the function of a protein is to characterize the consequences of its deletion. In the past, this has involved inactivation of the gene by homologous recombination either in the germline or later through conditional deletion. RNA interference (RNAi) provides an alternative way to knock down proteins, but both of these approaches have their limitations. Recently, the CRISPR/Cas9 system has suggested another way to selectively inactivate genes. We have now tested this system in postmitotic neurons by targeting two well-characterized synaptic proteins, the obligatory GluN1 subunit of the NMDA receptor and the GluA2 subunit of the AMPA receptor. Expression of CRISPR/Cas9 in hippocampal slice cultures completely eliminated NMDA receptor and GluA2 function. CRISPR/Cas9 thus provides a powerful tool to study the function of synaptic proteins.
INTRODUCTION
Historically, pharmacology provided most of the initial insight into function of central synapses. Selective antagonists for neurotransmitter receptors, which have the advantage of rapid and reversible actions, established the role of AMPA and NMDA receptors in synaptic transmission and plasticity (Collingridge et al., 1983; Hestrin et al., 1990; Watkins et al., 1990). For most of the myriad proteins in the postsynaptic density, however, a pharmacological approach has not been possible. The introduction of gene targeting in embryonic stem (ES) cells allowed the production of mice in which specific genes are deleted (Capecchi, 1989; Silva et al., 1992). This technology enabled many advances in vertebrate biology including neuroscience but suffers from two limitations. First, the gene deletion is global and in many cases lethal. Second, the deletion occurs in the germline, raising the possibility of compensation during development. The introduction of conditional knockout (KO) technology has circumvented some of these problems (Adesnik et al., 2008; Plück, 1996; Sauer and Henderson, 1988; Tsien et al., 1996). In this approach, the selective expression of Cre recombinase restricts gene deletion to those cells expressing Cre. This approach nonetheless requires the production of conditional KO mice, involving considerable time and expense. RNA interference (RNAi) provides a more facile way to knock down protein expression (Ehrlich and Malinow, 2004; Elias et al., 2006; Fire et al., 1998; Futai et al., 2007; Hannon, 2002) but has its own limitations. Off-target effects can occur (Alvarez et al., 2006; Persengiev et al., 2004). Although this problem can be addressed by rescuing the loss of function with an RNAi-resistant construct, the lack of effect in an animal lacking the gene is even more definitive. More important, however, RNAi rarely eliminates the protein, making it difficult to determine whether any residual function reflects residual protein or an unrelated process.
New genome editing approaches have recently been developed that have the potential to circumvent many of the problems associated with homologous recombination in ES cells and RNAi (Wijshake et al., 2014). These include TALENs, zinc finger nucleases, and more recently clustered regularly interspaced short palindromic repeats (CRISPR). Originally discovered in bacteria as an adaptive immune defense mechanism against viral attack, CRISPR has now been developed for gene editing, such as deleting, silencing, enhancing, or changing specific genes in eukaryotes (Canver et al., 2014; Cong et al., 2013; Hsu et al., 2014; Mali et al., 2013; Perez-Pinera et al., 2013; Qi et al., 2013; Yang et al., 2013). The expression of nuclease Cas9 along with specifically designed guide RNAs enables the genome to be cut at virtually any location (Ran et al., 2013b).
In previous work, we and others have used genetic manipulation in single cells of hippocampal slice culture to study the role of specific postsynaptic proteins in synaptic transmission (Adesnik et al., 2008; Hayashi et al., 2000; Schnell et al., 2002). This approach requires the sparse transfection of pyramidal neurons, by either biolistics or viral infection. After a period of time, the afferent fibers to a transfected cell and untransfected neighbor (as control) are stimulated and the postsynaptic responses compared. The direct simultaneous comparison of transfected and control cells increases the power of this system to identify functionally important differences.
To assess the ability of the CRISPR/Cas9 system to delete specific synaptic proteins, we focused on two proteins easily amenable to measurement. Excitatory synapses express two types of glutamate receptor: NMDA receptors (NMDARs) and AMPA receptors (AMPARs). NMDARs are composed of two essential GluN1 subunits and two GluN2 subunits. After the removal of GluN1 by CRISPR/Cas9, we indeed detected no NMDAR-mediated currents. AMPARs are composed of four subunits, GluA1–GluA4, that contribute to the formation of heterotetrameric receptors. Most AMPARs contain the GluA2 subunit, which has a profound effect on the biophysical properties of the receptor: in the presence of GluA2, the receptor is impermeable to calcium and the current-voltage (IV) relationship is linear; in the absence of GluA2, AMPA receptors are permeable to calcium and exhibit strong inward rectification (Jonas and Burnashev, 1995). We have thus used the I-V relationship to assess GluA2 subunit content after inactivation of GluA2 by CRISPR/Cas9.
RESULTS
Deletion of GluN1
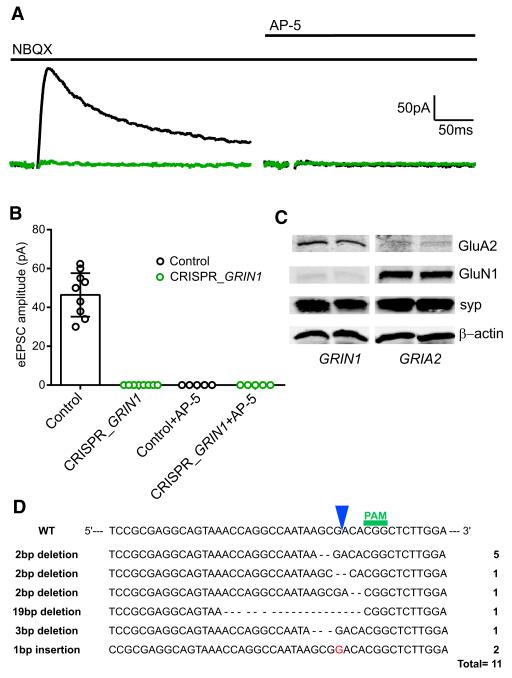
We first coated gold particles with a vector encoding Cas9 as well as guide RNAs (gRNAs) targeting the GluN1 subunit of the NMDAR (see Experimental Procedures). These particles were biolistically delivered to hippocampal slice cultures and recordings made 14 days later from a transfected cell and simultaneously from a neighboring control cell. The AMPAR antagonist NBQX was used so that NMDAR-mediated excitatory postsynaptic currents (NMDAR EPSCs) could be recorded in isolation. Figure 1A shows a sample pair of recordings. Stimulation elicited a large NMDAR EPSC in the control cell (black trace) but no current in the transfected cell (green trace). Subsequent application of the NMDAR antagonist AP-5 abolished the response in the control cell and had no further effect in the transfected cell, indicating that CRISPR had eliminated the NMDAR EPSC. A summary of all the recordings indicates that cells expressing CRISPR for 14 days invariably lacked NMDAR EPSCs (Figure 1B). The physiological records thus suggest that CRISPR has deleted the GluN1 protein from all these cells. To confirm the loss of NMDAR, we expressed CRISPR/Cas9 in dissociated neuronal cultures using a lentiviral construct and measured receptor expression in these neurons by western analysis 14 days later. As anticipated from the physiology, CRISPR/Cas9 effectively deleted the GluA2 and GluN1 proteins, but the expression of β-actin and synaptophysin was not affected (Figure 1C). We also analyzed the types and frequency of mutations obtained when targeting GRIN1 with CRISPR in our dissociated neuronal cultures. The most common (5 out of 11) mutation that we identified is a 2 bp deletion that occurred 1 bp upstream of the cleavage site. We also found two 1 bp insertions, two other 2 bp deletions, a 3 bp deletion, as well as a bigger 19 bp deletion. Interestingly, >90% of the mutations led to out-of-frame insertions or deletions (Figure 1D).
Figure 1. Deletion of NMDA Currents by CRISPR/Cas9 Knockout of GRIN1.
(A) Sample traces of NMDAR evoked EPSCs, from a transfected CRISPR/Cas9 cell (green trace) and a neighboring control cell (black trace) in the presence of NBQX (10 μM) and after addition of D AP-5 (50 μM). (B) Bar graph showing the averaged eEPSC amplitudes of control, 46.43 ± 4.22 pA, n = 9; CRISPR_GRIN1, 0 pA, n = 9; control + AP-5, 0 pA, n = 5; CRISPR_GRIN1 + AP-5, 0 pA, n = 5. (C) Western blot for the GluA2 subunit of the AMPAR and GluN1 subunit of the NMDAR of 18-day-old dissociated hippocampal neurons infected with lentiCRISPR_GRIN1 or lentiCRISPR_GRIA2 on day 4. (D) Types and frequency of insertions or deletions obtained after infecting dissociated hippocampal neurons with lentiCRISPR GRIN1. See also Figure S1.
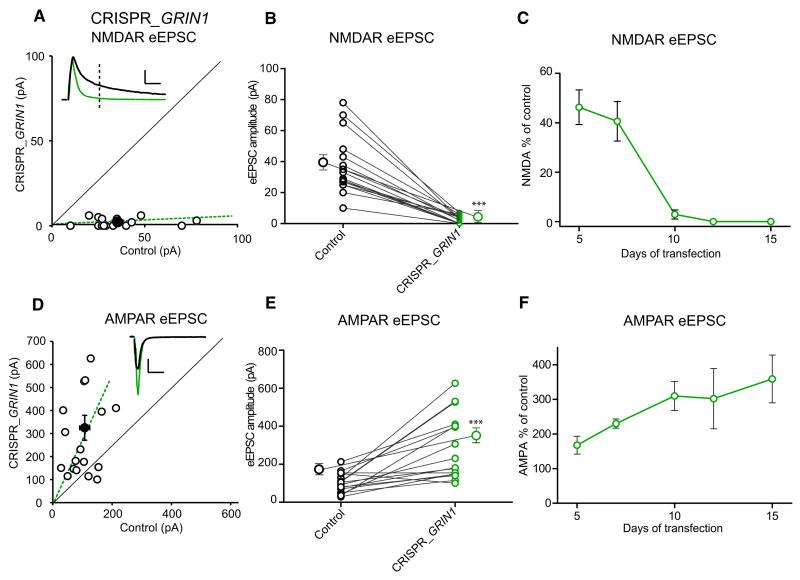
Deletion of GluN1 by Cre recombinase in conditional KO mice produces a substantial increase in AMPAR EPSCs (Adesnik et al., 2008; Gray et al., 2011; Ultanir et al., 2007). To determine whether the deletion of GluN1 by CRISPR reproduces this effect, we repeated the experiments as above but omitted NBQX to measure the AMPAR as well as NMDAR EPSC. To measure the AMPAR EPSC, we held the cell at −70 mV (Figure 2D), but for the NMDAR EPSC we held the cell at +40 mV, measuring the current 100 ms after the stimulus when the AMPAR EPSC had returned to baseline (Figure 2A, dotted line). As shown in Figure 1, the NMDAR component of the evoked EPSC was entirely blocked (Figures 2A and 2B). We also determined the time course of this effect and found that by 5 days, the NMDAR EPSC was reduced by 60%, and by 12 days, it was absent (Figure 2C). Over this period, the AMPAR EPSC increased approximately 2-fold (Figures 2D and 2E). However, the time course of this increase was delayed relative to the decline in NMDAR EPSC and continued to increase even at 15 days (Figure 2F). Finally, we examined paired-pulse facilitation, which is a sensitive measure for changes in the presynaptic probability of transmitter release (Figure S2 available online). As anticipated from previous work (Adesnik et al., 2008; Gray et al., 2011), the change in glutamate receptor expression did not affect release probability. To test the reproducibility of this approach, we also designed another gRNA that targets a different sequence of GluN1 and observed identical effects (Figure S1). The results show that deletion of GluN1 using CRISPR/Cas9 has effects identical to the deletion of GluN1 mediated by cre recombinase in conditional KO mice (Adesnik et al., 2008; Gray et al., 2011; Ultanir et al., 2007).
Figure 2. Deletion of NMDAR by CRISPR/Cas9 Increases AMPAR EPSCs.
(A) Scatterplot shows amplitudes of NMDA EPSCs for single pairs (open circles) and mean ± SEM (filled circle) for GRIN1 KO by CRISPR/Cas9. Data represent pairs of simultaneously recorded neurons in slice culture from CRISPR_GRIN1-transfected and neighboring control cells 14–15 days after transfection. Scale bar, 50 pA and 50 ms. (B) Paired average of single pairs from control and transfected cells. Mean ± SEM for control and CRISPR_GRIN1 are 37.53 ± 4.7 pA, n = 16 and 2 ± 0.8 pA, n = 16, respectively. ***p = 0.0005 Wilcoxon signed-rank test. The very small remaining current measured at 100 ms is due to a small residual tail of AMPAR current. (C) Time course for the changes in NMDAR EPSC amplitudes in hippocampal slice cultures after CRISPR_GRIN1 transfection. This includes data in the absence of NBQX (5 days and 7 days) and in the presence of NBQX (10 days, 12 days, and 15 days). The values represent the percentage of control for different days following transfection: 5 days 46.3% ± 7%, n = 10; 7 days 40.6% ± 8%, n = 8; 10 days 2.8% ± 1.8%, n = 8; 12 days 0% ± 0%, n = 8; 15 days 0% ± 0%, n = 9. (D) Scatterplot for AMPAR EPSC amplitudes for single pairs (open circles) and mean scatterplot (filled circle). Scale bar, 50 pA and 50 ms. (E) Paired average of single pairs from control and transfected CRISPR_GRIN1 cells. Mean ± SEM for control and transfected neurons are 105 ± 13.5 pA n = 16 and 281.2 ± 41 pA n = 16, respectively. ***p = 0.0001 Wilcoxon signed-rank test. (F) Time course for the changes in AMPAR EPSC amplitudes in hippocampal slices after CRISPR_GRIN1 transfection. Ratio of AMPAR EPSC: 5 days 167.7% ± 25.8%, n = 10; 7 days 230.2% ± 13.2%, n = 8; 10 days 310% ± 42%, n = 13; 12 days 302% ± 87%, n = 11; 15 days 359% ± 69%, n = 16. See also Figure S2.
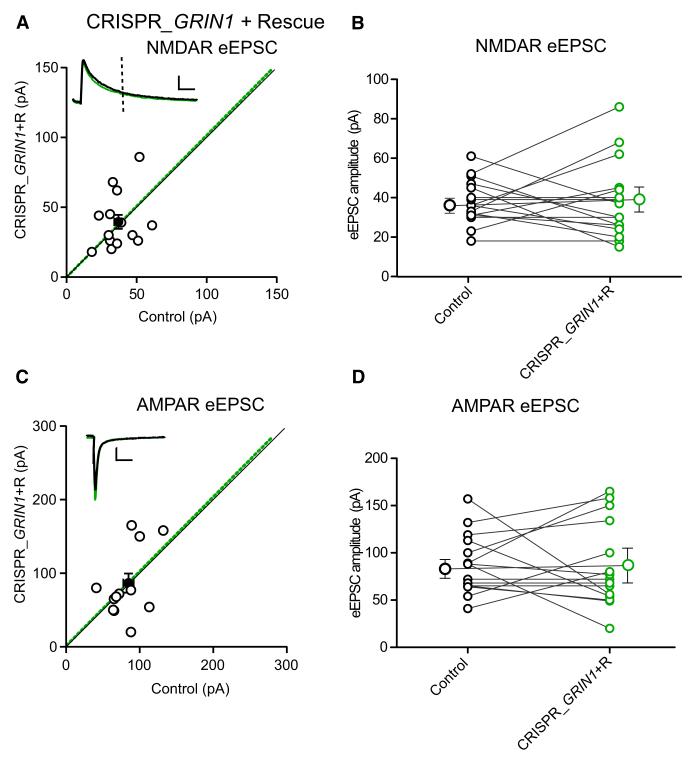
Since off-target effects are of concern with the CRISPR/Cas9 system (Fu et al., 2013; Pattanayak et al., 2013), we designed a gRNA that encompasses an intron-exon junction of the GluN1 coding region (see Experimental Procedures). This gRNA should not target the NMDAR cDNA and would thus enable simple rescue experiments using an NMDAR expression vector. Indeed, we found that expression of this gRNA sequence efficiently eliminated NMDAR EPSCs (Figure S1) and that cotransfection of NMDAR cDNA completely rescued the NMDAR EPSC (Figures 3A and 3B). It also eliminated the effect of GluN1 deletion on AMPAR EPSC (Figures 3B and 3C), showing that both of these effects are indeed due to the selective targeting of GluN1. Furthermore, to demonstrate that an unrelated cDNA does not rescue the effect of GluN1 CRISPR disruption unspecifically, we cotransfected a p-CAGG-IRES mCherry cDNA together with the FUGW GFP and CRISPR_GRIN1. Under these conditions, the GluN1 deletion is also complete, excluding the possibility that the GluN1 rescue is caused by a lower transfection efficiency of the Cas9/gRNA components (Figure S3). In Figure S3C, we also show confocal stack images of a neuron expressing both p-CAGG-mCherry and FUGW-GFP from a 14 day transfected slice. The morphology is that of a typical healthy pyramidal neuron. As a confirmation of the healthy state of the transfected neurons, we also measured the input resistance and found no difference between control and transfected neurons.
Figure 3. Rescue of NMDAR EPSC with GluN1 cDNA Resistant to Cas9.
(A) NMDAR EPSC amplitudes of control and neighboring rescued GluN1 cells, for single pairs (open circles) and mean ± SEM (filled circle). Scale bar, 50 pA and 50 ms. (B) Paired average of single pairs from control and transfected cells. Mean ± SEM for control and rescue are 37.8 ± 2.8 pA, n = 16 and 38.1 ± 4.9 pA, n = 16, respectively. p = 0.84 Wilcoxon signed-rank test. (C) AMPAR EPSC amplitudes for single pairs and mean ± SEM. Scale bar, 50 pA and 50 ms. (D) Paired average of single pairs from control and transfected cells. AMPAR EPSC amplitudes for control and rescue are 87.6 ± 8.1 pA, n = 15 and 86.53 ± 11.56 pA, n = 15, respectively. Mean ± SEM, p = 0.96 Wilcoxon signed-rank test. See also Figure S3.
We have also tried the double nicking approach using the D10A mutant nickase Cas9 together with a pair of offset gRNAs complementary to opposite strands of the target site, which has been proposed to improve specificity (Ran et al., 2013a). However, we found that this approach is not as efficient as the Cas9 endonuclease in our experimental conditions (Figures S1E and S1F).
Deletion of GluA2
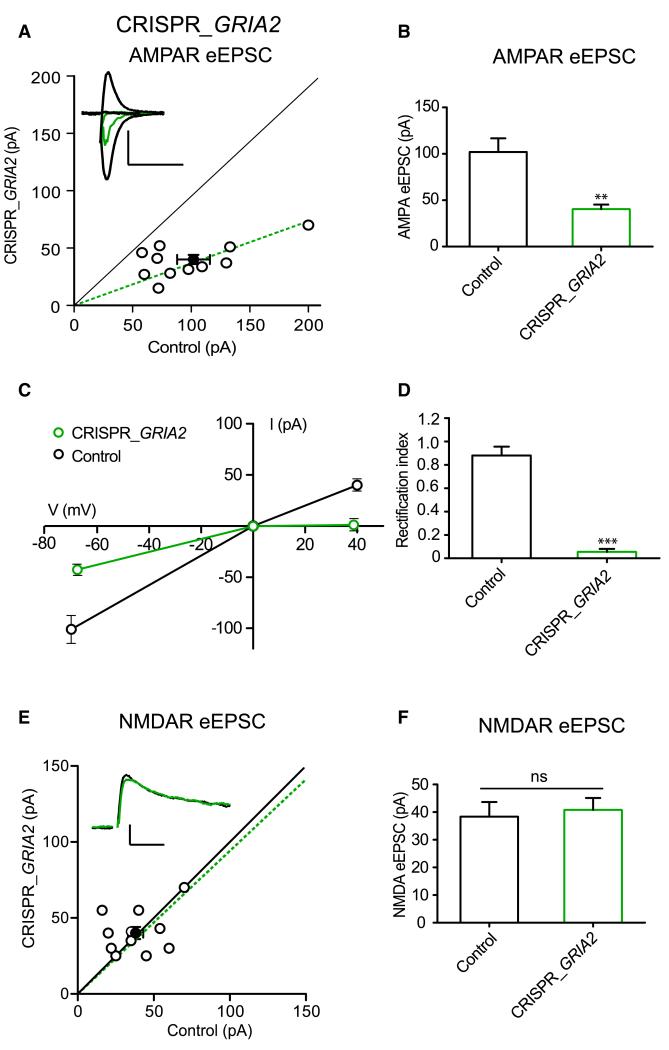
CRISPR/Cas9 can delete GluN1 using two independent sequences, but will the approach work for other synaptic proteins? AMPARs are heterotetramers that comprise various combinations of GluA1–Glu4 subunits. In CA1 hippocampal pyramidal cells, all receptors contain the GluA2 subunit (Lu et al., 2009). GluA2-containing receptors have a linear current-voltage (I-V) relationship such that the magnitude of current flow through the receptor is the same at negative and positive potentials. In contrast, receptors lacking the GluA2 subunit rectify strongly such that little outward current flows through the receptor at positive potentials. Thus, inward rectification provides a sensitive measure of GluA2 content in the AMPAR. Using two different gRNAs (GRIA2#1 and GRIA2#2, see Experimental Procedures), we found that CRISPR_GRIA2 knockout in transfected cells reduced AMPAR eEPSC (Figures 4A and 4B) and the remaining AMPAR currents were entirely inwardly rectifying (Figures 4C and 4D), confirming the results of GluA2 deletion by Cre recombinase in conditional KO mice (Lu et al., 2009). This result is expected from the strong reduction in GluA2 protein expression shown in Figure 1C. We also observed that NMDAR eEPSCs did not change (Figures 4E and 4F), further confirming the specificity of the GluA2 deletion.
Figure 4. CRISPR/Cas9 GRIA2 Knockout.
(A) AMPAR EPSC amplitudes of control and neighboring GRIA2 KO by CRISPR/Cas9. Scatterplot for single pairs (open circles) and mean ± SEM (filled circle). Scale bar, 50 pA and 10 ms. (B) Graph bars represent mean values of AMPAR EPSC amplitudes for control and CRISPR_GRIA2. Mean ± SEM are Cnt 101.6 ± 13 n = 11; CRISPR_GRIA2 39.7 ± 4.4 n = 11. **p = 0.001 Wilcoxon signed-rank test. (C) Graph representing I/V plots for control and CRISPR_GRIA2 cells: the AMPAR EPSC from the GluA2 KO cell is inwardly rectifying. (D) Mean ± SEM of rectification index values for control (0.88 ± 0.1) and CRISPR_GRIA2 cells (0.06 ± 0.03). ***p < 0.0001 Student’s unpaired t test. (E) NMDAR EPSC scatterplot for single pairs (open circles) and mean ± SEM (filled circle). Scale bar, 50 pA and 50 ms. (F) Graph bars represent mean values of NMDAR EPSC amplitudes for control and CRISPR_GRIA2. Mean ± SEM are Cnt 38.36 ± 5 n = 11; CRISPR_GRIA240.8 ± 4 n = 11. p = 0.77 Wilcoxon signed-rank test. See also Figure S4.
To address possible off-target effects of the CRISPR_GRIA2 knockout, we designed a rescue experiment. Similar to the GRIN1#2 gRNA, the GRIA2#1 gRNA encompasses an intron-exon junction of the GluA2 gene (see Experimental Procedures). Thus, we used a cDNA that expresses the edited form of the GluA2 subunit in order to rescue the inward rectification observed in the GRIA2 KO. Indeed, the cotransfection of CRISPR_GRIA2#1 and of the GluA2 cDNA eliminated the AMPAR eEPSC reduction observed in the GRIA2 KO (Figures S4A and S4B) and largely rescued the inward rectification index (Figures S4C and S4D).
DISCUSSION
We have evaluated the ability of the CRISPR/Cas9 system to delete synaptic proteins. For these experiments, we first focused on the GluN1 subunit. Since the GluN1 subunit is required for NMDAR function, the size of the NMDAR EPSC provides a sensitive measure of GluN1 expression. Indeed, we found that coexpression of gRNAs to GluN1 together with Cas9 in hippocampal pyramidal cells gradually reduced the size of the NMDAR EPSC and by 12 days after transfection no NMDAR function could be detected. Importantly, we completely rescued the NMDAR EPSC by expressing a GluN1 cDNA resistant to the action of intron-directed CRISPR/Cas9. To determine whether the CRISPR/Cas9 system could be used for other synaptic proteins, we turned to the GluA2 subunit of the AMPAR. AMPARs are heterotetrameric receptors and those expressed in CA1 pyramidal cells contain the GluA2 subunit. Using inward rectification to monitor the expression of synaptic GluA2 (Jonas and Burnashev, 1995), we found that coexpression of gRNAs to GluA2 along with Cas9 for 12 days resulted in strongly rectifying AMPAR EPSCs, indicating loss of the GluA2 subunit from AMPARs.
The CRISPR/Cas9 system has multiple advantages over previous methods to delete neuronal proteins. Conditional gene inactivation using cre recombinase provides a similarly complete loss of gene function. However, the associated genetic manipulation in embryonic stem cells followed by injection of these cells into embryos requires considerable time and effort, even with recent technical improvements. The resulting animals also require at least two generations of breeding before initial experiments can be performed, and longer if crossing to a line that expresses cre recombinase in specific tissues. The CRISPR/Cas9 system now enables the inactivation of genes in somatic cells, without the requirement for homologous recombination in the germline. In the case of GluN1, inactivation occurred within 2 weeks after transfection.
RNAi can also be performed on a relatively rapid timescale and has been used for high-throughput experiments such as genetic screens in vitro and in vivo (Fire et al., 1998; Hannon, 2002). However, a major problem associated with RNAi, at least in most mammalian systems, is incomplete gene silencing. RNAi rarely eliminates the protein targeted, and residual protein makes it very difficult to interpret the resulting phenotype. The phenotypes are often incomplete or variable, and the lack of phenotype can be particularly difficult to interpret in the presence of residual protein. We now find that the CRISPR/Cas9 system works much more efficiently, with essentially complete loss of the glutamate receptor subunits targeted. All of the transfected cells in hippocampal slice culture failed to express GluN1 after 2 weeks incubation, and the miniscule amounts of residual protein observed by western analysis from dissociated hippocampal cultures may simply reflect incomplete infection by the recombinant virus. The ability to introduce CRISPR/Cas9 into specific cells also circumvents problems such as lethality associated with global gene inactivation in knockout mice. The relatively acute effect of CRISPR/Cas9 further mitigates the potential for compensation. In addition, it is essential to perform rescue experiments that definitively rule out off-target effects as the cause of the observed phenotype. For the rescue, it is important to ensure that the rescue construct will not itself be targeted by CRISPR. This can be achieved either by using a cDNA with silent mutations introduced into the region recognized by the gRNA (and/or the PAM) or by using a cDNA from a different species if there is sufficient divergence in the sequence. In this study, we performed the rescue experiment using another, simpler approach. Indeed, we searched for a specific gRNA sequence encompassing an intron-exon junction such that the cleavage site (position −3) would be in the exon, but most of the gRNA sequence would lie within the intron. By using this very simple strategy, it is now possible to perform rescue experiments using unmodified cDNA, which greatly simplifies and accelerates the validation of the phenotype.
It is remarkable that CRISPR/Cas9 so effectively eliminates the expression of proteins, and this may specifically reflect the postmitotic nature of neurons. Our sequencing analysis of the GRIN1 locus identified a series of insertions or deletions that led to out-of-frame mutations in >90% of the case. This strongly suggests that the Cas9 nuclease, indeed, produces a double-strand break that triggers repair, and this nonhomologous end-joining repair results in gene inactivation. However, there is some chance that this repair will restore the original sequence. In dividing cells where the gRNAs and Cas9 are lost through repeated cell division, repair would prevent gene inactivation, particularly since only one properly repaired allele may suffice for a wild-type phenotype. In contrast, the persistent gRNAs and Cas9 nuclease in postmitotic neurons might continue to cleave any properly repaired sequence, resulting in eventual incorrect repair and inactivation of both alleles. Another possibility to consider is that the repair following the Cas9 double strained break, although not perfect, fails to silence the gene and Cas9 also fails to recognize the imperfectly repaired sequence. In this case, one would expect a transfected neuron without physiological phenotype. This scenario does not appear to have happened in our experimental sample. In a total of 107 experiments, we did not encounter a cell in which the function of the targeted protein remained unaltered after the expression of Cas9 and a gRNA. Regardless of mechanism, CRISPR/Cas9 appears remarkably effective in neurons and holds great promise for understanding the molecular underpinning of synaptic plasticity.
EXPERIMENTAL PROCEDURES
For details, please see Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. B.E. Herring, J.A. Gray, J. Levy, and N. Sheng in the R.A.N. lab for assistance, K. Bjorgan and M. Cerpas for technical assistance, Dr. S.L. Shipman for discussions on CRISPR, and Drs. J.S. Weissman and L.A. Gilbert for comments on the manuscript. We thank Dr. F. Zhang for pX330, px335, and lentiCRISPR. This work was supported by NIH grant MH-38256 to R.A.N. and MH096863 to R.H.E.
Footnotes
AUTHOR CONTRIBUTIONS
S.I. and C.S.A. performed and analyzed the experiments. R.A.N. and R.H.E. designed the study. R.A.N., R.H.E., C.S.A., and S.I. interpreted the results and wrote the paper.
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2014.07.043.
REFERENCES
- Adesnik H, Li G, During MJ, Pleasure SJ, Nicoll RA. NMDA receptors inhibit synapse unsilencing during brain development. Proc. Natl. Acad. Sci. USA. 2008;105:5597–5602. doi: 10.1073/pnas.0800946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez VA, Ridenour DA, Sabatini BL. Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J. Neurosci. 2006;26:7820–7825. doi: 10.1523/JNEUROSCI.1957-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. Characterization of Genomic Deletion Efficiency Mediated by Clustered Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 Nuclease System in Mammalian Cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Kehl SJ, McLennan H. Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. J. Physiol. 1983;334:33–46. doi: 10.1113/jphysiol.1983.sp014478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J. Neurosci. 2004;24:916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K, Kim MJ, Hashikawa T, Scheiffele P, Sheng M, Hayashi Y. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci. 2007;10:186–195. doi: 10.1038/nn1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R. Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science. 2000;287:2262–2267. doi: 10.1126/science.287.5461.2262. [DOI] [PubMed] [Google Scholar]
- Hestrin S, Nicoll RA, Perkel DJ, Sah P. Analysis of excitatory synaptic action in pyramidal cells using whole-cell recording from rat hippocampal slices. J. Physiol. 1990;422:203–225. doi: 10.1113/jphysiol.1990.sp017980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Burnashev N. Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron. 1995;15:987–990. doi: 10.1016/0896-6273(95)90087-x. [DOI] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plück A. Conditional mutagenesis in mice: the Cre/loxP recombination system. Int. J. Exp. Pathol. 1996;77:269–278. [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013a;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013b;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc. Natl. Acad. Sci. USA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Ultanir SK, Kim JE, Hall BJ, Deerinck T, Ellisman M, Ghosh A. Regulation of spine morphology and spine density by NMDA receptor signaling in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:19553–19558. doi: 10.1073/pnas.0704031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JC, Krogsgaard-Larsen P, Honoré T. Structure-activity relationships in the development of excitatory amino acid receptor agonists and competitive antagonists. Trends Pharmacol. Sci. 1990;11:25–33. doi: 10.1016/0165-6147(90)90038-a. [DOI] [PubMed] [Google Scholar]
- Wijshake T, Baker DJ, van de Sluis B. Endonucleases: new tools to edit the mouse genome. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbadis.2014.04.020. Published online April 30, 2014. http://dx.doi.org/10.1016/j.bbadis.2014.04.020. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.